Abstract
Infectious bursal disease virus (IBDV) is a nonenveloped avian virus with a two-segment double-stranded RNA genome. Its T=13 icosahedral capsid is most probably assembled with 780 subunits of VP2 and 600 copies of VP3 and has a diameter of about 60 nm. VP1, the RNA-dependent RNA polymerase, resides inside the viral particle. Using a baculovirus expression system, we first observed that expression of the pVP2-VP4-VP3 polyprotein encoded by the genomic segment IBDA results mainly in the formation of tubules with a diameter of about 50 nm and composed of pVP2, the precursor of VP2. Very few virus-like particles (VLPs) and VP4 tubules with a diameter of about 25 nm were also identified. The inefficiency of VLP assembly was further investigated by expression of additional IBDA-derived constructs. Expression of pVP2 without any other polyprotein components results in the formation of isometric particles with a diameter of about 30 nm. VLPs were observed mainly when a large exogeneous polypeptide sequence (the green fluorescent protein sequence) was fused to the VP3 C-terminal domain. Large numbers of VLPs were visualized by electron microscopy, and single particles were shown to be fluorescent by standard and confocal microscopy analysis. Moreover, the final maturation process converting pVP2 into the VP2 mature form was observed on generated VLPs. We therefore conclude that the correct scaffolding of the VP3 can be artificially induced to promote the formation of VLPs and that the final processing of pVP2 to VP2 is controlled by this particular assembly. To our knowledge, this is the first report of the engineering of a morphogenesis switch to control a particular type of capsid protein assembly.
Infectious bursal disease virus (IBDV) is the causative agent of a highly contagious disease of young chickens, bursal disease or Gumboro disease (3). The virus causes a severe immunosuppression by destroying B cells present in the bursa of Fabricius. The induced immunodepression leads to an increased susceptibility to other pathogens.
IBDV is a member of the Birnaviridae family (14). Birnaviruses are nonenveloped and contain two segments of double-stranded RNAs (A and B). The smaller segment, B, encodes the VP1 protein, which is the putative viral RNA-dependent polymerase, whereas the larger segment, A, contains two partially overlapping open reading frames. The smaller one encodes VP5, a nonstructural protein of 17 kDa (reference 16 and references therein). The larger one encodes a polyprotein precursor in the order NH2-pVP2-VP4-VP3-COOH. The polyprotein is cotranslationally processed through the proteolytic activity of VP4 to generate pVP2 (amino acids [aa] 1 to 512), VP4 (aa 513 to 755), and VP3 (aa 756 to 1012) (13, 19). pVP2 is further processed at its C terminus to become VP2, through the cleavage of at least three alanine-alanine bounds (positions 487-488, 495-496, and 501-502) (13). VP2 and VP3 form the outer and inner layers, respectively, of the virions, which contain several VP1 molecules and the genomic RNAs (1).
Preparations of purified IBDV virions were found to contain full and empty icosahedral virions and tubules with a diameter of about 60 nm (type I) or 24 to 26 nm (type II) (7). The type II tubules, which contain VP4, have also been identified in infected cells. Electron cryomicroscopy studies showed that the structure of the virion is based on a T=13 lattice formed by trimer-clustered subunits (1).
Recombinant expression of the IBDV polyprotein in heterologous cell systems has been extensively reported. Few of these studies showed the production of virus-like particles (VLPs) (5, 15). When the baculovirus-insect cell system was used to express the polyprotein, the production of VLPs was inefficient (4, 9, 11, 17, 21). Furthermore, the processing of pVP2 to VP2 was blocked (11, 17) and assembly products other than VLPs were observed, suggesting a defect in viral morphogenesis (17).
On this basis, we speculated that the charged amino acids present at the C terminus of VP3 might interfere with assembly in the absence of the viral genome. To modulate the effects of this amino acid stretch, we fused a large protein domain at its C terminus. We hypothesized that an extra protein could fit into the space occupied by VP1 and by the genome into the virions. Accordingly, we prepared a DNA construct encoding the chimeric polyprotein in which the IBDA polyprotein was fused after residue 1012 to a 7-aa long linker and the entire 238-aa green fluorescent protein (GFP). This addition promoted favorable protein arrangements, leading to the almost exclusive formation of VLPs and to processing of pVP2. The environment of the C-terminal domain of VP3 thus appears to be an important switch controlling the virus morphogenesis.
MATERIALS AND METHODS
Plasmids and recombinant baculovirus constructs.
Plasmid pUC-IBDA (13), constructed for the T7-driven expression of IBDA, was digested by EcoRI to isolate a 3.3-kb fragment containing the complete IBDA segment under the control of the T7 promoter. This fragment was then subcloned in pFastBac1 (Gibco BRL) into the EcoRI site to generate pFBIBDAwt. Then pFBIBDAwt was digested by PvuI and self-ligated. A recombinant plasmid with a 114-base deletion upstream of the polyprotein codon was then selected by nucleotide sequencing and named pFBΔIBDA. A NheI restriction site was placed behind the last VP3 codon in pUC-IBDA by mutagenesis using Pfu DNA polymerase with the QuickChange site-directed mutagenesis kit (Stratagene) to generate pUC-IBDANhe1. The EGFP gene was recovered from pEGFPC1 (Clontech) by restriction with NheI and KpnI. The fragment was ligated into pUC-IBDANhe1 that has been digested by NheI and KpnI, and the generated plasmid was named pUC-IBDAGFP. Then the IBDAGFP gene was recovered from pUC-IBDAGFP by restriction with EcoRI and KpnI for ligation in pFastBac1 that had been identically restricted. The recovered plasmid was then PvuI digested and self-ligated. A recombinant plasmid with a 114-base deletion upstream of the polyprotein codon was then selected by nucleotide sequencing and named pFBΔIBDAGFP.
