Abstract
The capsid proteins VP2 and VP3 of infectious bursal disease virus, a birnavirus, are derived from the processing of a large polyprotein: NH2-pVP2-VP4-VP3-COOH. Although the primary cleavage sites at the pVP2-VP4 and VP4-VP3 junctions have been identified, the proteolytic cascade involved in the processing of this polyprotein is not yet fully understood, particularly the maturation of pVP2. By using different approaches, we showed that the processing of pVP2 (residues 1 to 512) generated VP2 and four small peptides (residues 442 to 487, 488 to 494, 495 to 501, and 502 to 512). We also showed that in addition to VP2, at least three of these peptides (residues 442 to 487, 488 to 494, and 502 to 512) were associated with the viral particles. The importance of the small peptides in the virus cycle was assessed by reverse genetics. Our results showed that the mutants lacking the two smaller peptides were viable, although the virus growth was affected. In contrast, deletions of the domain 442 to 487 or 502 to 512 did not allow virus recovery. Several amino acids of the peptide 502 to 512 appeared essential for virus viability. Substitutions of the P1 and/or P1" position were engineered at each of the cleavage sites (P1-P1": 441-442, 487-488, 494-495, 501-502, and 512-513). Most substitutions at the pVP2-VP4 junction (512-513) and at the final VP2 maturation cleavage site (441-442) were lethal. Mutations of intermediate cleavage sites (487-488, 494-495, and 501-502) led to viable viruses showing different but efficient pVP2 processing. Our data suggested that while peptides 488 to 494 and 495 to 501 play an accessory role, peptides 442 to 487 and 502 to 512 have an unknown but important function within the virus cycle.
The birnaviruses are a family of small icosahedral viruses infecting insects, fish, and birds (15). Only five proteins, generally referred to as VP1, VP2, VP3, VP4, and VP5, are encoded by the viral genome. The T=13 icosahedral birnavirus capsids are made by the VP2 and VP3 proteins. They contain the two double-stranded RNA genomic segments (A and B) and the VP1 protein, a putative RNA-dependent RNA polymerase. Translation of genomic segment A yields a polyprotein, pVP2-VP4-VP3, and a small protein, VP5, of unknown function. The B segment encodes VP1. The polyprotein processing gives rise to VP4, the viral protease, and VP2 and VP3. VP2 carries all the neutralizing epitopes, suggesting that it is at least partly exposed at the outer surface of the capsid. VP3, which interacts with VP1 (16, 24), is thought to be located on the inner surface of the capsid (6). VP3 contains charged residues at its carboxy-terminal domain, a domain suggested to be involved in the genomic RNA interaction. As found for other virus families, the capsid assembly seems to be regulated by polyprotein processing.
The infectious bursal disease virus (IBDV), an avian birnavirus, is of major importance to the poultry industry. It causes an immunosuppressive disease in young chickens. After infection, IBDV multiplies rapidly in the B lymphocytes of the bursa of Fabricius, leading to increased susceptibility to other diseases. Very virulent strains have resulted in high rates of mortality in many countries.
The first step governing the IBDV capsid assembly is the autoproteolytic cleavage of the polyprotein (1,012 amino acids). This process generates pVP2, VP4, and VP3. The pVP2-to-VP2 conversion involves several proteolytic cleavages at the carboxy end of pVP2 (1, 14, 20). Based on mutagenesis studies, the putative cleavage site was proposed as defined by the (Thr/Ala)-X-Ala↓Ala motif; three potential sites are present in the C-terminal domain of pVP2 (14).
In the present study, we further analyzed the maturation process of VP2. By using mass spectrometry and N-terminal sequence analysis, we showed that VP2 and three (most probably four) peptides derived from the pVP2 are associated with the virus particle. These results confirmed the presence of the three cleavage sites previously proposed (14). To analyze the importance of these peptides to the virus cycle, we developed a reverse genetic system similar to the one previously described (5), based on a self-processing ribozyme cassette (7). We showed that the two 7-amino-acid long peptides are not essential for virus viability, in contrast to the two peptides located at the ends of the domain characteristic of pVP2. These essential peptides are likely to play an important role in the virus cycle.
MATERIALS AND METHODS
Virus purification.
A 100-ml volume of supernatants of IBDV strain CT-infected chicken embryo fibroblasts was ultracentrifugated at 40,000 rpm for 1 h in a 45Ti rotor (Beckman) at 4°C. The pellets were resuspended in 10 mM Tris (pH 7.4)-100 mM NaCl-1 mM EDTA and then treated with Freon 113 for purification by density gradient centrifugation in CsCl with a density of 1.33. Ultracentrifugation was carried out overnight at 35,000 rpm in an SW55 rotor (Beckman) at 4°C. The bands were collected and stored at −4°C.
Gel electrophoresis.
To analyze large peptides, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (16% acrylamide) was performed using a Mini-Protean II system (Bio-Rad, Paris, France) by the method of Schägger and von Jagow (21) with modifications (19). LMW and PMW calibration kits (Pharmacia) were used as molecular weight standards, and the proteins were stained with Serva blue G. To analyze proteins, aliquots were subjected to SDS-PAGE (10% acrylamide) (13) and the gels were dried and exposed for autoradiography.
Protein labeling and immunoprecipitation.
In vitro, polymerase T7-driven expression was carried out using the TNT Quick-Coupled transcription-translation system (Promega) as described by the manufacturer, except that the reactions were performed in a final volume of 11 μl. The DNA template (1 μg) was incubated for 1 h at 30°C. After incubation, aliquots of 2 to 3 μl were subjected to gel electrophoresis. In cell culture, 2 × 106 LSCC-BK3 cells were infected with 150 μl of supernatant of cells infected for 5 days. At 18 h postinfection, the cells were resuspended in 1.5 ml of medium-10 μl of Promix (Amersham). At 6 h later, the cell supernatants were half-diluted in RIPA buffer (50mM Tris [pH 8], 150 mM NaCl, 2% Triton X-100) and the cells were resuspended in 1 ml of the same buffer. After clarification by centrifugation, the cell or supernatant extracts were processed for immunoprecipitation. Extracts were incubated for 2 h at room temperature with 1 μl of undiluted hybridoma ascites fluid and 35 μl of a 1:1 slurry of protein A-Sepharose 4B (Pharmacia) under gentle agitation. The beads were washed four times with 1 ml of RIPA buffer, treated for 2 min at 100°C in Laemmli denaturing buffer plus 5% 2-mercaptoethanol, and centrifugated. The resulting supernatants were subjected to electrophoresis.
