Abstract
Poliovirus translation is initiated at the internal ribosome entry site (IRES). Most likely involving the action of standard initiation factors, this highly structured cis element in the 5" noncoding region of the viral RNA guides the ribosome to an internal silent AUG. The actual start codon for viral protein synthesis further downstream is then reached by ribosomal scanning. In this study we show that two of the secondary structure elements of the poliovirus IRES, domain V and, to a minor extent, domain VI, are the determinants for binding of the eukaryotic initiation factor eIF4B. Several mutations in domain V which are known to greatly affect poliovirus growth also seriously impair the binding of eIF4B. The interaction of eIF4B with the IRES is not dependent on the presence of the polypyrimidine tract-binding protein, which also binds to the poliovirus IRES. In contrast to its weak interaction with cellular mRNAs, eIF4B remains tightly associated with the poliovirus IRES during the formation of complete 80S ribosomes. Binding of eIF4B to the IRES is energy dependent, and binding of the small ribosomal subunit to the IRES requires the previous energy-dependent association of initiation factors with the IRES. These results indicate that the interaction of eIF4B with the 3" region of the poliovirus IRES may be directly involved in translation initiation.
When poliovirus, the paradigm of the family of picornaviruses, infects a susceptible cell, the positive-strand viral RNA can be used directly for translation of the viral polyprotein. In contrast to normal cellular mRNAs, translation of the picornaviral RNA is facilitated by an internal region of the viral RNA, the internal ribosome entry site (IRES). This strategy of internal initiation allows some picornaviruses to take advantage of the obvious opportunity and shut down the cap-dependent cellular translation, while synthesis of the picornaviral polyprotein is initiated cap independently from the IRES element (for an overview, see reference 3).
Picornavirus IRES elements are large cis-acting RNA regions that guide ribosomes to an internal site of the viral RNA (17, 37, 51). They are classified into three groups according to their primary and secondary structures: the type I elements of the enterovirus-rhinovirus group (including poliovirus), the type II elements of the cardiovirus-aphthovirus group, and the type III element of hepatitis A virus (HAV). The IRES elements consist of highly conserved RNA secondary structure domains and have a characteristic oligopyrimidine tract followed by a conserved AUG triplet at their 3" borders (8, 42, 44). In poliovirus, this AUG is important but silent for translation, while the actual initiation codon further downstream is reached by ribosomal scanning (20, 22, 25, 39). The small ribosomal subunit is guided to a starting window (43) containing the conserved AUG triplet, most likely by the action of initiation factors. In addition to the standard initiation factors, picornavirus IRES elements recruit other cellular RNA-binding proteins that enhance translation or modulate the balance between translation of the viral RNA and its replication, such as the 57-kDa polypyrimidine tract-binding protein (PTB) (13, 18, 31), the 52-kDa La protein (26), or the 39-kDa poly(rC) binding protein (6).
For the internal initiation of translation on picornaviral RNA, almost the complete set of the canonical eukaryotic translation initiation factors (eIFs) appears to be essential (2, 35, 38), with the exception of the actual cap-binding protein eIF4E. In cap-dependent initiation of normal cellular mRNAs, the large adapter protein eIF4G associates with eIF4E and with the RNA helicase eIF4A to form the cap-binding protein complex eIF4F, which guides the small ribosomal subunit to the 5" end of the mRNA. eIF4B appears to be involved in several interactions during translation initiation. It stimulates the RNA helicase activity of eIF4A (35), it can bind to 18S rRNA (27), and it interacts with the ribosome-bound eIF3 (28). Moreover, eIF4B has two RNA-binding domains and can catalyze the hybridization of two complementary single-stranded RNAs by an RNA annealing activity (1).
For the type II picornavirus IRES elements, the binding sites for these initiation factors have been roughly mapped, e.g., for encephalomyocarditis virus (EMCV) and for foot-and-mouth-disease virus (FMDV). A large Y-shaped RNA structure in the 3" region of these IRES elements (referred to as domain 4 in FMDV and J-K-L in EMCV) precedes the starting window with the conserved AUG. This RNA region is known to bind eIF4B (29, 33, 45) as well as eIF4G and eIF4A (19, 23, 38, 46).