The resulting plasmids, pFBIBDAwt, pFBΔIBDA, and pFBΔIBDAGFP, were used to generate the recombinant baculoviruses BacIBDAwt, BacΔIBDA, and BacΔIBDAGFP, respectively. Briefly, the three pFastBac derivatives were transformed into DH10Bac competent cells which contain the bacmid. Colonies containing recombinant bacmids were identified by disruption of the lacZα gene. High-molecular-weight mini-prep DNA was prepared from selected colonies, and this DNA was used to transfect Sf9 cells with Lipofectin. Recombinant baculovirus were prepared by standard procedures. Then high-titer viral stocks of the recombinant baculoviruses (108 PFU/ml) were prepared.
Viruses, cells, and MAbs.
The Gumboral CT IBDV vaccine strain (Mérial, Lyon, France) was kindly provided by N. Eterradossi (AFSSA, Ploufragan, France). IBDV was propagated in LSCC-BK3 cells (8) provided by J. Korb (Institute of Molecular Genetics, Prague, Czech Republic). The 1C6 monoclonal antibody (MAb) specific for pVP2/VP2 was provided by J.-F. Bouquet (Mérial), and MAb 20 specific for VP3 was provided by N. Eterradossi. MAb 66 specific for VP4 was produced by immunization of mice with preparations of VP4 produced in Escherichia coli (to be reported elsewhere).
Preparation of protein assembly specimens.
Sf9 cells were infected at a multiplicity of infection higher than 5 PFU/ml in the presence of the protease inhibitors leupeptin (0.5 μg/ml) and aprotinin (1 μg/ml), collected 100 h postinfection after addition of the same inhibitors at the identical concentrations, and then treated with Freon 113. Purification was carried out by density gradient centrifugation in a CsCl solution. The concentration of protein in the purified suspension was estimated by the method of Bradford using bovine serum albumin BSA as standard and UV spectrophotometry at 280 and 260 nm. From the ratio of absorbance at 260 and 280 nm, we determined that the contamination of all our samples with nucleic acid was less than 5%. As far as contamination is concerned, our preparations did not show any significant difference.
Optical microscopy.
VLPs tagged with GFP were vizualized with a Nikon Eclipse fluorescence microscope using an Omega XF116 filter. Images were collected with a charge-coupled device camera. Confocal microscopy analysis was carried out using the TCS NT confocal imaging system (Leica Instruments, Heidelberg, Germany), equipped with a 63× objective (plan apo; numerical aperture = 1.4). For enhanced GFP (EGFP) and tetramethylrhodamine isocyanate, an argon-krypton ion laser adjusted to 488 or 554 nm, respectively, was used. For comparaison purpose, we used FluoSpheres carboxylate-modified microspheres, 0.1 μm in diameter (2.7 × 1013 particles/ml), red fluorescent (580/605 nm), purchased from Molecular Probes Europe BV, Leiden, The Netherlands. For microscopy examination, EGFP-VLPs or latex bead suspension (1 μl) was put on a slide. The color adjustment was done in Photoshop v3.0 (Adobe Systems).
Electron microscopy.
(i) Isolated assemblies. Specimens were prepared from the appropriate CsCl gradient fractions containing the different assembled forms by desalting through Micro Bio-Spin chromatography columns (Bio-Rad) equilibrated with TN buffer. Samples of the suspensions were applied to an air-glow discharged carbon-coated grid and stained with a 2% uranyl acetate aqueous solution.
(ii) Cell in culture.
Cells grown on Thermanox cover slips (Miles, Elkhart, Ind.) were fixed for 1 h with 2.5% glutaraldehyde in 100 mM phosphate buffer (pH 6). They were postfixed for 1.5 h with 1% osmium tetroxide in water. After the specimens were rinsed, they were then maintained overnight in 2% uranyl acetate aqueous solution. The samples were then embedded in Epon. Thin sections were stained with uranyl acetate in acetone and lead citrate.
Protein analysis.
Sf9 cells (3 × 106) were infected (or mock infected) at a multiplicity of infection of 10 PFU per cell and maintained in 2 ml of Hinks medium supplemented with 10% fetal calf serum. At 48 h postinfection, the medium was discarded and the cells were lysed in RIPA buffer (50 mM Tris [pH 8], 150 mM NaCl, 2% Triton X-100) with a protease inhibitor cocktail (Boehringer). This material was centrifuged for 30 min at 13,000 × g. Aliquots of the supernatants were incubated for 2 h at room temperature under gentle agitation with 1 μl of ascites fluids of hybridomas and 35 μl of a 1:1 slurry of protein A-Sepharose beads (Pharmacia). The beads were washed four times with 1 ml of RIPA buffer, treated for 2 min at 100°C in Laemmli denaturating buffer plus 5% 2-mercaptoethanol, and centrifuged. The resulting supernatants were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis (10 or 12.5% polyacrylamide, 0.1% SDS), and proteins were detected by Coomassie blue staining. Protein Mr standards (Pharmacia) were used.
To reveal radiolabeled antigens by immunoprecipitation, the same procedure was carried out with some modifications. A 10-μl volume of Promix (Amersham) was added to the cell medium at 24 h postinfection for during 24 h. Polyacrylamide gels were processed for fluorography. 14C-methylated protein Mr standards (Amersham) were used.
Western blot analyses were performed using MAbs followed by appropriate secondary antibodies conjugated to alkaline phosphatase (Biosys, Compiègne, France) and nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate substrate.
RESULTS
Expression of IBDA polyprotein leads mainly to the formation of pVP2 tubules.