Amino acid sequence analysis.
The virus purified by CsCl density centrifugation (140 μl of 2.7 M CsCl) was diluted 10-fold with water and then adsorbed on a Prosorb device (polyvinylidene difluoride [PVDF] membrane) by low-speed centrifugation (500 × g at 15°C) in Hettich 16R (flow rate, 20 μl/min). Membrane washes were carried out with 10% methanol in water. For large peptides, electrophoretic samples were transferred onto the PVDF membrane by passive absorption as described previously (L. R. Zieske, S. Masiaiarz, S. Chamberlain, and J. McGovern, Bioinformatics Biomol. Technol: Linking Genomes Proteomes Biochem., poster, 1999). After the destaining step, the band of interest was excised and dried in a Speed-Vac for 30 min, and the gel piece was reswollen in 50 μl of 2% SDS in 0.2 M Tris-HCl (pH 8.5) for 30 min. After swelling, 200 μl of high-pressure liquid chromatography (HPLC) water was added and then a piece of prewet (methanol) PVDF membrane (4 by 4 mm [Problott]) was added to the solution. The procedure required 2 days at room temperature (23°C) with gentle vortexing. At the end of this transfer time, the gel piece and the solution were clear and the membrane was blue. The membrane was washed five times with 1 ml of 10% methanol with vortexing. After drying, the membranes were placed in the cartridge of the Perkin-Elmer Procise 494 HT protein sequencer. N-terminal amino acid sequence analyses were performed by automated Edman chemistry with methods and reagents recommended by the manufacturer.
Reversed-phase liquid chromatography of peptides.
The virus purified by CsCl density centrifugation (140 μl), desalted twice on Sephadex G-25, and dried in a Speed-Vac was extracted with 4 μl of 5% formic acid-10% acetonitrile in water for 2 h and diluted to 20 μl with solvent A (0.1% [vol/vol] Fluka formic acid and 4 mM Fluka ammonium acetate in Merck HPLC water). Reversed-phase liquid chromatography was run with an Applied Biosystems device (pump 140D and UV detector 785 with U-shaped fused silica tubing; 7-mm path length) on a C4 LC-Packings capillary column (0.3 by 150 mm; porosity, 300 Å) at a controlled temperature (40°C). The gradient was made by mixing solvent A with solvent B (90% Perkin-Elmer acetonitrile, 0.1% [vol/vol] Fluka formic acid, and 4 mM Fluka ammonium acetate in Merck HPLC water) at a flow rate of 4 μl/min. The acetonitrile gradient began at 4.5% and increased linearly to 85.5% in 90 min. After being monitored for absorbance at 215 nm, the flow was split between 0.67 μl/min for matrix-assisted laser desorption ionization (MALDI) interplate collection and 3.33 μl/min for manual collection.
Mass spectrometry analysis.
A 4-μl volume 50% (vol/vol) acetonitrile-0.3% (vol/vol) trifluoroacetic acid was added to the dry sample, and the solution was vortexed for 10 min. Next, the sample was desalted on ZipTip C4 (Millipore) and eluted with 0.5 μl of 70% (vol/vol) acetonitrile-0.3% (vol/vol) trifluoroacetic acid. The sample was directly spotted onto the MALDI plate and dried at room temperature, and then 0.5 μl of α-cyano-4-hydroxycinnamic acid (α-CHCA; 3 mg/ml) in 50% (vol/vol) acetonitrile-0.1% (vol/vol) trifluororacetic acid was added. Mass spectra were acquired on a Voyager-DE-STR time-of-flight mass spectrometer (Applied Biosystems, Framingham, Mass.) equipped with a nitrogen laser emitting at λ = 337 nm (Laser Science, Franklin, Mass.). The accelerating voltage used was 20 kV. All spectra were recorded in the positive reflector mode with a delayed extraction of 130 ns and a 62% grid voltage. The spectra were calibrated using an external calibration: Des-Arg bradykinin, (M + H)+ = 904.4681 Da; angiotensin, (M + H)+ = 1296.6853 Da; neurotensin, (M + H)+ = 1672.9175 Da; melittin, (M + H)+ = 2845.762 Da, and bovine insulin B chain, (M + H)+ = 3494.6513 Da.
Construction of pT7-A-HDR.
The complete genomic segment A of the Gumboral IBDV vaccine strain CT (Mérial, Lyon, France) was amplified by reverse transcriptase PCR and cloned into pUC19 using the EcoRI restriction site to generate pUC-IBDA as previously described (14). Thus, the 5" terminus of the IBDA segment (GGATACGATCG…) was placed just downstream of the T7 promoter. The pUC-IBDA plasmid was digested with BsrGT and treated with mung bean nuclease to generate a blunt end just at the 3" end of the IBDA segment. A plasmid carrying the hepatitis delta virus antigenomic ribozyme and the terminator T7 sequence was previously constructed (2), digested with SmaI and EcoRV for excision of the insert, and subcloned into the pUC-IBDA plasmid at the blunted BsrGI position. DNA sequencing revealed the unexpected codon AAG at position 841 of the polyprotein gene, which was modified to GAG by homologous substitution of an independent SacII-BglII IBDA segment (nucleotides 1756 to 3019). Sequence analysis was carried out to confirm the sequence of the exchanged segment. The resulting plasmid was named pT7-A-HDR.
Construction of pT7-B-HDR.
The complete IBDB segment of the CT strain was amplified by reverse transcriptase PCR with oligonucleotides 5"-AGAGAATTCTAATACGACTCACTATAGGATACGATGGGTCTGACCCTCT and 5"-GATGAATTCTGCAGCCCGGGGGCCCCCGCAGGCGAAGGCC and cloned into the EcoRI site of pUC19. Next, the pBluescript II SK(−) phagemid (Stratagene) was cut with SspI and HincII and self-ligated to delete the T7 promoter (plasmid SKΔ). The pUC-IBDB plasmid was digested with PstI and EcoRI, and the insert was cloned into SKΔ. SKΔ-IBDB was cut with SmaI for blunt-end ligation with the ribozyme terminator DNA segment excised SmaI-EcoRV. The complete sequencing of the insert was carried out, and an unexpected codon CCA was identified at position 773 of VP1. Codon 773 was modified to TCA by site-directed mutagenesis and homologous substitution of the NarI-BsrGI-mutated segment (nucleotides 2046 to 2536). Sequence analysis was carried out to verify the sequence of the substituted segment. The resulting plasmid was named pT7-B-HDR.
Construction of pT7-A-HDR derivatives.
