Abstract
The TOM1 gene of Arabidopsis thaliana encodes a putative multipass transmembrane protein which is necessary for the efficient multiplication of tobamoviruses. We have previously shown that mutations severely destructive to the TOM1 gene reduce tobamovirus multiplication to low levels but do not impair it completely. In this report, we subjected one of the tom1 mutants (tom1-1) to another round of mutagenesis and isolated a new mutant which did not permit a detectable level of tobamovirus multiplication. In addition to tom1-1, this mutant carried a mutation referred to as tom3-1. Positional cloning showed that TOM3 was one of two TOM1-like genes in Arabidopsis. Based on the similarity between the amino acid sequences of TOM1 and TOM3, together with the results of a Sos recruitment assay suggesting that both TOM1 and TOM3 bind tobamovirus-encoded replication proteins, we propose that TOM1 and TOM3 play parallel and essential roles in the replication of tobamoviruses.
A variety of host factors are suggested to be involved in the intracellular multiplication of viruses. For eukaryotic positive-strand RNA viruses, host proteins that are associated with viral RNA-dependent RNA polymerases and those which specifically bind virus-related RNAs have been identified and implicated in viral RNA replication (reviewed in references 6 and 14). More recently, host mutants have been isolated in which the intracellular multiplication of a virus is suppressed (10, 12, 15, 17, 19), and the wild-type genes corresponding to some of these mutations have subsequently been cloned. Such genetic approaches have led to the identification of Saccharomyces cerevisiae genes that are necessary for efficient RNA replication and translation of brome mosaic virus (7, 15, 17) and an Arabidopsis thaliana gene, TOM1, that is necessary for efficient multiplication of tobamoviruses (25). These accumulating data suggest that viruses recruit many host components for their own intracellular multiplication, including gene expression and replication.
Tobamovirus is a representative member of the alpha-like virus supergroup of positive-strand RNA viruses (6). It replicates in a membrane-bound replication complex containing the virally encoded protein with an approximate molecular weight of 130,000 (130K protein), the 180K protein (read-through product of the 130K protein), and host-derived proteins (20, 23). We have previously identified the Arabidopsis TOM1 gene, which encodes a putative multipass transmembrane protein necessary for the efficient multiplication of tobamoviruses (25). The 291-amino-acid TOM1 polypeptide likely interacts with the tobamovirus-encoded 130K-180K proteins via the conserved helicase domain and has been suggested elsewhere to function as a membrane anchor for the tobamovirus replication complex in vivo (25). The function of TOM1 in the host remains to be revealed.
Despite the anticipated important role of TOM1 in tobamovirus multiplication, none of the three tom1 mutations (tom1-1 is a point mutation at a splice acceptor site, tom1-2 is a nonsense mutation at amino acid residue 68, and tom1-3 is a frameshift mutation at amino acid residue 147) repressed tobamovirus multiplication completely (Fig. 1) (25). This residual activity supporting tobamovirus multiplication might depend on one or more TOM1-like genes in Arabidopsis. Alternatively, TOM1 might function primarily to enhance the replication of tobamoviruses but might not be absolutely required for it. In the study reported in this paper, we mutagenized one of the tom1 mutants and isolated a new double mutant in which the multiplication of tobamoviruses could not be detected. Positional cloning identified TOM3 as the causal mutation gene, which is one of the two TOM1-like genes in Arabidopsis. The results suggested that TOM1 and TOM3 have parallel and essential roles in the multiplication of tobamoviruses.
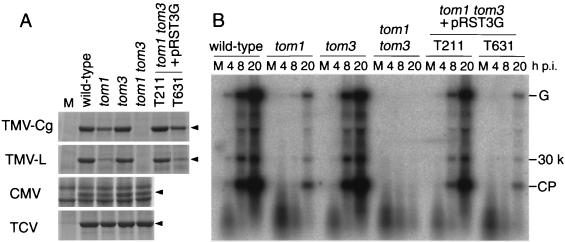
FIG. 1.