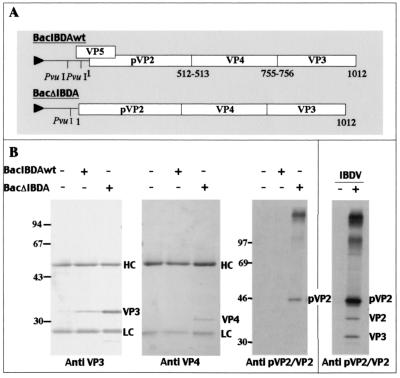
The expression products of the two recombinant baculoviruses carrying either the complete IBDA segment (BacIBDAwt) or only the polyprotein open reading frame (BacΔIBDA) were first analyzed (Fig. 1A). Sf9 cells were infected with the recombinant viruses, and cell extracts were collected 48 h postinfection. The presence of the IBDA proteins VP3, VP4, and pVP2/VP2 was analyzed by immunoprecipitation and SDS-PAGE. Figure 1B shows that the expression levels of VP3, VP4, and pVP2 proteins were higher with BacΔIBDA than with BacIBDAwt. Therefore, part of the 5" end of IBDA upstream of the polyprotein initiation codon appeared to be deleterious for polyprotein expression. It should also be noted that pVP2 was easily detectable only with BacΔIBDA and only after immunoprecipitation of radioactive cell labeling, suggesting that pVP2 was less immunoreactive than VP3 and VP4 in this assay. Consequently, the following experiments were carried out with BacΔIBDA and additional derived baculovirus recombinants were constructed with the same 5"-end deletion. As described previously (11), Fig. 1B shows that pVP2 (48 kDa) was not further processed into the mature form of VP2. We also immunoprecipitated IBDV-infected cells with an anti-pVP2/VP2 antibody. While pVP2 is the major form, VP2 (40 kDa) is detectable, as well as VP3, which is coprecipitated, because they form the viral capsid.
FIG. 1.
IBDA polyprotein-baculovirus recombinants. (A) Map of the two constructs derived from IBDA. In BacIBDAwt, the complete IBDA segment was placed under the control of the polyhedrin promoter, whereas in BacΔIBDA, part of the 5" end upstream of the polyprotein initiation codon was deleted. Numbers indicate the coordinates of P1-P"1 amino acids cleaved by the viral protease VP4, and the triangle upstream of the IBDA sequence indicates the polyhedrin promoter. (B) Immunoprecipitation analyses using an anti-VP3 antibody, an anti-VP4 antibody, and an anti-pVP2/VP2 antibody. Sf9 cells were infected by recombinant baculovirus, and LSCC-BK3 cells were infected by IBDV (right panel). +, infected cells; −, mock-infected cells. Immune complexes were analyzed by SDS-PAGE (10% polyacrylamide) under reducing conditions. The gels were stained with Coomassie blue (left panels) or fluorographed for pVP2/VP2 immunoprecipitations (right panels). The relative Mrs (shown in thousands) were determined by reference to marker proteins. HC and LC indicate the positions of the heavy and light chains of the immunoglobulins, respectively. Note the presence of a VP3 band that coimmunoprecipitated with VP2 in BK3 cells infected by IBDV, an observation consistent with the presence of viral particles inside the cells.
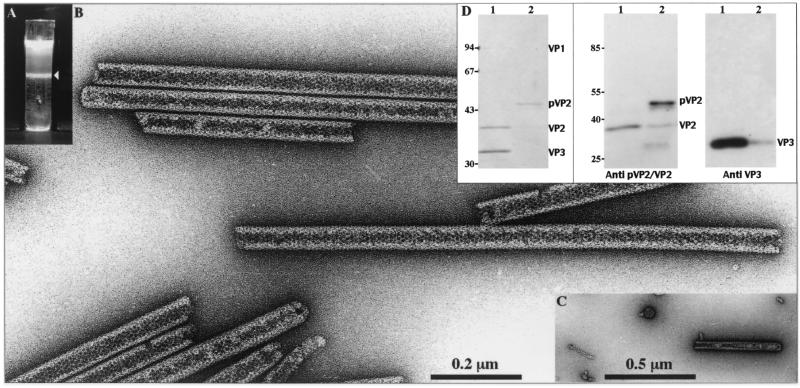
To analyze the structures which were generated by IBDA polyprotein expression, extracts of BacΔIBDA-infected cells were Freon extracted and subjected to density gradient centrifugation. A single band at a density of 1.30 was observed in the gradient (Fig. 2A). Electron microscopy observations revealed large numbers of rigid tubules (Fig. 2B) with a diameter of about 50 nm, as recently described with a baculovirus expressing the IBDA polyprotein (17). Some very rare VLPs and tubules with a diameter of 25 nm were also detected in this band (Fig. 2C). Analysis of the protein content present in the band was carried out by SDS-PAGE followed by Coomassie blue staining and by immunoblotting (Fig. 2D). After calibration with purified IBDV virions (lanes 1), the main band (48 kDa) was found to be reactive with the anti-pVP2/VP2 antibody and was identified as pVP2. Very small amounts of mature VP2 and VP3 were also detected in this preparation. Therefore, baculovirus-driven expression of the polyprotein results in the efficient assembly of rigid tubules, composed mainly of pVP2. The low efficiency of VLP assembly is not due to a possible deleterious mutation in the IBDA cDNA sequence since this DNA insert was successfully used for the recovery of infectious IBDV in reverse genetics experiments (3a).
FIG. 2.
Analysis of the structures produced by BacΔIBDA. Sf9 cells were infected with BacΔIBDA and treated with Freon 113 as described in Materials and Methods. (A) After on 18-h centrifugation in CsCl at 100,000 × g, the gradient was illuminated with white light and photographed. (B and C) Material collected from the band present in the gradient was negatively stained with 1% uranyl acetate. Large numbers of tubules with an apparent diameter of 52 nm (B) and some icosahedral VLPs and tubules with a diameter of 25 nm (C) were observed. (D) The left panel shows SDS-PAGE analysis and Coomassie blue staining of purified IBDV (lane 1) and of the material isolated from the band present in the CsCl gradient (lane 2). The right panels show Western blot analyses of purified IBDV (lanes 1) and of the BacΔIBDA band present in the CsCl gradient (lanes 2) with an anti-pVP2/VP2 antibody or with an anti-VP3 antibody. The relative Mrs (in thousands) determined by reference to marker proteins are indicated on the left.