In contrast, for poliovirus as well as for the related rhinoviruses, the site of binding of the canonical initiation factors is not yet known. However, reasonable assumptions can be made about the IRES region possibly involved. In the poliovirus IRES, the domain V resembles the Y-shaped domain 4 (or J-K-L) of the type II IRES elements with regard to its relative position in the array of IRES secondary structures and its position directly upstream of the conserved cryptic AUG. Consistent with this idea, domain V has been found to be the most important structure of the poliovirus IRES, since nearly any mutation affecting its structure is deleterious for IRES activity (7, 11, 21, 36, 50).
In this study, we investigated the interaction of eIF4B with the poliovirus IRES. We mapped the determinants that cause the binding of eIF4B in the poliovirus IRES to reside in domains V and VI. Moreover, we analyzed the association of eIF4B with the IRES in the course of the formation of ribosomal initiation complexes and the possible interaction of eIF4B with a noncanonical RNA binding protein, PTB.
MATERIALS AND METHODS
Plasmids.
pMPolio contains, downstream of an SP6 RNA polymerase promoter, the complete poliovirus type 1 (Mahoney) IRES sequence obtained from plasmid pT7XLmyr(−) (kindly provided by C. U. T. Hellen). The authentic poliovirus initiator AUG (nucleotides 743 to 745) was fused to the luciferase coding sequence from plasmid pM12 (33). Plasmid pMPolio-Hind was derived from pMPolio by PCR mutagenesis. The poliovirus sequence in domain VI (582-GCUUAUGU-589, with the conserved AUG at nucleotide 586 represented in bold face) was replaced by AAGCUUGG, mutating the silent AUG and introducing an artificial HindIII site. In plasmids 5"-NC-X482, -X515, -X521, -X529, -X539, -X543, and -X585 (kindly provided by B.L. Semler), poliovirus sequences of eight nucleotides are replaced by an eight-nucleotide linker sequence (CCUCGAGG) (11). All mutations are in the context of the otherwise complete poliovirus IRES. For construction of pQE-PTB, the human PTB sequence was excised from pGEX-2TKhuPTB (kindly provided by M. Garçia-Blanco) and inserted into pQE-9 (Qiagen).
Preparation of RNAs.
To obtain full-length poliovirus IRES RNA, plasmid pMPolio was linearized with BsiWI 156 bp downstream of the luciferase ATG (compare Fig. 2A). The resulting RNA contained 9 nucleotides of linker sequence, the complete poliovirus 5" nontranslated region from nucleotide 1 to 742, and 156 nucleotides of the luciferase coding sequence. For obtaining shorter RNAs, pMPolio was linearized with AgeI (poliovirus IRES, nucleotide 343), BsmI (nucleotide 460), or MscI (nucleotide 629). pMPolio-Hind was linearized at the artificial HindIII site at the beginning of domain VI. Plasmid 5"-NC-X482 and related plasmids were linearized with MscI at nucleotide 629 downstream of the IRES domain VI sequence. Labeled RNAs were synthesized as described previously (32) by using SP6 RNA polymerase in the presence of 2.5 μM [α-32P]CTP, -ATP, -GTP, or -UTP (400 Ci/mmol; Amersham) as indicated with 15 μM nonradioactive labeling nucleotide added. RNAs were separated from unincorporated nucleotides by gel filtration on Sephadex G-50 columns (Pharmacia).