Tobamovirus multiplication in the tom1-1 tom3-1 double mutant. (A) Accumulation of viral CPs in inoculated plants. Wild-type (Col-0), mutant, and T3 transgenic line T211 and T631 plants were inoculated with the viruses TMV-Cg, TMV-L, CMV-Y, and TCV-B as indicated. T211 and T631 were derived from a tom1-1 tom3-1 double mutant transformed with the T-DNA clone pRST3G, and each carried the transgene at a single locus homozygously. At 16, 21, 10, and 10 days after inoculation with TMV-Cg, TMV-L, CMV-Y, and TCV-B, respectively, total protein was prepared from aerial parts of the inoculated plants, separated by SDS-PAGE, and stained with Coomassie blue. M, mock-inoculated wild-type plants. Arrowheads show the positions of viral CPs. (B) Accumulation of TMV-Cg-related RNA in protoplasts. Protoplasts were inoculated with TMV-Cg virion RNA by electroporation and cultured for 4, 8, or 20 h, and the accumulation of TMV-Cg-related RNAs was examined by Northern blot hybridization. The positions of the genomic RNA (G) and the subgenomic mRNAs for 30-kDa protein (30 k) and CP are indicated at the right. M, mock-inoculated protoplasts harvested at 20 h postinoculation (h p.i.). The quality of protoplasts in each preparation was confirmed by similar levels of accumulation of TCV-B-related RNAs after inoculation (data not shown). A representative result of three independent repeats is shown.
MATERIALS AND METHODS
Plants, viruses, and DNA clones.
A. thaliana (L.) Heynh. ecotype Columbia (Col-0) was used as the wild-type strain. The tom1-1 and tom1-3 mutants have been previously described (12, 25). TMV-Cg, a crucifer-infecting tobamovirus (24), and TMV-L, a tomato mosaic tobamovirus (18), were used for the inoculation of Arabidopsis plants. The former virus is closely related to Chinese rape mosaic virus (1), which was independently characterized. Cucumber mosaic virus (CMV) strain Y and turnip crinkle virus (TCV) strain B (12) were also used in this study. Tobamoviruses and CMV belong to the alpha-like virus supergroup, whereas TCV belongs to the carmo-like virus supergroup (6). The conditions of plant growth, inoculation with the viruses, mutagenesis of A. thaliana seeds with ethyl methanesulfonate, and screening for mutants with reduced TMV-Cg multiplication were set or performed as described previously (12). Isolation of the tom3-1 single mutant from the tom1-1 tom3-1 double mutant was carried out by first crossing the tom1-1 tom3-1 double mutant with wild-type Col-0. Genomic DNA was then separately purified from the F2 plants and used for derived cleaved amplified polymorphic sequence analysis (16) to determine the genotype at the TOM1 and TOM3 loci (T. Yamanaka, S. Naito, and M. Ishikawa, unpublished data). The tom3-1 single mutant (wild type at TOM1 locus) was selected and self-pollinated, and the resulting F3 seeds were used for analysis. Yeast artificial chromosome (YAC) and bacterial artificial chromosome (BAC) Arabidopsis genomic clones and expressed sequence tag (EST) clones were obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus).
Isolation of TOM3 and THH1 cDNAs and sequence analysis.
For TOM3, we screened an A. thaliana 5"-STRETCH cDNA library (Clontech) containing 105 plaques and identified cDNA clones hybridizing with the EST clone 86D11 (GenBank accession no. T20522). Inserts of the cDNA clones were amplified by PCR and sequenced. Primers used to amplify and sequence the cDNA inserts were 5"-CGCCTCCATCAACAAACTTTCTTG-3" and 5"-GTTCTGGTAAAAAGCGTGGTC-3". For THH1, the mRNA sequence covering the coding region was identified by assembling the sequence of the EST clone 701681346 (GenBank accession no. AI997527) together with that determined by 5" rapid amplification of cDNA ends (RACE) using a SMART PCR cDNA synthesis kit (Clontech). A 377 DNA sequencer (Applied Biosystems) and BigDye Terminator Sequencing kit (Perkin-Elmer) were used for sequencing.
Complementation analysis of the tom3-1 mutation.
The T-DNA clone pRST3G was constructed by inserting the 7.4-kb XbaI-XhoI fragment encompassing the TOM3 mRNA region from the BAC genomic clone F5O4 (GenBank accession no. AC005936) into the T-DNA vector pCLD04541 (8). pRST3G was electroporated into Agrobacterium tumefaciens C58C1(pGV2260) and was used to transform A. thaliana plants by vacuum infiltration (5).
Protein and RNA analysis.
The accumulation of viral coat proteins (CPs) was examined by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) followed by Coomassie blue staining or immunoblot analysis as previously described (12). Preparation of protoplasts, introduction of viral RNAs into the protoplasts by electroporation, and subsequent RNA analysis were performed as described previously (11). For the analysis of TOM3 mRNA, total RNA was extracted and purified from frozen plant tissues by using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Northern blotting and hybridization were performed as described previously (11). To prepare a probe, the open reading frame (ORF) sequence of TOM3 was amplified from Arabidopsis SMART cDNA by PCR and cloned into pET3b (Novagen, Madison, Wis.). 32P-labeled DNA probe was prepared with a gel-purified DNA fragment corresponding to the TOM3 ORF sequence with a Multiprime or Megaprime DNA labeling system (Amersham Pharmacia Biotech). Preparation of a DNA probe for 18S rRNA has been described previously (25).