Expression of pVP2 leads to the formation of isometric particles.
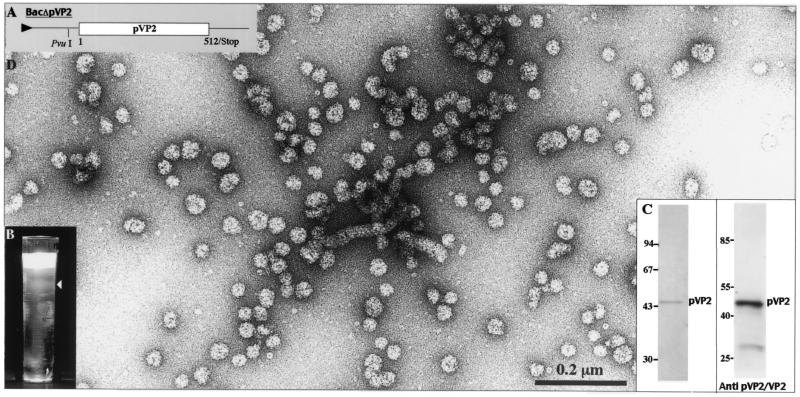
To determine the role of VP3 and VP4 in the formation of pVP2 rigid tubules, we constructed a recombinant baculovirus driving the expression of a truncated form of the IBDA polyprotein (Fig. 3A). Since primary cleavage at the pVP2-VP4 junction occur-ed between Ala512 and Ala513, BacΔpVP2 encoding the pVP2 precursor (residues 1 to 512) was engineered. Extracts of BacΔpVP2-infected cells were Freon extracted and subjected to density gradient centrifugation. A single fuzzy band was observed at a density of 1.29 (Fig. 3B). Analysis of the protein present in the band was carried out by SDS-PAGE followed by Coomassie blue staining (Fig. 3C). A unique band at 48 kDa was revealed. Western blot analysis revealed this band to be the pVP2 protein and a probable minor degradation product. Electron microscopy observation showed rather isometric particles with a diameter of about 30 nm (Fig. 3D). Very few flexible tubules were also identified in these preparations, but we failed to identify rigid tubules as were observed when the IBDA polyprotein was expressed. Therefore, we concluded that assembly of pVP2 into rigid tubules when expressed as part of the IBDA polyprotein is probably controlled by the presence of the capsid protein VP3 or possibly by the VP4 protease.
FIG. 3.
Analysis of the structures produced by BacΔpVP2. (A) Map of the construct encoding pVP2. Sf9 cells were infected with BacΔpVP2 and treated with Freon 113 as described in Materials and Methods. (B) After an 18-h centrifugation in CsCl at 100,000 × g, the gradient was illuminated with white light and photographed. (C) The left panel shows SDS-PAGE analysis and Coomassie blue staining of the material isolated from the fuzzy band present in the CsCl gradient. The right panel shows a Western blot analysis with an anti-pVP2/VP2 antibody. (D) Material collected from the band present in the gradient was negatively stained with 1% uranyl acetate. Large numbers of small capsids and some flexible tubes were vizualized.
Rationale for construction of the IBDA-GFP chimeric polyprotein and expression of the fusion polyprotein.
VP3, and most probably its C-terminal domain, could be responsible for the nonassembly of pVP2/VP2 into VLPs in insect cells. This domain, which is composed mainly of charged residues and is thought to be involved in genome encapsidation, may disturb the formation of VLPs in the absence of genome and associated protein (VP1). We hypothesized that the fusion of a protein at the C terminus of the polyprotein may suppress the negative effect of VP3 terminal residues on VLP formation. Accordingly, we engineered the baculovirus BacΔIBDA-GFP, encoding the complete IBDA polyprotein fused at its C terminus with a 7-aa linker and the entire 238-aa EGFP (Fig. 4A). EGFP was used because proper folding is required for its fluorescence activity, which would facilitate the detection of the structures associated with VP3.
FIG. 4.
Expression of the fusion polyprotein IBDA-GFP in insect cells. (A) Schematic representation of the construct expressing the IBDA polyprotein fused at its C terminus with EGFP. A 7-aa linker was added between the two partners, and its sequence is reported using the single-letter code. (B) Expression of the fusion polyprotein IBDA-GFP in insect cells analyzed by immunoprecipitation with specific antibodies and by SDS-PAGE. Sf9 cells were infected with BacIBDA-GFP, and LSCC-BK3 cells were infected by IBDV (right panel). +, infected cells; −, mock-infected cells. Immune complexes were analyzed by SDS-PAGE (10% polyacrylamide) under reducing conditions. HC and LC indicate the positions of the heavy and light chains of the immunoglobulins, respectively. The gels were stained with Coomassie blue (left panels) or fluorographed for pVP2/VP2 immunoprecipitation (right panels). The relative Mrs (in thousands) were determined by reference to marker proteins, and positions of the molecular weight markers are indicated on the left. (C) Visualization of IBDA-GFP expression in Sf9-infected cells. The cells were examined under a optical microscope with UV light excitation 48 h postinfection. The left panel shows mock-infected cells, and the right panel shows infected cells.