The mutations were introduced by using the Pfu DNA polymerase with the QuickChange site-directed mutagenesis kit (Stratagene) as described by the manufacturer. Nucleotide sequence analyses were carried out to confirm nucleotide substitutions. The corresponding DNA stretch defined by restriction sites Eco52i (nucleotide 1291) and NruI (nucleotide 1689) was excised and subcloned into pT7-A-HDR restricted by Eco52i and NruI. Next, pT7-A-HDR derivatives were validated by sequence analysis of the complete [Eco52i-NruI] insert using the primers 5"-GGTGAGCAACTTCGAGCTGATCCCA and 5"-ACTGAGTGAGAGGGACCGTCTTG.
Generation of recombinant viruses.
To generate infectious IBDV from cDNA clones, a strategy similar to that previously described by Boot et al. (5) was used. For virus rescue experiments, we used the primary hepatocellular carcinoma epithelial cell line LMH (12) or the quail pectoralis fibrosarcoma cell line QT6 (17) grown in RPMI medium supplemented with 10% Fetal calf serum, 1% chicken serum, 2 mM glutamine, and 1 mM sodium pyruvate and complemented with tryptose phosphate broth (Gibco-BRL) for QT6 cells. Cells at 90% confluence in P12 wells were infected with MVA-T7 (27) at a multiplicity of infection of 1. After a 1-h adsorption, the cells were rinsed with the RPMI medium. In the meantime, pT7-A-HDR derivatives and pT7-B-HDR were mixed (0.5 μg for each plasmid) with 3.5 μl of LipofectAMINE (GIBCO-BRL) in 350 μl of OptiMEM and were kept at room temperature for 30 min. The cells were again rinsed in OptiMEM and incubated with the DNA-LipofectAMINE mixture at 37°C for 5 h. Subsequently, 0.5 ml of OptiMEM was added to the cells for overnight incubation. Next, 1.5 ml of complemented RPMI medium was added to each well. Recombinant mutant viruses were recovered 48 or 72 h posttransfection after filtration through a 0.22-μm-pore-size filter. At least four independent transfection experiments were carried out to analyze each pT7-A-HDR derivative. The viruses were amplified on LSCC-BK3 cells, a B-lymphoid cell line highly permissive for IBDV (25).
FACS analysis.
IBDV-infected LSCC-BK3 cells were analyzed 3 or 5 days postinfection. Briefly, the cells were fixed with 2.5% paraformaldehyde in phosphate-buffered saline for 30 min at room temperature and permeabilized by incubation for 10 min in 0.1% Triton X-100. Fixed cells were incubated with a 1:250 dilution of an anti-VP3 monoclonal antibody. Next, the cells were rinsed and incubated with an anti-immunoglobulin mouse fluorescein isothiocyanate conjugate in phosphate-buffered saline-0.05% Tween. The cells were rinsed three times and subjected to FACScan (Becton Dickinson) analysis using the Cell Quest software.
RESULTS
Identification of several IBDA polyprotein-derived peptides as components of the IBDV virions.
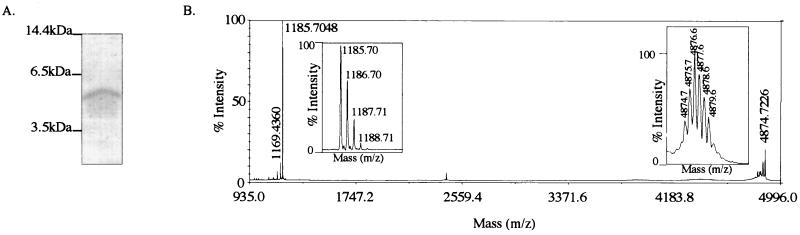
VP2, one of the protein capsids of IBDV, arises from pVP2 through proteolytic processing. To gain information on the maturation of pVP2, we investigated the possible presence in the viral particles of pVP2-derived peptides. IBDV particles were purified by density gradient centrifugation. Three main bands were observed in the gradient (Fig. 5, in the accompanying paper [6a]). All the bands displayed virus particules when analyzed by electron microscopy, and they displayed the structural proteins VP1, VP2, and VP3 when analyzed by SDS-PAGE. It should be noted that type I and type II tubules, made of pVP2 and VP4, respectively (10), were not revealed by electron microscopy in our preparations. Experiments were first carried out with the material present in the lower band; other bands gave similar results. In addition to the capsid proteins, we identified a polypeptide with a relative mass of about 5 kDa by using SDS-PAGE (16% polyacrylamide) (Fig. 1A). The first 15 amino acids of the polypeptide were determined through N-terminal sequencing: NH2-Phe-Gly-Phe-Lys-Asp-Ile-Ile-Arg-Ala-Leu-Arg-Arg-Ile-Ala-Val-COOH. This sequence was identical to the IBDA polyprotein sequence starting at Phe-442. To investigate the presence of this peptide, mass spectrometry analysis was carried out on the viral particles. A peptide with an [M + H]+ mass (23) of 4,874.72 Da was indeed detected (Fig. 1B). No other peptide with the same mass range was detected. This [M + H]+ mass fits well the mass of a peptide extending from Phe-442 to Ala-487, which is 4,874.67 Da.
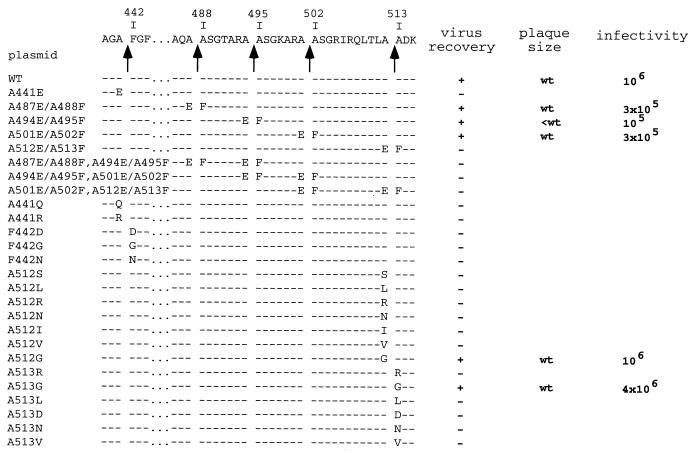
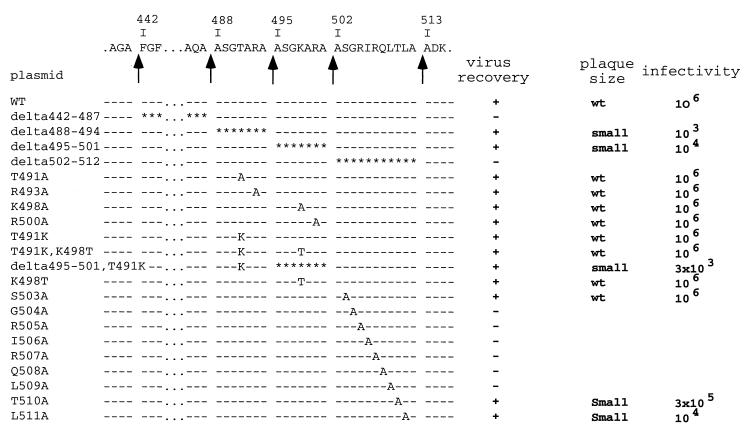
FIG. 5.