The Sos recruitment system.
The S. cerevisiae cdc25-2 strain and plasmids to express TOM1, 5"Sos-CgHel (referred to as “5"Sos-Hel” in reference 25), TOM1-5"Sos, and p110-5"Sos, have been previously described (4, 25). TOM3-5"Sos is a fusion of the full-length TOM3 (amino acids 1 to 303) to the N terminus of 5"Sos (amino acids 1 to 1066). 5"Sos-LHel is a fusion of the C-terminal part of the TMV-L 130K protein (amino acids 648 to 1116) (18) to the C terminus of 5"Sos. The S. cerevisiae ADH1 promoter (3) was used to drive expression on 2μm plasmid vectors (9).
Nucleotide sequence accession number.
The sequences reported in this paper have been deposited in the GenBank database (accession no. AB036427 and AB057678).
RESULTS
Isolation of an Arabidopsis mutant in which tobamovirus multiplication is undetectable.
In order to investigate why tobamovirus multiplication was not completely impaired by mutations severely destructive to TOM1, tom1-1 seeds were mutagenized with ethyl methanesulfonate and the M2 plants were screened for mutants in which the multiplication of TMV-Cg was further reduced. TMV-Cg multiplication was determined by using the dot enzyme-linked immunosorbent assay method to measure the accumulation of TMV-Cg CP at 12 days after inoculation (12). From a total of 6,100 M2 plants, one mutant was isolated in which the multiplication of TMV-Cg was undetectable. Genetic analyses suggested that the mutant harbored a recessive or semidominant mutation in addition to tom1-1 (data not shown). This new mutation was referred to as tom3-1. The tom1-1 tom3-1 double mutant grew a little slower than, and the plants were slightly smaller than, the wild-type plants. The CP of TMV-Cg was not detected in inoculated tom1-1 tom3-1 double mutant plants by either Coomassie blue staining (Fig. 1A) or immunoblot analysis (data not shown) of total proteins separated by SDS-PAGE. When tom1-1 tom3-1 double mutant protoplasts were inoculated with TMV-Cg RNA by electroporation, neither genomic nor subgenomic TMV-Cg RNA could be detected by Northern analysis (Fig. 1B). Comparison with serial dilutions of an RNA sample from TMV-Cg-inoculated wild-type protoplasts indicated that the level of TMV-Cg RNA accumulation in the double mutant protoplasts was less than 0.1% of the level detected in the wild-type protoplasts at 20 h postinoculation (data not shown).
The multiplication of TMV-L, a tomato mosaic tobamovirus, was also undetectable in the tom1-1 tom3-1 double mutant plants (Fig. 1A). On the other hand, CMV (a cucumovirus) and TCV (a carmovirus) both multiplied in the tom1-1 tom3-1 double mutant plants as efficiently as in the wild-type plants (Fig. 1A). In tom3-1 single mutant plants, which were obtained by back-crossing the double mutant with wild-type Col-0, TMV-Cg, TMV-L, CMV, and TCV all multiplied as efficiently as in the wild-type plants (Fig. 1A). Accumulation of TMV-Cg RNA in tom3-1 single mutant protoplasts was also similar to that in wild type (Fig. 1B).
Identification of TOM3.
The tom3-1 mutation was first roughly mapped by selecting 12 F2 lines carrying a tom1 tom3 double mutation from a cross between a tom1-3 single mutant (ecotype Wassilewskija background) and the tom1-1 tom3-1 double mutant (Col-0 background). Genotypes of DNA markers were determined for each line, and this indicated that the tom3-1 mutation was located on chromosome 2 within a region covered by the YAC clone CIC11A4 (Fig. 2) (The Arabidopsis Information Resource [http://www.arabidopsis.org/]).
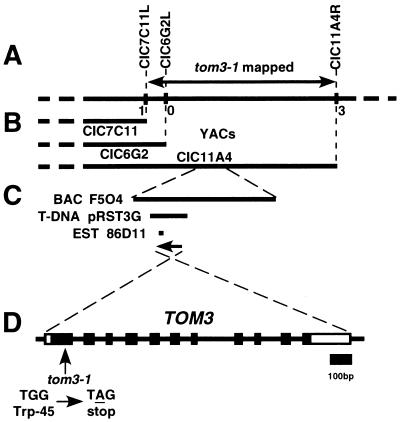
FIG. 2.