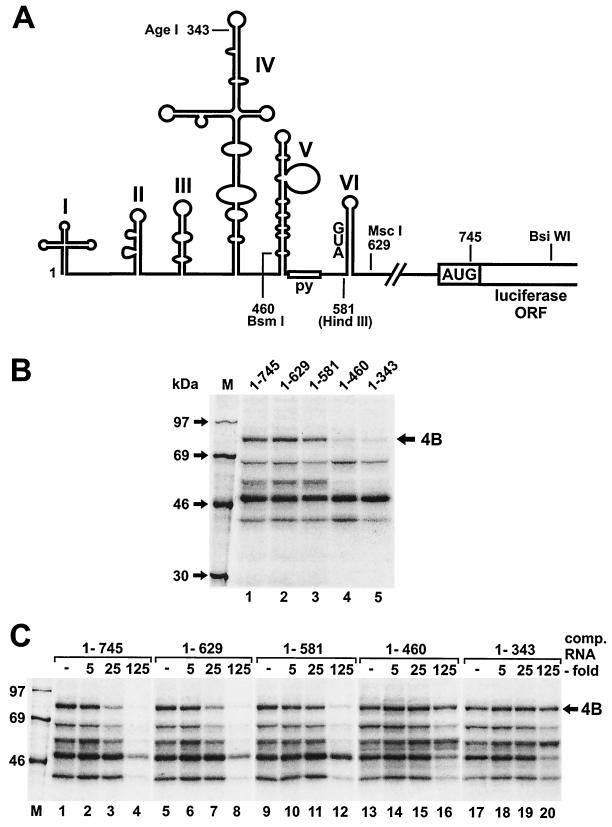
FIG. 2.
Mapping of the eIF4B binding site in the poliovirus IRES. (A) Schematic representation of the poliovirus IRES. The IRES domains are marked with Roman numerals as described previously (8). Enzymes used for linearization of the templates for in vitro transcription are indicated together with the positions of the last nucleotide of the poliovirus sequence present in the respective RNAs. (B) UV cross-linking assay using RRL and [α-32P]CTP-labeled poliovirus IRES RNAs. The nucleotide numbers of the poliovirus sequences are indicated on the top. RNA 1-745 contains the authentic poliovirus initiator AUG at nucleotides 743 to 745 plus 156 additional nucleotides of luciferase sequence at the 3" end (up to the BsiWI site). Molecular masses of marker proteins are indicated. (C) Competitions. [α-32P]CTP-labeled poliovirus IRES RNA 1-745 was used in all lanes. Unlabeled competitor RNAs are indicated on the top together with the molar concentrations used for competition. Molecular masses of marker proteins are indicated. 4B, eIF4B.
UV cross-linking assays, immunoprecipitations, and immunoblots.
UV cross-linking assays were performed with 3.3 μl of nuclease-treated rabbit reticulocyte lysate (RRL) (Promega) and 0.2 pmol of labeled IRES RNA in a volume of 10 μl. Unlabeled competitor RNAs were added if indicated. Reactions were incubated for 10 min at 30°C and were irradiated with UV light for 10 min. Excess RNA was digested with RNase A at 0.1 mg/ml at 37°C for 60 min, and proteins were separated on sodium dodecyl sulfate (SDS)-10% polyacrylamide gels and analyzed by autoradiography. RRL depleted of endogenous PTB was prepared as described previously (31). Recombinant PTB with an N-terminal His6 tag was expressed and purified according to the supplier (Qiagen). Recombinant La protein was kindly supplied by M. Bachmann (16) and T. Heise. Anti-La antibodies 4B6 and 3B9 were kindly supplied by M. Bachmann. Anti-ribosomal protein S6 antibody was purchased from Cell Signaling Technology, Beverly, Mass. Anti-eIF4B antibodies against recombinant human eIF4B were elicited in mice and used for immunoprecipitations (33) and for immunoblot detection of eIF4B in sucrose gradient fractions. Immunoblotting was performed as described previously (46). Cellular poly(A)+ RNAs (capped mRNAs) used for competition were extracted from mouse liver using the RNeasy and Oligotex kits from Qiagen. Uncapped competitor RNA was transcribed from plasmid pGEM2-2β (34) and linearized with BfrI and contained nucleotides 1 to 417 of the eIF2 β subunit mRNA (29).
Initiation complex formation, sedimentation analyses, and UV cross-linking.
Initiation complex formation and separation were performed essentially as described previously (32, 33). RRL devoid of ribosomes was prepared as described previously (33). Briefly, RRL was adjusted to 250 mM K acetate and ribosomes were pelleted for 2 h at 200,000 × g and 4°C. The supernatant was saved, and ribosomes were resuspended in 80 μl of 15 mM Tris-HCl (pH 7.5)-0.5 mM MgCl2-1 mM dithiothreitol.