Virus recovery for the pVP2 cleavage site mutants. The amino acid sequence of the pVP2-specific domain and the different cleavage sites (arrows) are indicated at the top of the figure. The mutated amino acid(s) is indicated for each construct in the single-letter code. Depending on the virus recovery results, the plasmids are indicated by a + (recovery) or − (nonrecovery). The relative plaque size at 48 h after infection of confluent LMH cell monolayers is indicated. wt, wild-type size; small, smaller than wild-type size. Rescued viruses were amplified on LSCC-BK3 cells, and virus titers were determined 3 days postinfection on LMH cells.
FIG. 1.
Characterization of peptides present in IBDV particles. (A) SDS-PAGE (16% polyacrylamide) analysis of purified virus. A peptide of about 5 kDa was identified and excised from the gel for N-terminal sequence analysis. Its N terminus was mapped at residue 442. Molecular mass markers are indicated on the left. (B) Mass spectrometry analysis of IBDV particles. Two main signals were identified on the mass/charge window ranging from 935 to 4,996. Magnified signals showing the isotopic pattern are inserted. The [M + H]+ values for the peptides are 1,185.7 and 4,874.7 Da, respectively.
Another peptide with an [M + H]+ mass of 1,185.70 Da was also revealed (Fig. 1B). To purify the peptide from the virus particle preparations, we performed HPLC purification (data not shown). Two major peaks were detected. Mass spectrometry was carried out on each fraction. A peptide with a mass of 1,185.82 Da eluted in the first peak. The complete sequence of this peptide was determined as NH2-Ala-Ser-Gly-Arg-Ile-Arg-Gln-Leu-Thr-Leu-Ala-COOH. This sequence was identical to the IBDA polyprotein sequence from residues 502 to 512. Since cleavage at the pVP2-VP4 junction occurs between amino acids 512 and 513, this peptide is indeed the C terminus of pVP2. Note that the theoretical [M + H]+ mass of this peptide was 1,185.71 Da, fitting well with its experimental determinations. The second peak that eluted from the gradient had a mass of 28,791 Da and corresponded to VP3.
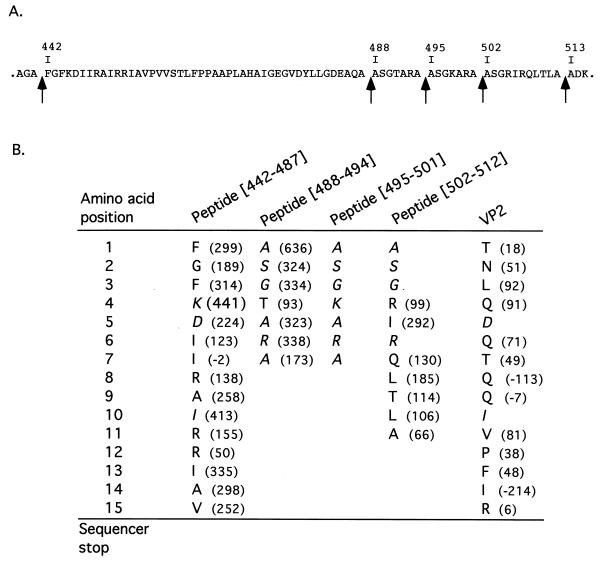
The presence of the two peptides from residues 442 to 487 ([442-487]) and from 502 to 512 ([502-512]) in the viral particles prompted us to locate a putative peptide(s) deriving from the 14-amino-acid sequence extending from residues 488 to 501 (Fig. 2A). The sequence surrounding the two alanines at positions 494 and 495 had a strong homology to the cleavage sites identified at positions 487-488 and 501-502, thus suggesting the possible generation of two 7-amino-acid peptides. To identify the putative peptide(s), we carried out an N-terminal sequencing on the purified virus. Because the main capsid proteins VP2 and VP3 were reported to be blocked (9), only peptides were expected to be sequenced. The results are presented in Fig. 2B. Large numbers of Ala, Ser, and Gly residues were revealed at positions 1, 2, and 3, a sequence which was present at the N terminus of peptide [502-512] but also at the N terminus of the two putative 7-amino-acid peptides. Importantly, the sequence Thr-Ala-Arg-Ala was identified at positions 4 to 7, thus demonstrating the presence of an additional peptide with an N terminus at position 488. No Ala, Ser, or Gly residues were detected at positions 8, 9, or 10, respectively, showing that the 7-amino-acid peptides were analyzed. Our results demonstrated that the viral particles contain the peptide extending from residues 488 to 494. The presence of the fourth peptide mapping from residues 495 to 501 inside the viral particles was not demonstrated, since each of its amino acids was redundant with a residue present in the three other peptides; however, its presence cannot be ruled out. Finally, we also observed that the N terminus of the VP2 protein was only partially blocked, allowing identification of the N terminus of VP2 as the Thr immediatly following the Met initiation codon. In conclusion, at least three (probably four) peptides, which correspond to amino acid stretches 442 to 487, 488 to 494, 495 to 501, and 502 to 512, were detected in viral particles. Note that the three shorter peptides have sequence identity: all of them have (i) the triplet Ala-Ser-Gly at their N termini, (ii) an Ala residue at their C termini, and (iii) generally positively charged residues at positions 4 and 6 (Fig. 2).
FIG. 2.
N-terminal sequencing of the purified virus. (A) Sequence of the pVP2-specific domain from amino acids 442 to 512. The single-letter code is used to indicate amino acids. Arrows indicate the cleavage sites. (B) The nature of the amino acids and the amount (differential picomoles) revealed at each Edman degradation cycle (amino acid position) are indicated. Sequences of the peptides (and of the mature VP2) have been aligned on the identified residues. Residues which are potentially present in more than one peptide are indicated in italics.