Genetic mapping and positional cloning of TOM3. (A) Genetic map around the TOM3 locus on chromosome 2. Vertical bars represent YAC end probes. Each number represents the number of recombined chromatids between the marker and the TOM3 locus among the 12 tom1 tom3 F2 mapping lines (see Materials and Methods). (B) YAC contig around TOM3 locus. (C) The positions of the BAC clone F5O4, T-DNA clone pRST3G, and EST clone 86D11. The T-DNA clone pRST3G was tested for complementation of the TMV-Cg multiplication phenotype in the tom1 tom3 double mutant. The position and orientation of the TOM3 gene are shown by the arrow. (D) Intron-exon organization of the TOM3 gene and the tom3-1 mutation. Exons are indicated by boxes. Open boxes indicate noncoding regions, and closed boxes indicate coding regions. The tom3-1 mutation is shown below the intron-exon structure.
A BLAST search (2) of the Arabidopsis genome database (The Arabidopsis Information Resource [http://www.arabidopsis.org/]) revealed two expressed genes closely related to TOM1. One of the EST clones, 86D11 (GenBank accession no. T20522), was interestingly derived from the region of chromosome 2 covered by both the BAC clone F5O4 (GenBank accession no. AC005936) and the YAC clone CIC11A4, in which the tom3-1 mutation was mapped. cDNA cloning using 86D11 DNA as a probe and the 5"- and 3"-RACE methods revealed a 1,229-nucleotide mRNA sequence containing an ORF encoding a 303-amino-acid protein (GenBank accession no. AB036427) (Fig. 3). The tom3-1 mutant had a nonsense mutation in the first exon of this gene (Fig. 2). Accumulation of the mRNA corresponding to this gene was reduced in tom3-1 and tom1-1 tom3-1 mutant plants compared to that in plants carrying the wild-type TOM3 gene (Fig. 4), possibly reflecting a nonsense-mediated mRNA decay-like phenomenon. These results suggested that this gene was TOM3.
FIG. 3.
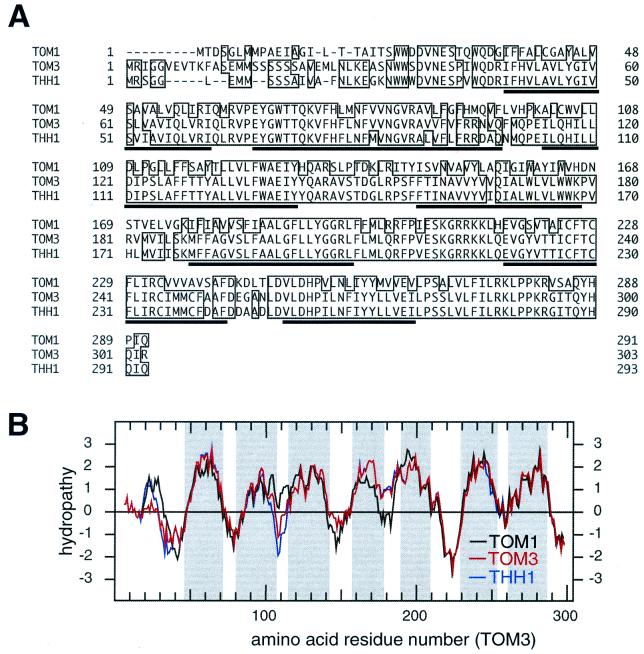
Structure of TOM3. (A) An alignment of the deduced amino acid sequences of TOM1, TOM3, and THH1. Underlined amino acid residues represent those predicted by the TMAP program (21) to be in the membrane-spanning regions. Identical amino acid residues are boxed. (B) Hydropathy plot for the deduced amino acid sequences of TOM1 (black), TOM3 (red), and THH1 (blue). The hydropathy plot was created using the method described by Kyte and Doolittle (13). The regions predicted to be the membrane-spanning regions in panel A are shadowed.
FIG. 4.
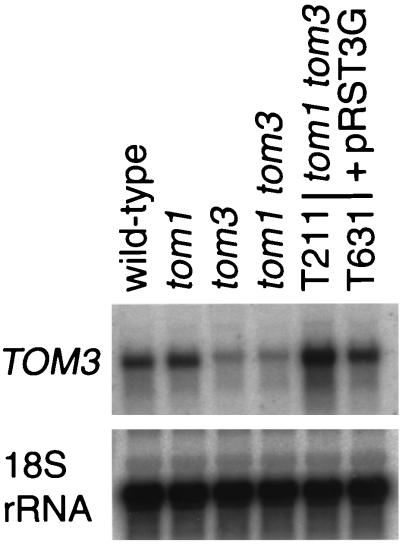
Northern blot hybridization analysis of the mRNA for TOM3. The accumulation of TOM3 mRNA and 18S rRNA in wild-type and mutant plants was examined. Total RNA was extracted from aerial tissues of 26-day-old noninoculated wild-type (Col-0), mutant (tom1-1 and tom3-1), and transgenic (lines T211 and T631) plants. Details of the transgenic lines T211 and T631 are given in the Fig. 1 legend. For the detection of TOM3 mRNA and 18S rRNA, 10 and 3 μg of the total RNA samples, respectively, were denatured by glyoxal, separated by 1% agarose gel electrophoresis, and blotted onto a nylon membrane. Blots were probed with 32P-labeled DNAs hybridizing with either TOM3 or 18S rRNA sequences. To prepare the TOM3-specific probe, a DNA fragment corresponding to the predicted ORF was amplified by PCR from cDNA clones and gel purified. This probe did not cross-hybridize with TOM1 RNA but did cross-hybridize slightly with THH1 RNA. However, the extent of cross-hybridization is negligible for the interpretation of the result (data not shown). The hybridization signals were detected with a Bio Imaging Analyzer (BAS1000; Fuji Photo Film, Tokyo, Japan). Results were confirmed in an independent experiment.