Binding reaction mixes contained 50 μl of RRL, 15 mM Tris-Cl (pH 7.5), 0.4 mM MgCl2, 10 mM dithiothreitol, and 40 mM KCl in a volume of 150 μl. In reactions containing translation inhibitors, either 4 mM 5"-guanylylimidodiphosphate (GMP-PNP) or 0.17 mM anisomycin (Sigma) was added if indicated, and the reaction was preincubated for 5 min at 30°C. To start the binding reaction, 2 to 5 pmol of IRES RNA was added. The samples were incubated for 10 min at 30°C for initiation complex formation and then irradiated for 10 min at 0°C with UV light (254 nm). RRL devoid of ribosomes was used as before in a binding reaction for 10 min at the temperature indicated. After UV cross-linking at 0°C for 5 min, 4 μl of the resuspended ribosomes was added and incubated for another 10 min at either 0 or 30°C as indicated.
After the binding reactions and irradiation, protease inhibitors were added as described previously (32). The sample was loaded onto a 10 to 35% sucrose gradient, centrifuged for 4.5 h at 200,000 × g at 4°C in a Beckman SW 41 Ti rotor, and fractionated into 24 fractions of 500 μl. Four-microliter aliquots of the resulting fractions were subjected to scintillation counting. The fractions were treated with RNase A at 0.1 mg/ml at 37°C for 60 min. Proteins were precipitated in the presence of 10% trichloroacetic acid (TCA) and 5 μg of gelatin for 30 min. After centrifugation, pellets were washed twice with ethanol, air dried, and redissolved in protein sample buffer containing 3 M urea. The samples were separated on SDS-10% polyacrylamide gels and analyzed by autoradiography.
For localization of ribosomal subunits in the gradients, proteins from aliquots of the gradient fractions were separated on SDS-polyacrylamide gels and subjected to immunoblotting using a ribosomal protein S6 antibody. Another aliquot of each gradient fraction was phenol-chloroform extracted, and nucleic acids were precipitated in the presence of 0.3 M Na acetate and 75% ethanol, separated on 1% agarose gels, and stained with ethidium bromide.
RESULTS
eIF4B binds to the poliovirus IRES.
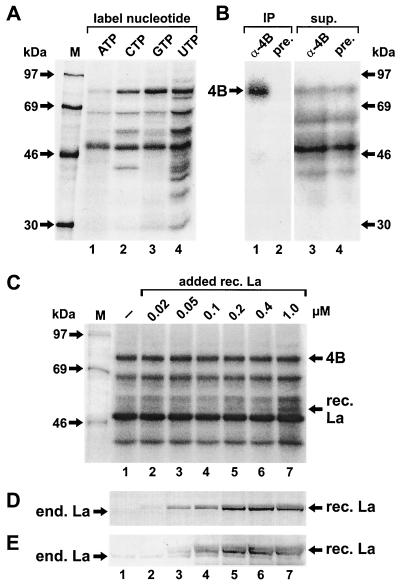
In order to detect the binding of eIF4B to the poliovirus IRES, RNAs comprising the complete IRES of poliovirus type 1 (Mahoney) were transcribed in vitro and radioactively labeled with either [α-32P]ATP, -CTP, -GTP or -UTP. Proteins from RRL binding to the IRES were then analyzed by the UV cross-linking assay. With all four differently labeled RNAs, a protein with an apparent molecular mass of about 80 kDa, as expected for eIF4B, was labeled (Fig. 1A). This 80-kDa protein was identified as eIF4B by immunoprecipitation. With an antiserum directed against human eIF4B, the 80-kDa band was immunoprecipitated after a UV cross-linking reaction with RRL (Fig. 1B, lane 1), whereas no band was detected with preimmune serum (lane 2).