Identification of several IBDA polyprotein-derived peptides as components of the IBDV VLPs.
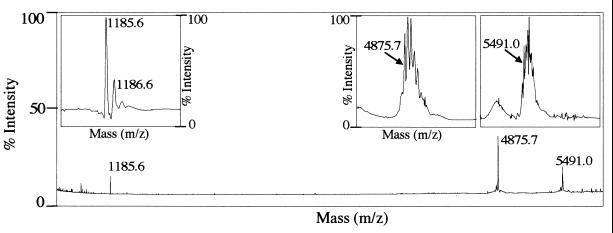
We showed that the final maturation process converting pVP2 into the mature form of VP2 occurs during the generation of virus-like particles (VLPs) in the baculovirus expression system (6a). We therefore carried out a mass spectrometry analysis on VLPs. Three peptides, with [M + H]+ masses of 1,185.6, 4,875.7 and 5,491.0 Da, were detected (Fig. 3). The first two peaks were identical to those identified on virions and corresponded to the [442-487] and [502-512] peptides. This result showed that the processing is similar in the virus and in VLPs, although not identical, as shown by the presence of a third peak. The mass of the third peak (5,491.0 Da) corresponded to the sum of the masses of the [442-487] and [488-494] peptides minus the mass of a water molecule (4874.67 + 632.32 − 18.01). These results suggested that the kinetics of pVP2 maturation is slower during VLP formation than for virions.
FIG. 3.
Mass spectrometry analysis of IBDV VLPs. Three main signals were identified in the mass/charge window. Magnified signals showing the isotopic pattern are inserted. The [M + H]+ values for the peptides are 1,185.6, 4,875.7 and 5,491.0 Da, respectively.
The amino acid stretch from 442 to 512 constitutes the pVP2-specific domain.
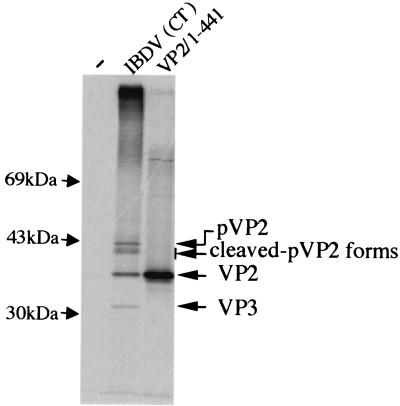
The existence of the four peptides suggests that the amino acid stretch from 442 to 512 represents the domain which is processed during pVP2 maturation. To determine if the pVP2-processed domain involves residus upstream of residue 441, we engineered a construct driving expression of the polyprotein with a stop codon at position 442 to compare the electrophoretic mobility of the mature VP2 with this truncated form. Because pVP2 processing is not observed in the in vitro expression system, full-size protein production was expected under these conditions. As shown in Fig. 4, the mature VP2 and the in vitro-expressed VP2-stop442 comigrated, suggesting that the pVP2 characteristic domain does not extend upstream of residue 441.
FIG. 4.
Comparison of the electrophoretic mobilities of the different forms of VP2. Mock-infected (−) or IBDV-infected [IBDV(CT)] cells were 35S labeled and subjected to immunoprecipitation with an anti-VP2 monoclonal antibody. Denatured immune complexes were run side by side on an SDS-PAGE gel with an in vitro translation product of the mutated IBDA polyprotein having a stop codon at position 442 (VP2/1-441). The gel was subjected to autoradiography. Molecular mass markers are indicated on the left.
Effect of mutations at the pVP2 cleavage sites.
To study the pVP2 processing, we constructed a reverse genetic system. Engineered plasmids pT7-A-HDR and pT7-B-HDR were constructed as described in Materials and Methods. Cotransfections of these plasmids into T7 polymerase-expressing cells resulted in the production of a viable virus. We analyzed the effects of amino acid substitutions at the P1 and P1" positions of each cleavage site. Two groups of mutants characterized by double or single substitutions were constructed (Fig. 5). The first group was made up of eight mutants in which the P1 and the P1" residues were replaced by Glu and Phe, respectively. The second group was made up of 18 point mutants to analyze amino acid specificity at the P1 or P1" positions of the pVP2-VP4 cleavage site (position 512-513) and the VP2 maturation cleavage site (position 441-442).
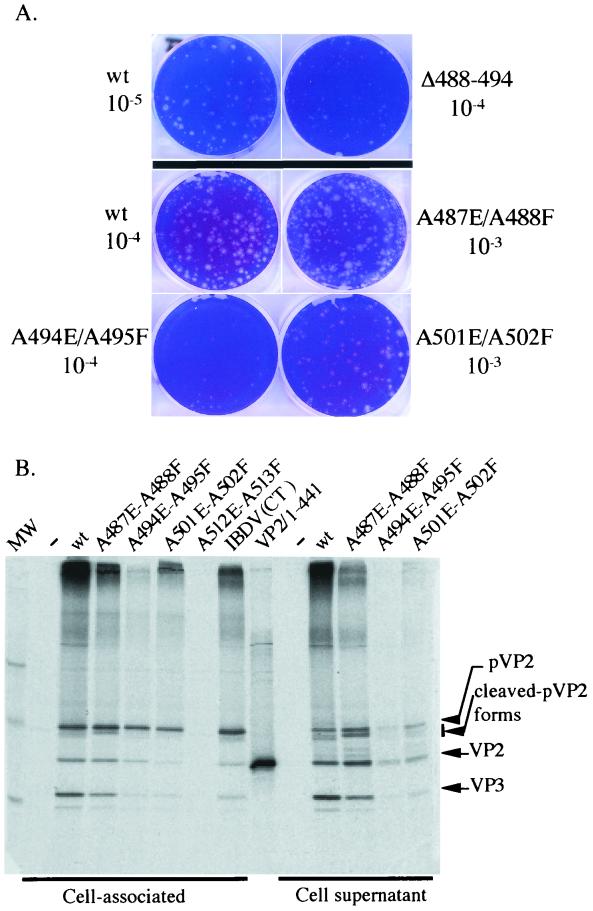
With the first group of mutants, the viruses were consistently obtained on transfection with the pT7-A-HDR derivatives A487E/A488F, A494E/A495F, and A501E/A502F (Fig. 5). No virus was recovered in the cells transfected with the A512E/A513F plasmid or with plasmids harboring cumulative double substitutions engineered at two pVP2 cleavage sites.