To further confirm whether the above mRNA sequence represented TOM3, a 7.4-kb wild-type genomic DNA fragment from the BAC clone F5O4 encompassing the mRNA sequence (Fig. 2C) was subcloned into a T-DNA vector and used to stably transform the tom1-1 tom3-1 double mutant. In the resulting 15 T2 plants, TMV-Cg multiplied to various degrees, ranging up to a level observed in the wild-type plants. In the transgenic line T211, TMV-Cg multiplied almost as efficiently as in the wild-type plants, whereas in the line T631 the multiplication level was similar to the level in the tom1-1 single mutant plants (Fig. 1). These two transgenic lines carried the transgene at a single locus homozygously. Northern blot hybridization analyses showed that the accumulation level of TOM3 mRNA in T211 and T631 was higher than and similar to that in the wild-type plants, respectively (Fig. 4). These results confirmed that the TOM1-like gene on BAC F5O4 is TOM3 and demonstrated that overexpression of TOM3 fully complements the tom1-1 tom3-1 double defect.
Characterization of the other TOM1-like gene, THH1.
The other EST encoding a TOM1-like sequence was 701681346 (GenBank accession no. AI997527), which was derived from a region on chromosome 1 covered by the BAC clone T5E2 (GenBank accession no. AC005936). Reverse transcription-PCR and 5"-RACE analyses, combined with the sequence data for the EST clone, identified a 1,218-nucleotide mRNA sequence (GenBank accession no. AB057678) containing an ORF encoding 293 amino acids (Fig. 3). We named this gene THH1 (for TOM three homolog), because the predicted amino acid sequence of THH1 resembles that of TOM3 more than that of TOM1. Based on the alignment shown in Fig. 3A, TOM1 and TOM3 share 56% identity, TOM1 and THH1 share 58% identity, and TOM3 and THH1 share 88% identity.
The amino acid sequences of TOM1, TOM3, and THH1 contained several highly hydrophobic regions (Fig. 3). A computer program that predicts transmembrane segments based on multiple sequence alignment (TMAP [21]) suggested that these three proteins were seven-pass transmembrane proteins (Fig. 3A). As was the case for TOM1 (25), database searches for proteins with amino acid sequences similar to that of TOM3 or THH1 did not list significant scores with any other proteins whose function has already been revealed.
Possible interaction between TOM3 and the tobamovirus-encoded replication proteins.
In our previous report, we demonstrated a possible interaction between the membrane-associated TOM1 protein and the helicase domain polypeptide of the TMV-Cg-encoded replication proteins by using the Sos recruitment system in yeast (25) (Fig. 5). This system is based on the observation that the N-terminal fragment of the Ras guanine nucleotide exchange factor (5"Sos), the human homolog of Drosophila melanogaster Son of Sevenless protein, suppresses temperature-sensitive growth caused by the cdc25-2 mutation in S. cerevisiae if 5"Sos is targeted to the plasma membrane in the vicinity of Ras (Fig. 5B) (4). To investigate the membrane association of TOM3 and possible interactions with the replication proteins of tobamoviruses, the same test was performed for TOM3 in this study.