Immunoprecipitations of BacΔIBDA-GFP-infected Sf9 cell extracts were carried out for SDS-PAGE analysis and Coomassie blue staining (Fig. 4B). A band with an apparent molecular mass of 64 kDa, corresponding to the expected size for the chimeric VP3-GFP fusion protein, was reactive with both the anti-VP3 and anti-GFP antibodies. The levels of expression of VP3-GFP, pVP2, and VP4 were comparable to those of VP3, pVP2, and VP4 expressed with BacΔIBDA. Note that in contrast to what was observed in IBDV-infected cells, mature VP2 and VP3 were not detectable by this assay in BacΔIBDA-GFP-infected cells. Direct fluorescence analysis of Sf9 cells infected with ΔIBDA-GFP showed a strong labeling of cells, suggesting that the GFP was correctly folded (Fig. 4C). Therefore, all the components of the fusion protein, pVP2, VP4, and VP3-GFP, appeared to be well expressed.
Efficient assembly of IBDA-GFP into VLPs and maturation of pVP2 to VP2 on VLPs.
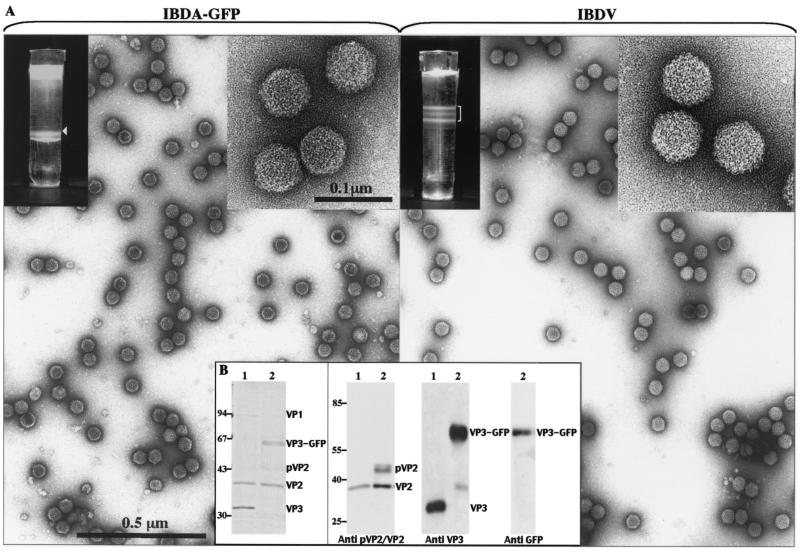
To analyze the type of assembly driven by the chimeric molecule IBDA-GFP, we expressed the molecule and subjected it to Freon extraction and density gradient centrifugation. In parallel, IBDV was purified using the same procedure, except for the gradient density, which was assigned at 1.33. As shown in Fig. 5A, a fluorescent band was revealed by illumination with both white and UV light with BacΔIBDA-GFP. The IBDV bands, corresponding to viruses containing different amounts of genomic material (3a), were not visible under UV light (data not shown). Electron microscopy of negatively stained preparations revealed the presence of large numbers of VLPs with a diameter of about 60 nm, looking similar to viral particules. Most VLPs appeared as full particles, suggesting that the VP3-fused GFP resides inside the particles. Very small numbers of 50-nm-diameter tubules were also identified. To confirm the assembly of pVP2/VP2 and VP3-GFP into the VLPs, the protein composition of the ΔIBDA-GFP band was investigated by SDS-PAGE followed by Coomassie blue staining or Western blot analysis using MAbs specific for GFP, VP3, and pVP2/VP2. Figure 5B shows that the VLPs contain the VP3-GFP fusion, pVP2, and the mature form of VP2. In contrast to previous results, the VP2 mature form was the major pVP2/VP2 species associated with VLPs. These results indicate that the final conversion of pVP2 to VP2 occurred efficiently during VLP assembly.
FIG. 5.
Analyses of the structures expressed by BacΔIBDA-GFP. (A) Sf9 and LSCC-BK3 cells were infected with BacΔIBDA-GFP (left) and IBDV (right), respectively, for treatment with Freon 113 as described in Materials and Methods. A gradient with a CsCl density of 1.30 or 1.33 was used for purification of structures produced by BacΔIBDA-GFP and IBDV, respectively. The gradients were illuminated with white light and photographed. Material collected from the bands present in the gradients was negatively stained with 1% uranyl acetate. For the IBDV gradient, the heavier band was selected for electron microscopy. The insets show VLP-GFP and virions at a higher magnification. (B) Polypeptide identification. The left panel shows SDS-PAGE analysis and Coomassie blue staining of purified IBDV (lane 1) and of the material isolated from the BacΔIBDA-GFP band present in the CsCl gradient (lane 2). The right panels show Western blot analysis of purified IBDV (lanes 1) and of the material present in the BacΔIBDA-GFP sample (lanes 2) with an anti-pVP2/VP2, an anti-VP3, and an anti-GFP antibody. The relative Mrs (in thousands) determined by reference to marker proteins are indicated on the left.
Fluorescence microscopy of VLP-GFP.
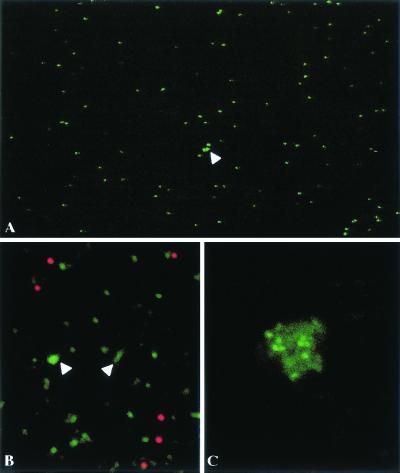
With a standard epifluorescence microscope, observation of 1 μl of a suspension of VLPs revealed green spots that looked like a myriad of stars on a dark night (Fig. 6). The number of spots in a field varied as a function of the dilution (data not shown). The intensities of the spots were fairly homogenous, with some exceptions that could correspond to small aggregates (Fig. 6A). To confirm that each spot corresponds to a single VLP, we observed a mixture of 0.1-μm-diameter red-fluorescent microspheres and VLP-GFP. It appeared under confocal microscopy that the spots had similar sizes, suggesting that the smaller green spots correspond to single VLPs (Fig. 6B). Addition of an anti-VP2 antibody led to VLP aggregation (Fig. 6C).