Viral growth of the three viable mutants was quantified by performing a plaque assay (Fig. 6A) and by measuring virus production at 3 days postinfection (Fig. 5). While the mutant viruses A487E/A488F and A501E/A502F showed wild-type growth characteristics, virus A494E/A495F produced only 10 to 20% of the amount of the wild-type virus and displayed a small-plaque phenotype. pVP2 processing was also studied by SDS-PAGE for all mutants. Figure 6B shows the VP2 band pattern in infected cells and in their cell medium. These mutations modulated the processing of pVP2. For instance, whereas two faint bands of cleaved pVP2 forms were observed with the wild-type virus, a unique main cleaved pVP2 form was identified with the virus A487E/A488F. To quantify the efficiency of the pVP2-to-VP2 conversion, autoradiograms were subjected to densitometry. The ratio between the pVP2 and mature VP2 was not significantly modified in these mutants.
FIG. 6.
Phenotypes of several mutants. (A) Typical aspect of plaques formed by the indicated mutants. The dilutions used for the assay are indicated. (B) Analysis of the pVP2 processing in cells infected with the indicated mutants. Left lanes show VP2 immunoprecipitations of the cell extracts; right lanes show VP2 immunoprecipitations of the cell supernatants. In the middle is shown mature VP2 (VP2/1-441) expressed in vitro. IBDV (CT) indicates the results of a regular (nonrescued) infection. Lane A512E-A513F, no rescued virus with the corresponding plasmid.
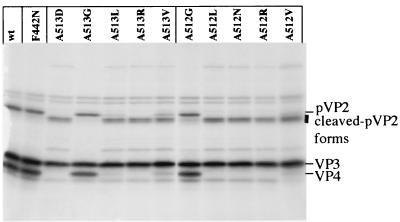
To analyze the cleavage specificity, point mutations were engineered at the pVP2-VP4 cleavage site (position 512-513) and at the VP2 maturation cleavage site (position 441-442) (Fig. 5). For these sites, no substitution was tolerated in general at the P1 or P1" position. However, replacements of Ala-512 and Ala-513 by Gly allowed virus recovery. The Ala-513Gly mutant even appeared to replicate better than the wild type in terms of rescue efficiency and virus production (Fig. 5). The in vitro processing of the different engineered polyprotein mutants at position 512-513 was then analyzed, and the results correlated with those of the rescue experiments. With the Ala512Gly and Ala513Gly substitutions, cleavage occurred efficiently, at the right position (Fig. 7). Small amounts of cleaved forms of pVP2 were nevertheless observed with the Ala512Gly mutant. In contrast, all other substitutions resulted in the generation of smaller forms of pVP2. In conclusion, rescue efficiency and correct cleavage at the pVP2-VP4 junction were well correlated. Note, that as already mentioned for the Ala512Glu-Ala513Phe mutant (14), VP4 was no longer detectable in in vitro expression assays when the cleavage between pVP2 and VP4 was incorrect.
FIG. 7.
In vitro processing of different P1 or P1" mutants. The autoradiograph shows the results obtained with pT7-IBDA-HDR (wt) and a set of P1 or P1" mutants produced in a rabbit reticulocyte expression system. Expression products were analyzed by SDS-PAGE (10% polyacrylamide), and the gel was processed for autoradiography. The positions of the viral polypeptides are indicated on the right.
Effect of deletions of peptides [442-487], [488-494], [495-501], and [502-512].
To investigate the importance of peptides [442-487], [488-494], [495-501], and [502-512] in the virus cycle, we constructed mutants with one of these peptides deleted (Fig. 8). The virus was consistently recovered on transfection of cells with pT7-A-delta[488-494]-HDR or pT7-A-delta[495-501]-HDR in our reverse genetic system. However, these deletions led to a small-plaque phenotype (Fig. 6A). The processing of the deleted pVP2s was analyzed by radioimmunoprecipitation and SDS-PAGE. The ratio between the pVP2s and the VP2 mature forms was not significantly modified with these deletion mutants (data not shown). When pT7-A-delta[442-487]-HDR or pT7-A-delta[502-512]-HDR was transfected, the virus was not recovered through our reverse genetic system. These results demonstrate the importance of the [442-487] and [442-487] peptides in the virus cycle.
FIG. 8.
Virus recovery of the pVP2 mutants. The amino acid sequence of the pVP2-specific domain and the different cleavage sites (arrows) are indicated at the top of the figure. Deleted amino acids are indicated by an asterisk, and substitutions are indicated for each plasmid in the single-letter code. Depending on the virus recovery results, the plasmids are indicated by + (recovery) or − (nonrecovery). Plaque size and virus production were analyzed as described in the legend to Fig. 5.
Effects of point mutations in the [488-494], [495-501], and [501-512] domains.
The point mutations in the two peptides [488-494] and [495-501] did not prevent virus rescue in our reverse genetic system, which is in agreement with the fact that deletions of these domains did not strongly affect virus recovery (Fig. 8). Most point mutations in the [502-512] domain were lethal, which is in agreement with the fact that the deletion of the domain did not allow virus rescue. The in vitro polyprotein processing of the lethal mutants was then analyzed. The mutations did not inactivate the cleavage site at the pVP2-VP4 junction (data not shown). These results confirmed the importance of the [502-512] peptide and particularly the amino acid stretch from residues 504 to 509.
DISCUSSION
The first step of the IBDV polyprotein processing is its autocatalytic cleavage by the VP4 viral protease to generate pVP2, VP4, and VP3. Cleavage at the pVP2-VP4 junction occurs between two alanines located at positions 512 and 513 (14, 20). The main objective of this study was to delineate the maturation process of pVP2.
In this study, we identified at least three (most probably four) peptides derived from the pVP2-specific domain and showed that these peptides are associated with the viral particles. Their presence in the viral particles was revealed using different biochemical approaches, including N-terminal sequencing and mass spectrometry. These peptides were made up of different amino acid stretches, 442 to 487, 488 to 494, 495 to 501, and 502 to 512, and, together, they defined the domain associated with the cleavage maturation of pVP2.
We also developed a reverse genetic approach to examine the importance of these peptides in the virus cycle. First, we tried to rescue mutants in which one of the four peptides was deleted. Deletions of the amino acid stretches from 488 to 494 and from 495 to 501 did allow production of infectious virus, even though these mutants appeared to replicate less efficiently than the wild-type. Substitutions at different positions of these two peptides were found to be permissive for virus replication. These results showed that these two peptides with nearly identical sequences have the same, but accessory, function in the virus cycle.