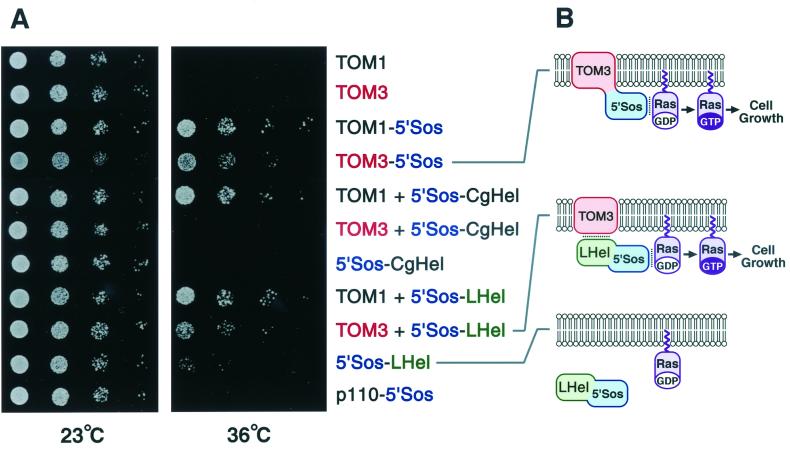
FIG. 5.
Interaction between TOM3 and the TMV-L-encoded replication proteins. (A) cdc25-2 yeast strains harboring plasmids designed to constitutively express the indicated proteins were diluted in sterile water to absorbances at 600 nm of 0.2, 0.025, 0.003, and 0.0004 (eightfold serial dilutions). Two microliters of each dilution was spotted onto yeast extract-peptone-dextrose plates and cultured at 23°C for 56.5 h or at 36°C for 54.5 h. Part of p110 β, a subunit of phosphatidylinositol-3-phosphate kinase, fused to 5"Sos (p110-15"Sos), was used as a negative control for suppression of cdc25-2 temperature sensitivity (4). For comparison, similar constructs with TOM1 in place of TOM3 were simultaneously analyzed. (B) Models explaining the results of panel A for TOM3. Lipid bilayers indicate plasma membranes with the lower sides cytoplasmic. Noncovalent interactions are indicated by dotted lines. Covalent linkage of 5"Sos polypeptide with TOM3 or a noncovalent interaction between TOM3 and the helicase domain of TMV-L-encoded replication proteins in 5"Sos-LHel recruits 5"Sos to the plasma membrane to activate Ras signaling.
Expression of a TOM3-5"Sos fusion protein but not the TOM3 protein alone suppressed the cdc25-2 temperature sensitivity in S. cerevisiae (Fig. 5A), confirming that TOM3 is actually expressed and that it is targeted, at least in part, to the plasma membrane in yeast cells with its C terminus exposed to the cytoplasm (Fig. 5B). The extent of suppression was slightly weaker than that observed for TOM1-5"Sos, suggesting that the amount of TOM3-5"Sos expressed and subsequently localized to the plasma membrane was less than that of TOM1-5"Sos. Alternatively, it is also possible that the position of the 5"Sos moiety in TOM3-5"Sos on the plasma membrane was not suitable for full complementation of the cdc25-2 mutation. Unlike TOM1, TOM3 did not exhibit an activity to enhance cell growth of cdc25-2 yeast expressing a fusion protein between 5"Sos and the helicase domain of TMV-Cg (5"Sos-CgHel [Fig. 5A]). In contrast, the growth of cdc25-2 yeast expressing a fusion protein between 5"Sos and the corresponding helicase domain of TMV-L (5"Sos-LHel) was enhanced by the expression of TOM3, although it was still weaker than the growth enhanced by the expression of TOM1 (Fig. 5A). This enhancement was not observed for a TOM3 derivative with an internal deletion (data not shown). Taken together, it is likely that TOM3 is able to interact with the helicase domain polypeptide of TMV-L (Fig. 5B).
DISCUSSION
Here, we have shown that simultaneous mutations in the two homologous host genes TOM1 and TOM3, likely encoding seven-pass transmembrane proteins, completely suppress tobamovirus multiplication in Arabidopsis. The combination of mutations in two different genes sometimes results in a more enhanced phenotype than that caused by the mutation in either one of the genes. Such genetic interaction, referred to as “synthetic enhancement,” is known to be caused by numerous mechanisms but is often observed when the genes are associated with parallel or related pathways controlling the same function (22). Since the predicted amino acid sequences of TOM1 and TOM3 share a high degree of similarity (56% identical), it is plausible that they do share a parallel and essential function in tobamovirus multiplication. In keeping with this possibility, overexpression of TOM3 in the tom1-1 tom3-1 double mutant restored tobamovirus multiplication to the wild-type level.
Multiple host factors are known to be involved in the replication process of a virus (reviewed in references 6 and 14), and the data presented in this report show that some of these host factors, such as TOM1 and TOM3, are not only involved but also essential for this replication process. Based on our observations of tobamovirus multiplication in the tom1-1, tom3-1, and tom1-1 tom3-1 mutants, we propose the following model regarding the effect on overall virus multiplication efficiency if the activity of an essential host factor is altered by mutation or another process. If the activity of an essential host factor is lower than a certain level (hereafter referred to as threshold I), host defense functions or degradation of virus genome will nullify the production of virus genome and no substantial amplification of the virus genome will occur. On the other hand, if the activity is higher than another specific level (hereafter threshold II), virus multiplication will no longer be affected because the activity of another essential host factor will limit the overall efficiency of multiplication. If the activity is between thresholds I and II, the overall efficiency of virus multiplication will be dependent on the activity of the host factor in question.