FIG. 1.
eIF4B binds to the poliovirus IRES RNA. (A) Detection of eIF4B in RRL. UV cross-linking assays were performed with poliovirus IRES RNA labeled with either [α-32P]ATP, -CTP, -GTP or -UTP as indicated. Proteins were resolved on SDS-10% polyacrylamide gels and visualized by autoradiography. Molecular masses of marker proteins are indicated. (B) Immunoprecipitation of eIF4B. The binding reaction was performed with RRL and [α-32P]CTP-labeled IRES RNA. After the UV cross-linking reaction, eIF4B was immunoprecipitated (IP) with anti-eIF4B antiserum (α-4B) (lane 1). Preimmune serum was used as negative control (pre.) (lane 2). Ten percent of the proteins from the supernatants (sup.) (right panel) was concentrated by TCA-precipitation (lanes 3 and 4), resolved on SDS-10% polyacrylamide gels, and visualized by autoradiography. 4B, eIF4B. (C) No effect of La protein on eIF4B binding. Poliovirus IRES RNA was used in the UV cross-linking assay by using RRL either without (lane 1) or after adding increasing amounts of recombinant La protein (rec. La) as indicated (lanes 2 to 7). (D and E) Immunoblots with anti-La antibodies 4B6 (D) and 3B9 (E). The bands representing endogenous La (end. La) and added recombinant La (rec. La) are indicated.
The use of [α-32P]CTP-, GTP- and UTP-labeled IRES RNA (Fig. 1A, lanes 2 to 4) resulted in strong labeling of eIF4B. In addition, a band of about 50 kDa was labeled strongly, and several other proteins were labeled, including a 57-kDa protein (lanes 2 and 4) which is identical with the PTB (12, 13). For all further experiments, we used [α-32P]CTP-labeled IRES RNAs, since this label allowed the simultaneous detection of eIF4B and other bands, including that of the PTB (which is not well labeled with GTP). UTP was not chosen because an additional band migrating close to eIF4B was detected with this label.
The translation of poliovirus RNA in RRL has been reported to be stimulated and corrected by the addition of the 52-kDa La protein (26). Therefore, we investigated whether the addition of recombinant La to the RRL could possibly affect the binding of eIF4B to the poliovirus IRES and thus reveal a molecular mechanism of the action of La. However, eIF4B binding was not affected when increasing amounts of recombinant La were added to the RRL (Fig. 1C, lanes 2 to 7; compare with lane 1). The preparation of recombinant La protein used is functional in terms of RNA binding (T. Heise, personal communication), and the integrity of the recombinant La protein was checked using two different anti-La antibodies (Fig. 1D and E). These immunoblots also showed that only minute amounts of endogenous La are present in the RRL. The level of 1 μM La protein which was added (Fig. 1C, lane 7) is similar to the level (about 1.5 μM) at which La stimulates the correct translation of the poliovirus P1 precursor approximately tenfold (48). In the same way, the addition of a cytoplasmic HeLa extract to the RRL did not affect the binding of eIF4B to the poliovirus IRES (data not shown).
Thus, irrespective of the reported effects of La protein on poliovirus translation, the binding of eIF4B appears not to be affected by La. Accordingly, normal RRL (i.e., not supplemented with La protein) was used in all further experiments.
Mapping the eIF4B binding site.
For delimiting the IRES region required for the binding of eIF4B, we used RNAs covering different regions of the poliovirus IRES (Fig. 2). When sequences downstream of domain VI, including the nonstructured spacer and the authentic initiator AUG, were removed (RNA 1-629), eIF4B still bound well to the IRES (Fig. 2B, lane 2), and the pattern of proteins detected did not change compared with the full-length IRES RNA 1-745 (lane 1). Thus, the initiator AUG at nucleotide 743 as well as the preceding nonstructured spacer sequence are not required for binding of eIF4B. When most of domain VI, including the conserved silent AUG at nucleotide 586, was removed (RNA 1-581), binding of eIF4B to the poliovirus IRES was slightly weaker (lane 3). This indicates that domain VI and possibly the cryptic AUG may contribute to eIF4B binding.