FIG. 6.
Visualization of individual fluorescent VLPs (A and B) A1-μl volume of purified VLPs was directly examined under a optical microscope (A) or mixed with red-fluorescent latex beads and examined under a confocal microscope (B). (C) VLPs were examined, after addition of an anti-VP2 antibody under a confocal microscope.
Vizualization of the assembly in the Sf9 cells.
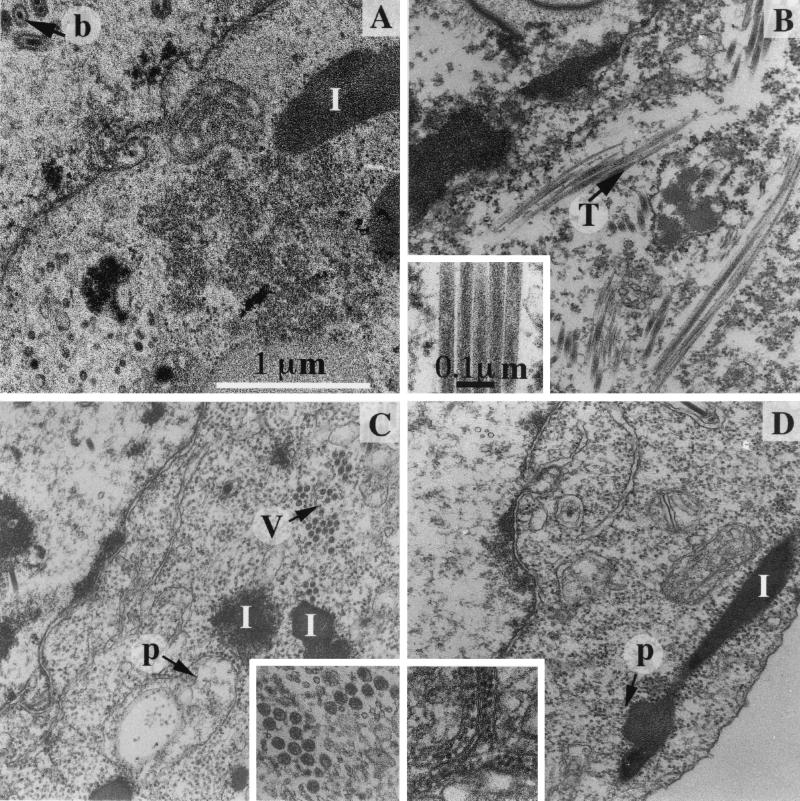
Baculovirus infection induces considerable morphological changes in the Sf9 nucleus as well as in the cytoplasm. In particular, large inclusions having more or less regular shapes accumulated in the cytoplasm (Fig. 7A). Infection with IBDA-expressing recombinant baculovirus led to the vizualization of 50-nm-diameter tubules in Sf9 cells (Fig. 7B). When the GFP was fused to VP3, infected cells showed numerous VLPs (Fig. 7C). When pVP2 was expressed without any other IBDV protein, small isometric particles were observed, which were identified as pVP2 assemblies (Fig. 7D). In many cases, they appeared strikingly associated with internal membranes.
FIG. 7.
Micrographs of thin sections of Sf9 cells infected by different recombinant baculoviruses. (A) The polyhedrin-negative baculovirus. Baculoviruses are assembled in the nucleus (b). In the cytoplasm, the main consequence of the baculovirus infection is the accumulation of inclusions with a more or less regular shape (I). (B) BacΔIBDA; (C) BacΔIBDA-GFP; (D) BacΔpVP2. The characteristic structures formed by the polyprotein-derived constructs are inserted in the corresponding micrographs: 50-nm-diameter tubules (T), VLPs (V), and isometric particles often associated with membranes (p). Tubules are occasionally observed in cells infected with BacΔIBDA-GFP (data not shown). Small numbers of isometric particles are also identified in cells infected by BacΔIBDA and BacΔIBDA-GFP, suggesting that pVP2 may self-assemble in all cases. Magnifications are identical in all micrographs and inserts.
DISCUSSION
We have discovered a morphogenesis switch controlling the assembly of VLPs in vivo. When expressed in baculovirus, the IBDA polyprotein pVP2-VP4-VP3 is not able to induce the self-assembly of the capsid proteins into VLPs. Polyprotein expression results mainly in the production of pVP2, VP3, and VP4 (11) and leads to the self-assembly of pVP2 into tubules (17) (see above). These tubules, which have a diameter of about 50 nm, are likely to be homologous to the type I tubules identified in IBDV-infected cells (7). Very few VLPs and tubules with a diameter of about 25 nm, made up of the viral protease VP4, were also identified in our preparations. In contrast, when an exogeneous protein was fused at the C terminus of VP3, we showed that the capsid proteins are able to self-assemble into VLPs. These particles have the same diameter and apparent geometry as the virions, and, remarkably, full processing of VP2 occurred during the formation of these structures. Our results have several implications for the identification of key events in IBDV morphogenesis. First, the fusion of GFP at the C terminus of VP3 triggers efficient capsid formation in insect cells expressing the hybrid polyprotein. We believe that the anchorage of the GFP molecules through a linker at the proximity of the charged residues of the C terminus of VP3 simulates the interaction of VP3 with genomic RNA and accounts for the correct VP3 assembly. We are carrying out further structural studies to validate this hypothesis. Second, the processing of pVP2 into the mature form VP2 appears to be dependent on proper assembly of the particle. While the processing is almost nonexistent when pVP2 assembles into tubules, it is very efficient during the formation of the capsids containing GFP. A similar maturation process has been identified for several viruses (6, 10, 18). In most cases, maturation occurs only on the viral particles and is an autocatalytic process. In our case, most of the processing is carried out by the VP4 protease. We indeed identified three target sites of VP4 in the carboxyl domain of pVP2 (13). Third, pVP2 self-assembles into different forms depending on the presence of VP3. While expression of the wild-type polyprotein leads to the self-assembly of pVP2 into tubules, expression of pVP2 alone gives rise to 30-nm-diameter isometric particles. Thus, VP3 plays a crucial role in IBDV assembly. From these findings, we propose a simple model for the virus morphogenesis, which is shown in Fig. 8. Our assembly model has two main characteristics. First, VP3 needs to be activated to promote virus particle assembly. In the virus cycle, the activation is carried out by either genomic RNA or VP1 (or both). In VLP assembly, the GFP activates VP3. In the absence of activation, VP3 can only nucleate pVP2 tube growth. Second, the maturation of VP2 occurs only during viral particle assembly.