In accordance with the above model, the activity of genomic TOM1 (plus THH1) would be above threshold II, since tobamovirus multiplication was normal (wild-type level) in the tom3-1 single mutant. On the other hand, the genomic TOM3 activity (plus THH1) would be between thresholds I and II, since the tom1 single mutations reduced the tobamovirus multiplication to a low but still detectable level. The fact that tobamovirus multiplication was not detected in the tom1-1 tom3-1 double mutant suggests that the activity of genomic THH1 would be below threshold I.
Considering the nature of TOM1 as a membrane protein and its ability to interact with tobamovirus-encoded replicase components, it seems likely that TOM1 participates in the formation of the replication complex in the infected cells by serving as a membrane anchor (25). In the Sos recruitment assay, coexpression of TOM3 enhanced the growth of cdc25-2 yeast harboring 5"Sos-LHel at 36°C, suggesting that TOM3, as well as TOM1, interacts with the helicase domain of TMV-L replication proteins. Although coexpression of TOM3 did not enhance the growth of cdc25-2 yeast cells expressing 5"Sos-CgHel, the demonstrated in vivo activity of TOM3 to support both TMV-Cg and TMV-L multiplication in place of TOM1 in Arabidopsis plants, together with the amino acid sequence similarity between TOM1 and TOM3, suggests that an interaction does likely occur between TOM3 and TMV-Cg replication proteins in plant cells.
We have previously found that coinoculation with CMV results in reduced multiplication of TMV-Cg in tom1-1 protoplasts but not in wild-type protoplasts (11). In contrast, coinoculation with TCV did not reduce TMV-Cg multiplication in either tom1-1 or wild-type protoplasts (11). A possible explanation for these observations is that CMV may also utilize TOM3 for its multiplication and compete with TMV-Cg for TOM3 in the coinoculated tom1-1 protoplasts. If this is true, CMV may also utilize THH1. The normal multiplication of CMV in the tom1-1 tom3-1 double mutant could therefore indicate that the activity of THH1 to support CMV multiplication is above threshold II. Inactivation of THH1 in the tom1-1 tom3-1 double mutant will test this possibility.
To our knowledge, this is the first demonstration of a complete inhibition of intracellular virus multiplication by inactivation of host genes essential for the multiplication of a virus in eukaryotes. While more than 10 host proteins have so far been implicated in the multiplication of positive-strand RNA viruses, most of them also have essential roles in maintaining viability of the hosts (7, 14, 15, 17). Thus, although virus multiplication could be inhibited by suppressing the expression of host genes essential for virus multiplication, it might be a difficult prerequisite to lower the activity of the target gene to at least below threshold II, preferably below threshold I, without affecting host viability. In this sense, the tom1-1 tom3-1 double mutant represents a unique case where mutations in host genes seriously affect tobamovirus multiplication with only a minimal, if any, effect on plant growth. We have recently identified TOM1- and TOM3-like genes in tobacco and tomato plants (unpublished data), raising the possibility that immunity against tobamovirus infection could also be engineered in these plant species by simultaneous inactivation of these genes. Although the effect of the tom1-1 tom3-1 double mutation is limited to tobamovirus multiplication, this is a significant finding in that it also indicates that a similar antivirus strategy could be applied to other viruses once appropriate host genes essential for the multiplication of the respective viruses are identified.
Acknowledgments
We are grateful to D. B. Goto for critical reading of the manuscript, to M. Yoshii and Y. Tsujimoto for experimental advice and assistance, to K. Fujiwara for general assistance, and to A. Aronheim and the Arabidopsis Biological Resource Center for materials. We used the facilities of the Biopolymer Analysis Laboratory in the Faculty of Agriculture, Hokkaido University, and the Research Center for Molecular Genetics at Hokkaido University.
This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan to M.I. (08680733) and S.N. (06278102) and by a grant from the Japan Society for the Promotion of Science to M.I. (RFTF96L00603).