In contrast, deletions of the amino acid stretches from 442 to 487 and from 502 to 512 did not allow the rescue of infectious virus. The importance of the [502-512] peptide was also demonstrated by site-directed mutagenesis. Each of the six residues of the amino acid stretch from 504 to 509 was shown to be essential for virus recovery. Further experiments are nevertheless needed to address the contribution of these two peptides to the assembly process of the virus particles or other critical steps of the virus cycle.
The presence in infected cells of cleaved forms of pVP2 with higher mobilities than pVP2 in gel electrophoresis suggested that the trimming of pVP2 to VP2 was directional and involved a cascade of proteolytic cleavages starting at the COOH terminus of pVP2 progressing toward the final maturation cleavage site at position 441-442. Four target cleavage sites were involved in the pVP2 maturation process. We previously proposed three of them (501-502, 494-495, and 487-488) as targets of the VP4 protease (14). These sites with the primary cleavage at the pVP2-VP4 junction were defined by the motif (T/A)XA↓A. This consensus sequence shared some homology to the sequence AGA↓F surrounding the maturation cleavage site at position 441-442, suggesting that VP4 was probably involved in the last cleavage, generating the mature VP2. However, it cannot be definitively ruled out that another protease (cellular or virus encoded) may cleave at this position. The fact that the Phe442Gly substitutions do not allow virus replication, in contrast to what is observed with the Ala513Gly substitution, provides some support for this hypothesis.
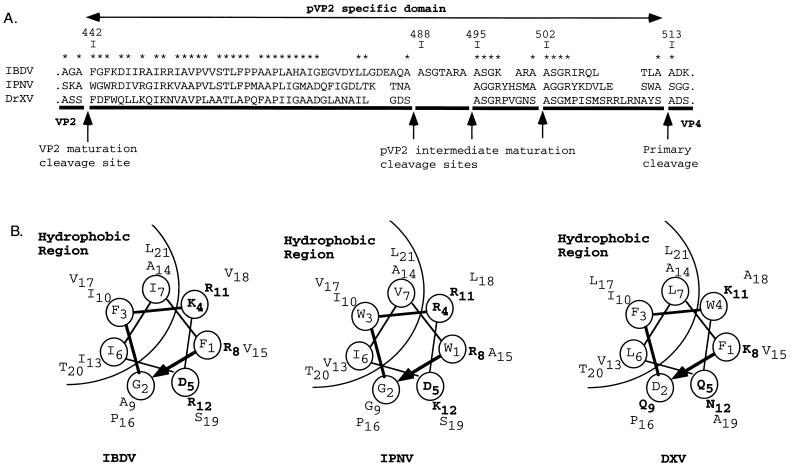
Several lines of evidence support the view that these peptides have homologs in other birnaviruses (Fig. 9A). (i) The pVP2 sequences of different birnaviruses (infectious pancreatic necrosis virus and Drosophila X virus) have strong similarities, mainly in the [442-487] peptide. In the short peptides, a basic residue is often identified at positions 4 (and 6). (ii) the IBDV cleavage site motif [T/A]XA↓A has homologs in infectious pancreatic necrosis virus ([S/T]XA↓A) (18) and in Drosophila X virus ([A/G]XS↓A) (8). (iii) The sequences surrounding the final maturation cleavage site at position 441-442 have sequence similarity. It should also be noted that for infectious pancreatic necrosis virus and Drosophila X virus, only three peptides have been predicted. Preliminary experiments carried out on purified infectious pancreatic necrosis virus agreed with this prediction (data not shown). We concluded that the pVP2 maturation is a characteristic process of birnaviruses.
FIG. 9.
The pVP2-specific domain of different birnavirus polyproteins. (A) Sequence alignment of the IBDV, infectious pancreatic necrosis virus (IPNV), and Drosophila X virus (DXV) pVP2-specific domains. The alignment is anchored to the multiple cleavage sites identified on IBDV or proposed for infectious pancreatic necrosis virus (18) and to the cleavage site at the pVP2-VP4 junction of Drosophila X virus (8). Stars indicate residues common to two or more sequences. (B) Helical-wheel representation of the putative amphipathic α-helices of the N-terminal domains of the [442-487] IBDV peptide and its infectious pancreatic necrosis virus and Drosophila X virus homologs.
In nodaviruses, another family of nonenveloped viruses, the maturation of the viral particles generates the mature coat protein beta (363 residues) and the gamma peptide (44 residues). This peptide plays a key role in the passage of the viral genomic material through the cell membrane. Indeed, while its C-terminal region drives specific packaging of the genomic RNA (22), the N-terminal region could alter membrane structure and then mediate the translocation of the viral genome (3, 11). Structural and biophysical information showed that the gamma N-terminal domain forms an α-helix inside the virus particle and lipid bilayer but adopts a random-coil structure in aqueous solution (4, 26). As with nodaviruses, the N-terminal half of the [442-487] peptide could form an α-amphipathic helix, as shown in Fig. 9B. We could therefore speculate that this peptide played a role in RNA-packaging specificity and in the disruption of the endosomal membrane during virus entry into the target cell.
In conclusion, we have shown for the first time that viral capsids contain short peptides (7 to 46 residues long) that are derived from the maturation processes of their structural proteins. These peptides may play a crucial role in capsid assembly, genome encapsidation, and genome entry into the target cell.
Acknowledgments
We thank Nicolas Eterradossi for providing the IBDV strain CT virus used for RT-PCR cloning and the anti-VP3 monoclonal antibody, Jean-François Bouquet (Mérial) for providing the IBDV-CT virus used for peptide characterization and the anti-pVP2/VP2 monoclonal antibody, Bernard Moss for providing the MVA-T7 virus, Michel Bremont for providing the plasmid encoding the hepatitis ∂ ribozyme, Valérie Bézirard for help in gel electrophoresis, Wendy Brand-Williams for revising the English manuscript, and Jean Cohen for helpful discussions.