In contrast, when the domain V and all downstream sequences were removed (RNA 1-460), binding of eIF4B to the IRES was nearly completely abolished (Fig. 2B, lane 4), whereas binding of the 39-, 50-, and 68-kDa proteins was not affected by this deletion. However, the 57-kDa band of PTB disappeared, suggesting that domain V is also a major determinant for the binding of PTB. When further upstream sequences, including the 3" half of the domain IV sequence, were removed (RNA 1-343), the same results were obtained (lane 5).
To exclude the possibility that the loss of detection of eIF4B was due only to the loss of a cross-link site and not to the loss of binding of the protein, we performed competition assays (Fig. 2C). Unlabeled competitor RNAs 1-745 and 1-629 competed efficiently with the binding of eIF4B and of other proteins to radiolabeled RNA 1-745 (lanes 3 to 4 and 7 to 8, respectively). RNA 1-581 competed only slightly more weakly (compare lane 11 with lane 3). However, the RNA regions comprising nucleotides 1 to 460 and 1 to 343 (lanes 13 to 20) could not compete successfully with the binding of eIF4B to the full-length IRES RNA (compare lanes 16 or 20 to lane 4). Thus, the loss of detection of eIF4B with the respective IRES deletions in Fig. 2B is actually due to the loss of eIF4B binding but not due to the loss of cross-link sites alone.
Taken together, the above results indicate that the major determinants for the binding of eIF4B reside in domain V of the poliovirus IRES, whereas domain VI contributes less to the binding of eIF4B.
Mutations within IRES domains V and VI affect eIF4B binding.
For a more detailed analysis of the eIF4B binding site, a series of linker-scanning mutations covering most of domain V and the beginning of domain VI (11) (Fig. 3A) in the context of the otherwise complete IRES was used. In mutant X482, the apical stem of the domain V was affected. In mutants X515 and X521, the predicted large unpaired bulge was mutated, and in mutants X529, X539, and X543, the lower portion of the domain V was affected.
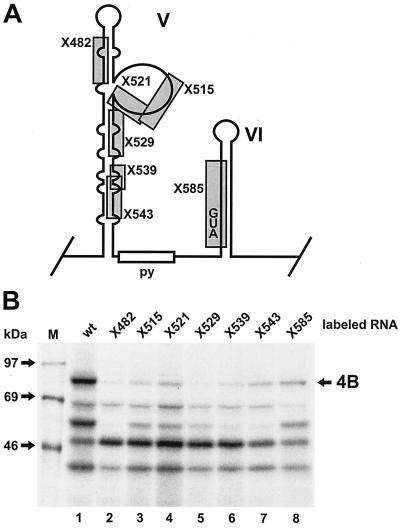
FIG. 3.
Effects of domain V and VI mutants on binding of eIF4B. (A) Schematic representation of the poliovirus IRES mutants used. Each boxed region of the wild-type poliovirus sequence is replaced by a linker sequence (CCUCGAGG) in the mutant (11). The drawing does not take into account possible changes in the secondary structure that may be caused by each mutation. All mutants are in the context of an otherwise complete IRES. (B) UV cross-linking assay using RRL and different [α-32P]CTP-labeled poliovirus IRES linker scanning mutants (11) as shown in panel A. Templates for RNA synthesis were linearized with BsiWI as in Fig. 2. The position of eIF4B (4B) is indicated. Molecular masses of marker proteins are indicated.
All of the mutations in domain V severely affected the binding of eIF4B to the poliovirus IRES (Fig. 3B), with those mutations located in the upper and middle stem (X482 and X529) having the greatest effect. Mutations affecting the lower part of the stem (X543) or the bulge (X515 and X521) also severely affected but did not completely abolish eIF4B binding. Thus, all parts of domain V are important for eIF4B binding, including the upper stem structure, which is predicted to be a relatively stable structure, the predicted unpaired bulge, and the lower part of domain V. Together with that of eIF4B, the binding of the 57-kDa protein PTB was also affected by all mutations, most severely by those affecting the stem. In contrast, the binding of the 50-kDa protein was not only not reduced but was in some cases even enhanced, possibly due to the loss of binding competition. When domain VI was mutated (Fig. 3B, X585, lane 8), binding of eIF4B was affected less severely. This confirmed that domain VI contributes to the binding of eIF4B to a lesser extent (compare with Fig. 2).