FIG. 8.
Model for the assembly of VLPs (A), virions (B), and tubules (C). We hypothesized that the VP3 protein has two different conformations in thermodynamic equilibrium: a relaxed state (pink) and an activated one (red). Only the activated VP3 is supposed to self-assemble. Quaternary interactions involving VP3 and GFP (A) or VP3 and genomic RNA-VP1 (B) shift the equilibrium toward the activated form, favoring the spontaneous assembly of VP3 subviral particles. The proteolytic maturation of pVP2 into VP2 requires interaction with assembled VP3. When the equilibrium is not shifted, the activated VP3 forms nuclei only for the polymerization of pVP2 into tubules (C). The existence of the VP3 nuclei for the polymerization of pVP2 has been proposed by Martinez-Torrecuadrada et al. (17) and agrees with the fact that pVP2 alone self-assembles not into tubules but into isometric particles.
An efficient self-assembly of pVP2/VP2 and VP3 into VLPs has been recently reported using the polyprotein expressed in a vaccinia virus-HeLa cells system (5). One obvious difference between the two expression systems is the different intracellular pH, which is around 6.1 for Sf9 cells and 7.3 for HeLa cells (20). The assembly process of IBDV capsid proteins may be pH dependent. To test this hypothesis, BacΔIBDA Sf9-infected cells (normally grown at pH 6.3) were grown in a medium buffered at pH 7.3 for protein assembly analysis. Expression of the wild-type polyprotein leads to the formation of large numbers of pVP2 tubules and a few VLPs (data not shown). Therefore, we did not induce morphological changes by increasing the extracellular pH. We did not measure the value of the intracellular pH. We therefore cannot ruled out the possibility that the capsid assembly is not pH independent. Understanding the differences between the expression systems calls for further studies.
These results have additional consequences. Since large proteins such as GFP can be well processed and packaged into chimeric birnavirus VLPs, we postulate that VLPs could be used for cell-specific delivery of drugs. IBDV VLP carriers present two major advantages: encapsidation of the drug and specific targeting. Chimeric VLPs could be used as vaccine carriers for natively displayed proteins. It is likely that strong antibody responses can be evoked against native protein antigens, similar to what was observed when GFP was fused to capsid proteins of hepatitis B virus or rotavirus (2, 12).
A perspective for structural studies is that proteins fused inside IBDV VLPs might become amenable for high-resolution cryoelectron microscopy analyses. This possibility was also investigated for hepatitis B virus VLP with a GFP protein on the outside of the capsid (12). In this case, the GFP insert flanked on both sides by Gly-rich inserts was inserted at the tips of the surface spikes. GFP was clearly visible in the cryoelectron micrographs, but the three-dimensional reconstruction revealed a partially ordered structure. In this respect, a possible interest of the IBDV chimeric VLPs is that the foreign protein should be more rigidly fixed inside the particles since it would trigger VLP assembly. Concerning specific applications of the GFP-VLPs, they might be used to correlate cell IBDV permissivity and virus binding and also to trace subsequent steps of virus infection.
In conclusion, we demonstrated that addition of a protein to the inner VP3 capsid protein of IBDV triggered VLP assembly in a recombinant expression system. A strong correlation between the efficient VLP formation and the maturation of pVP2 and VP2 was demonstrated, suggesting that pVP2-to-VP2 cleavage is driven by initial preassembly of pVP2 into premature virions. This work opens the possibility of developing new vectors by producing chimeric VLPs. Attractive foreign candidates would be protein antigens implicated in immune protection against important microorganisms.
Acknowledgments
We thank Philippe Fontange for assistance with the confocal microscopy work; Jean Cohen and Annie Charpilienne for helpful discussions; Cynthia Jaeger for the DNA sequencing work; Nathalie Castagné, Yann Benureau, and Nathalie Lejal for technical help; and Yves Gaudin and Felix Rey for critical reading of the manuscript.