REFERENCES
- 1.Aguilar, I., F. Sanchez, M. A. Martin, D. Martinez-Herrera, and F. Ponz. 1996. Nucleotide sequence of Chinese rape mosaic virus (oilseed rape mosaic virus), a crucifer tobamovirus infectious on Arabidopsis thaliana. Plant Mol. Biol. 30:191-197. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ammerer, G. 1983. Expression of genes in yeast using the ADC1 promoter. Methods Enzymol. 101:192-201. [DOI] [PubMed] [Google Scholar]
- 4.Aronheim, A., E. Zandi, H. Hennemann, S. J. Elledge, and M. Karin. 1997. Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol. Cell. Biol. 17:3094-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bechtold, N., and G. Pelletier. 1993. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82:259-266. [DOI] [PubMed] [Google Scholar]
- 6.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diez, J., M. Ishikawa, M. Kaido, and P. Ahlquist. 2000. Identification and characterization of a host protein required for efficient template selection in viral RNA replication. Proc. Natl. Acad. Sci. USA 97:3913-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flanders, D. J., S. Weng, F. X. Petel, and J. M. Cherry. 1998. AtDB, the Arabidopsis thaliana database, and graphical-web-display of progress by the Arabidopsis genome initiative. Nucleic Acids Res. 26:80-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 10.Ishikawa, M., J. Diez, M. A. Restrepo-Hartwig, and P. Ahlquist. 1997. Yeast mutations in multiple complementation groups inhibit brome mosaic virus RNA replication and transcription and perturb regulated expression of the viral polymerase-like gene. Proc. Natl. Acad. Sci. USA 94:13810-13815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishikawa, M., S. Naito, and T. Ohno. 1993. Effects of the tom1 mutation of Arabidopsis thaliana on the multiplication of tobacco mosaic virus RNA in protoplasts. J. Virol. 67:5328-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishikawa, M., F. Obata, T. Kumagai, and T. Ohno. 1991. Isolation of mutants of Arabidopsis thaliana in which accumulation of tobacco mosaic virus coat protein is reduced to low levels. Mol. Gen. Genet. 230:33-38. [DOI] [PubMed] [Google Scholar]
- 13.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 14.Lai, M. M. C. 1998. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology 244:1-12. [DOI] [PubMed] [Google Scholar]
- 15.Lee, W.-M., M. Ishikawa, and P. Ahlquist. 2001. Mutation of host Δ9 fatty acid desaturase inhibits brome mosaic virus RNA replication between template recognition and RNA synthesis. J. Virol. 75:2097-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neff, M. M., J. D. Neff, J. Chory, and A. E. Pepper. 1998. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J. 14:378-392. [DOI] [PubMed] [Google Scholar]
- 17.Noueiry, A. O., J. Chen, and P. Ahlquist. 2000. A mutant allele of essential, general translation initiation factor DED1 selectively inhibits translation of a viral mRNA. Proc. Natl. Acad. Sci. USA 97:12985-12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohno, T., M. Aoyagi, Y. Yamanashi, H. Saito, S. Ikawa, T. Meshi, and Y. Okada. 1984. Nucleotide sequence of the tobacco mosaic virus (tomato strain) genome and comparison with the common strain genome. J. Biochem. 96:1915-1923. [DOI] [PubMed] [Google Scholar]
- 19.Ohshima, K., T. Taniyama, T. Yamanaka, M. Ishikawa, and S. Naito. 1998. Isolation of a mutant of Arabidopsis thaliana carrying two simultaneous mutations affecting tobacco mosaic virus multiplication within a single cell. Virology 243:472-481. [DOI] [PubMed] [Google Scholar]
- 20.Osman, T. M. A., and K. W. Buck. 1997. The tobacco mosaic virus RNA polymerase complex contains a plant protein related to the RNA-binding subunit of yeast eIF-3. J. Virol. 71:6075-6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Persson, B., and P. Argos. 1996. Topology prediction of membrane proteins. Protein Sci. 5:363-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sherman, F. 1997. Yeast genetics, p. 302-325. In R. A. Mayers (ed.), The encyclopedia of molecular biology and molecular medicine. VCH Publishers, Weinheim, Germany.
- 23.Watanabe, T., A. Honda, A. Iwata, S. Ueda, T. Hibi, and A. Ishihama. 1999. Isolation from tobacco mosaic virus-infected tobacco of a solubilized template-specific RNA-dependent RNA polymerase containing a 126K/183K protein heterodimer. J. Virol. 73:2633-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamanaka, T., H. Komatani, T. Meshi, S. Naito, M. Ishikawa, and T. Ohno. 1998. Complete nucleotide sequence of the genomic RNA of tobacco mosaic virus strain Cg. Virus Genes 16:173-176. [DOI] [PubMed] [Google Scholar]
- 25.Yamanaka, T., T. Ohta, M. Takahashi, T. Meshi, R. Schmidt, C. Dean, S. Naito, and M. Ishikawa. 2000. TOM1, an Arabidopsis gene required for efficient multiplication of a tobamovirus, encodes a putative transmembrane protein. Proc. Natl. Acad. Sci. USA 97:10107-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]