The interaction of eIF4B with the poliovirus IRES is independent of PTB.
Since binding of both eIF4B and PTB was affected by the same mutations in domain V, both factors appear to bind to the same RNA regions of the domain V, raising the question of whether binding of eIF4B to the IRES could be dependent on binding of the PTB. In order to investigate the possibility of an interaction of PTB and eIF4B that would thus help to explain the molecular mechanism of PTB action on picornavirus IRES elements, we used a modified reticulocyte lysate. The endogenous PTB, together with another approximately 68-kDa protein (Fig. 4, lane 2), had been removed from this lysate by treatment with poly(U)-Sepharose (32).
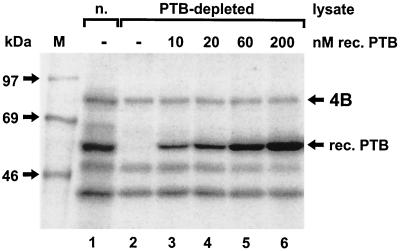
FIG. 4.
Binding of eIF4B to the poliovirus IRES in the presence of different PTB concentrations. Either normal (n.) (lane 1) RRL or RRL depleted of endogenous PTB (lanes 2 to 6) was used for UV cross-linking assays with poliovirus IRES RNA 1-745. In lanes 3 to 6, the indicated concentrations of recombinant PTB were added before adding the RNA. Molecular masses of marker proteins are indicated. 4B, eIF4B; rec. PTB, recombinant PTB.
In this PTB-depleted lysate (Fig. 4, lane 2), binding of eIF4B to the poliovirus IRES was only slightly reduced compared with that of the untreated lysate (lane 1). When increasing amounts of purified recombinant PTB were added to the depleted lysate, the PTB band reappeared with increasing intensity, while the eIF4B band remained unaffected (lanes 3 to 6). Thus, (i) binding of eIF4B is not dependent on PTB, and (ii) binding of increasing amounts of PTB does not interfere with eIF4B binding, although both proteins bind to the same IRES domain.
eIF4B remains bound to the poliovirus IRES in complete 80S ribosomes.
In order to investigate at which stages eIF4B is involved in the formation of ribosomal initiation complexes with the poliovirus IRES, we used the UV cross-linking assay in combination with sucrose gradient sedimentation of ribosomal initiation complexes. By using the UV cross-linking assay with the radiolabeled IRES RNA as a probe, we selectively detected the subfraction of interest of the eIF4B molecules in the RRL, namely, those eIF4B molecules that are actually directly bound to the IRES RNA. In contrast, a Western blot is not suitable for this purpose, since it would detect all of the eIF4B molecules in the lysate, including those molecules that are not bound at all to the IRES RNA.
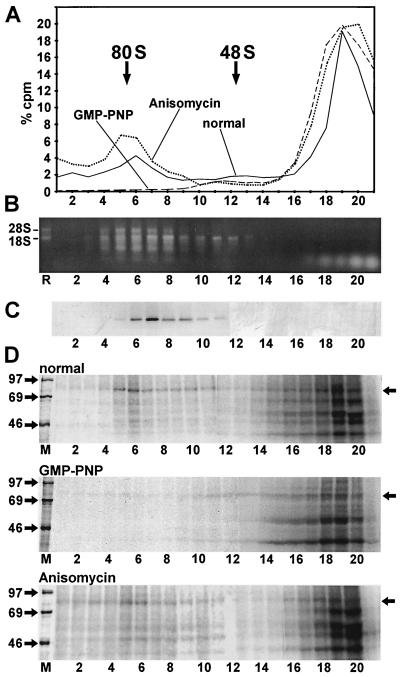
For analyzing the presence of eIF4B in initiation complexes with the poliovirus IRES, the RNA was incubated with RRL for 10 min at 30°C to allow the binding of initiation factors and ribosomal subunits to the IRES (32, 33). Following the formation of initiation complexes, the reaction product was transferred to 0°C and irradiated with UV light. The initiation complexes were then separated by differential sucrose gradient centrifugation. The 80S initiation complexes formed in large numbers with the poliovirus IRES RNA (Fig. 5A, fractions 4 to 8, solid line). The amounts of 48S complexes detected were lower (fractions 11 to 13). This may be due to the fact that binding of an RNA to the 43S complex is thought to be rate limiting during the formation of complete 80S ribosomes with several RNAs. After entering the RNA, the small ribosomal subunit may then be rapidly transferred to the initiator AUG and may associate with the 60S ribosomal subunit. Therefore, the actual amounts of 48S complexes detected at a given time may be very low. At the top of the gradient (fractions 15 to 21), free RNA and RNA protein aggregates not associated with ribosomal subunits are detected (32, 33).