REFERENCES
- 1.Azad, A. A., M. N. Jagadish, M. A. Brown, and P. J. Hudson. 1987. Deletion mapping and expression in Escherichia coli of the large genomic segment of a birnavirus. Virology 161:145-152. [DOI] [PubMed] [Google Scholar]
- 2.Biacchesi, S., Y. X. Yu, M. Bearzotti, C. Tafalla, M. Fernandez-Alonso, and M. Bremont. 2000. Rescue of synthetic salmonid rhabdovirus minigenomes. J. Gen. Virol. 81:1941-1945. [DOI] [PubMed] [Google Scholar]
- 3.Bong, D. T., C. Steinem, A. Janshoff, J. E. Johnson, and M. R. Ghadiri. 1999. A highly membrane-active peptide in Flock House virus: implications for the mechanism of nodavirus infection. Chem. Biol. 6:473-481. [DOI] [PubMed] [Google Scholar]
- 4.Bong, D. T., A. Janshoff, C. Steinem, and M. R. Ghadiri. 2000. Membrane partitioning of the cleavage peptide in flock house virus. Biophys. J. 78:839-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boot, H. J., A. A. H. M. ter Huurne, B. P. H. Peeters, and A. L. J. Gielkens. 1999. Efficient rescue of infectious bursal disease virus from cloned cDNA: evidence for involvement of the 3"-terminal sequence in genome replication. Virology 265:330-341. [DOI] [PubMed] [Google Scholar]
- 6.Böttcher, B., N. A. Kiselev, V. Y. Stel'Mashchuk, N. A. Perevozchikova, A. V. Borisov, and R. A. Crowther. 1997. Three-dimensional structure of infectious bursal disease virus determined by electron cryomicroscopy. J. Virol. 71:325-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Chevalier, C., J. Lepault, I. Erk, B. Da Costa, and B. Delmas. 2002. The maturation process of pVP2 requires assembly of infectious bursal disease virus capsids. J. Virol. 76:2384-2392. [DOI] [PMC free article] [PubMed]
- 7.Chowrira, B. M., P. A. Pavco, and J. A. McSwiggen. 1994. In vitro and in vivo comparison of hammerhead, hairpin, and hepatitis delta virus self-processing ribozyme cassettes. J. Biol. Chem. 269:25856-25864. [PubMed] [Google Scholar]
- 8.Chung, H. K., S. Kordyban, L. Cameron, and P. Dobos. 1996. Sequence analysis of the bicistronic drosophila X virus genome segment A and its encoded polypeptides. Virology 225:359-368. [DOI] [PubMed] [Google Scholar]
- 9.Dobos, P. 1995. The molecular biology of infectious pancreatic necrosis virus (IPNV). Annu. Rev. Fish Dis. 5:25-54. [Google Scholar]
- 10.Granzow, H., C. Birghan, T. C. Mettenleiter, J. Beyer, B. Köllner, and E. Mundt. 1997. A second form of infectious bursal disease virus-associated tubule contains VP4.J. Virol. 71:8879-8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janshoff, A., D. T. Bong, C. Steinem, J. E. Johnson, and M. R. Ghadiri. 1999. An animal virus-derived peptide switches membrane morphology: possible relevance to nodaviral transfection processes. Biochemistry 38:5328-5336. [DOI] [PubMed] [Google Scholar]
- 12.Kawaguchi, T., K. Nomura, Y. Hirayama, and T. Kitagawa. 1987. Establishment and characterization of a chicken hepatocellular carcinoma cell line, LMH. Cancer Res. 47:4460-4464. [PubMed] [Google Scholar]
- 13.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 14.Lejal, N., B. Da Costa, J.-C. Huet, and B. Delmas. 2000. Role of Ser-652 and Lys-692 in the protease activity of infectious bursal disease virus VP4 and identification of its substrate cleavage sites. J. Gen. Virol. 81:983-992. [DOI] [PubMed] [Google Scholar]
- 15.Leong, J. C., D. Brown, P. Dobos, F. S. B. Kibenge, J. E. Ludert, H. Müller, E. Mundt, and B. Nicholson. 2000. Family Birnaviridae, p. 481-490. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, D. J. McGeoch, J. Maniloff, M. A. Mayo, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Seventh Report of the International Committee on the Taxonomy of Viruses. Academic Press, Inc., San Diego, Calif.
- 16.Lombardo, E., A. Maraver, J. R. Caston, J. Rivera, A. Fernandez-Arias, A. Serrano, J. L. Carrascosa, and J. F. Rodriguez. 1999. VP1, the putative RNA-dependent RNA polymerase of infectious bursal disease virus, forms complexes with the capsid protein VP3, leading to efficient encapsidation into virus-like particles. J. Virol. 73:6973-6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moscovici, C., M. G. Moscovici, H. Jimenez, M. M. Lai, M. J. Hayman, and P. K. Vogt. 1977. Continuous tissue culture cell lines derived from chemically induced tumors of Japanese quail. Cell. 11:95-103. [DOI] [PubMed] [Google Scholar]
- 18.Petit, S., N. Lejal, J.-C. Huet, and B. Delmas. 2000. Active residues and viral substrate cleavage sites of the protease of the birnavirus infectious pancreatic necrosis virus. J. Virol. 74:2057-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sallantin, M., J.-C. Huet, C. Demarteau, and J.-C. Pernollet. 1990. Reassessment of commercially available molecular weight standards for peptide sodium dodecyl sulfate-polyacrylamide gel electrophoresis using electroblotting and microsequencing. Electrophoresis 11:34-36. [DOI] [PubMed] [Google Scholar]
- 20.Sanchez, A. B., and J. F. Rodriguez. 1999. Proteolytic processing in infectious bursal disease virus: identification of the polyprotein cleavage sites by site-directed mutagenesis. Virology 262:190-199. [DOI] [PubMed] [Google Scholar]
- 21.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 22.Schneemann, A., and D. Marshall. 1998. Specific encapsidation of nodavirus RNAs is mediated through the C terminus of capsid precursor protein alpha. J. Virol. 72:8738-8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spengler, B. 2001. The basics of matrix-assisted laser desorption, ionisation time-of-flight mass spectroscopy and post-source decay analysis, p. 33-53. In P. James (ed.), Proteome research: mass spectroscopy. Springer-Verlag KG, Berlin, Germany.
- 24.Tacken, M. G., P. J. Rottier, A. L. Gielkens, and B. P. Peeters. 2000. Interactions in vivo between the proteins of infectious bursal disease virus: capsid protein VP3 interacts with the RNA-dependent RNA polymerase, VP1. J. Gen. Virol. 81:209-218. [DOI] [PubMed] [Google Scholar]
- 25.Tsukamoto, K., T. Matsumura, M. Mase, and K. Imai. 1995. A highly sensitive, broad-spectrum infectivity assay for infectious bursal disease virus. Avian Dis. 39:575-586. [PubMed] [Google Scholar]
- 26.Wery, J. P., V. S. Reddy, M. V. Hosur, and J. E. Johnson. 1994. The refined three-dimensional structure of an insect virus at 2.8 Å resolution. J. Mol. Biol. 235:565-586. [DOI] [PubMed] [Google Scholar]
- 27.Wyatt, L. S., B. Moss, and S. Rozenblatt. 1995. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology 210:202-205. [DOI] [PubMed] [Google Scholar]