REFERENCES
- 1.Böttcher, B., N. A. Kiselev, V. Y. Stel'Mashchuk, N. A. Perevozchikova, A. V. Borisov, and R. A. Crowther. 1997. Three-dimensional structure of infectious bursal disease virus determined by electron cryomicroscopy. J. Virol. 71:325-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charpilienne, A., M. Berois, M. Nejmeddine, G. Trugnan, E. Neumann, E. Hewat, and J. Cohen. 2001. Vizualization of individual rotavirus-like-particle containing 120 molecules of green fluorescent protein (in living cells). J. Biol. Chem. 276:29361-29367. [DOI] [PubMed] [Google Scholar]
- 3.Cosgrove, A. S. 1962. An apparently new disease of chicken—avian nephrosis. Avian Dis. 6:385-389. [Google Scholar]
- 3a.Da Costa, B., C. Chevalier, C. Henry, J.-C. Huet, S. Petit, J. Lepault, H. Boot, and B. Delmas. 2002. The capsid of infectious bursal disease virus contains several small peptides arising from the maturation process of pVP2. J. Virol. 76:2393-2402. [DOI] [PMC free article] [PubMed]
- 4.Dybing, J. K., and D. J. Jackwood. 1997. Expression of MD infectious bursal disease viral proteins in baculovirus. Avian Dis. 41:617-626. [PubMed] [Google Scholar]
- 5.Fernandez-Arias, A., C. Risco, S. Martinez, J. P. Albar, and J. F. Rodriguez. 1998. Expression of ORF A1 of infectious bursal disease virus results in the formation of virus-like particles. J. Gen. Virol. 79:1047-1054. [DOI] [PubMed] [Google Scholar]
- 6.Gallagher, T. M., and R. R. Rueckert. 1988. Assembly-dependent maturation cleavage in provirions of a small icosahedral insect ribovirus. J. Virol. 62:3399-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Granzow, H., C. Birghan, T. C. Mettenleiter, J. Beyer, B. Kollner, and E. Mundt. 1997. A second form of infectious bursal disease virus-associated tubule contains VP4. J. Virol. 71:8879-8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hihara, H., H. Yamamoto, K. Arai, W. Okazaki, and T. Shimizu. 1980. Conditions for successful cultivation of tumor cells from chickens with avian lymphoid leukosis. Avian Dis. 24:971-979. [PubMed] [Google Scholar]
- 9.Hu, Y. C., W. E. Bentley, G. H. Edwards, and V. N. Vakharia. 1999. Chimeric infectious bursal disease virus-like particles expressed in insect cells and purified by immobilized metal affinity chromatography. Biotechnol. Bioeng. 63:721-729. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, J. E. 1996. Functional implications of protein-protein interactions in icosahedral viruses. Proc. Natl. Acad. Sci. USA 93:27-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kibenge, F. S., B. Qian, E. Nagy, J. R. Cleghorn, and D. Wadowska. 1999. Formation of virus-like particles when the polyprotein gene (segment A) of infectious bursal disease virus is expressed in insect cells. Can. J. Vet. Res. 63:49-55. [PMC free article] [PubMed] [Google Scholar]
- 12.Kratz, P. A., B. Bottcher, and M. Nassal. 1999. Native display of complete foreign protein domains on the surface of hepatitis B virus capsids. Proc. Natl. Acad. Sci. USA 96:1915-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lejal, N., B. Da Costa, J.-C. Huet, and B. Delmas. 2000. Role of Ser-652 and Lys-692 in the protease activity of infectious bursal disease virus VP4 and identification of its substrate cleavage sites. J. Gen. Virol. 81:983-992. [DOI] [PubMed] [Google Scholar]
- 14.Leong, J. C., D. Brown, P. Dobos, F. S. B. Kibenge, J. E. Ludert, H. Müller, E. Mundt, and B. Nicholson. 2000. Family Birnaviridae, p. 481-490. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, D. J. McGeoch, J. Maniloff, M. A. Mayo, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Seventh Report of the International Committee on the Taxonomy of Viruses. Academic Press, Inc., San Diego, Calif.
- 15.Lombardo, E., A. Maraver, J. R. Caston, J. Rivera, A. Fernandez-Arias, A. Serrano, J. L. Carrascosa, and J. F. Rodriguez. 1999. VP1, the putative RNA-dependent RNA polymerase of infectious bursal disease virus, forms complexes with the capsid protein VP3, leading to efficient encapsidation into virus-like particles. J. Virol. 73:6973-6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lombardo, E., A. Maraver, I. Espinosa, A. Fernandez-Arias, and J. F. Rodriguez. 2000. VP5, the non structural polypeptide of infectious bursal disease virus, accumulates within the host plasma membrane and induces cell lysis. Virology 277:345-357. [DOI] [PubMed] [Google Scholar]
- 17.Martinez-Torrecuadrada, J. L., J. R. Caston, M. Castro, J. L. Carrascosa, J. F. Rodriguez, and J. I. Casal. 2000. Different architectures in the assembly of infectious bursal disease virus capsid proteins expressed in insect cells. Virology 278:322-331. [DOI] [PubMed] [Google Scholar]
- 18.Rossmann, M. G., E. Arnold, J. W. Erickson, E. A. Frankenberger, J. P. Griffith, H. J. Hecht, J. E. Johnson, G. Kamer, M. Luo, A. G. Mosser, R. R. Rueckert, B. Sherry, and G. Vriend. 1985. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature (London) 317:145-153. [DOI] [PubMed] [Google Scholar]
- 19.Sanchez, A. B., and J. F. Rodriguez. 1999. Proteolytic processing in infectious bursal disease virus: identification of the polyprotein cleavage sites by site-directed mutagenesis. Virology 262:190-199. [DOI] [PubMed] [Google Scholar]
- 20.Vachon, V., M. J. Paradis, M. Marsolais, J-.L. Schwartz, and R. Laprade. 1995. Endogenous K+/H+ exchange activity in the Sf9 insect cell line. Biochemistry 34:15157-15164. [DOI] [PubMed] [Google Scholar]
- 21.Vakharia, V. N., D. B. Snyder, J. He, G. H. Edwards, P. K. Savage, and S. A. Mengel-Whereat. 1993. Infectious bursal disease virus structural proteins expressed in a baculovirus recombinant confer protection in chickens. J. Gen. Virol. 74:1201-1206. [DOI] [PubMed] [Google Scholar]