FIG. 5.
Detection of eIF4B in ribosomal initiation complexes with poliovirus IRES RNA. (A) Radioactivity profile of the gradients. [α-32P]CTP-labeled poliovirus IRES 1-745 RNA was incubated with RRL for 10 min at 30°C either in the absence of translation inhibitors (solid line), in the presence of 4 mM GMP-PNP (broken line), or in the presence of 0.17 mM anisomycin (dotted line). Samples were irradiated with UV, and initiation complexes were separated on a 10 to 35% sucrose gradient. Fractions were collected from the bottom (fraction 1), and aliquots were used for scintillation counting. The amount of radioactivity in each fraction relative to the total amount of radioactivity in the entire gradient (% cpm) is plotted versus the fraction number. The ribosomal 80S and 48S peaks are indicated. (B and C) Identification of ribosomal subunits in gradients run in parallel with the gradients in panel A. (B) Nucleic acids isolated from gradient fractions were heated, separated on an 1% agarose gel, and stained with ethidium bromide. The positions of 18S and 28S rRNAs from a preparation of ribosomal RNAs (R) are labeled on the left. (C) Immunoblot of gradient fractions with an antibody directed against ribosomal protein S6. (D) UV cross-linked proteins from the gradients shown in panel A. Fractions were treated with RNase A, and proteins were precipitated with TCA, resolved on SDS-polyacrylamide gels, and visualized by autoradiography. Molecular masses of marker proteins (M) are given in kilodaltons. The position of eIF4B is indicated by arrows on the right.
The identity of the ribosomal initiation complexes was confirmed using stage-specific translation inhibitors. The GTP analogue GMP-PNP competes with GTP for incorporation into the ternary complex (eIF2-GTP-tRNAi); it inhibits release of eIF2 from the 40S subunit and by so doing prevents the subsequent 60S subunit joining. Accordingly, 80S complex formation was completely abolished by GMP-PNP (Fig. 5A, broken line). In contrast, anisomycin inhibits the ribosomal peptidyl-transferase and freezes the 80S ribosomes on the RNA immediately after the association of the 60S subunit. With this antibiotic, the amount of RNA associated with 80S complexes was increased (Fig. 5A, dotted line). The identity and localization of ribosomal subunits in the gradients were further checked by staining the ribosomal RNAs (Fig. 5B). 18S rRNA contained in the small ribosomal subunit appeared in fractions corresponding to 48S and 80S initiation complexes, whereas 28S rRNA appeared only in the 80S complexes, which contain the large ribosomal 60S subunit. 40S subunit localization was also confirmed with an antibody against ribosomal protein S6 (Fig. 5C).
Proteins bound to the IRES RNA were analyzed by UV cross-linking, RNase treatment, and SDS-polyacrylamide gel electrophoresis (Fig. 5D, upper panel). In the top fractions of the gradient, eIF4B bound to the poliovirus IRES, as did proteins of about 39, 50, 57, and 68 kDa (fractions 15 to 21). eIF4B is also associated with the poliovirus IRES RNA in the ribosomal 48S complexes (fractions 11 to 13) and, remarkably, in the complete 80S ribosomes as well (fractions 4 to 8). In the gradients with translation inhibitors, the intensity of the eIF4B band correlates with the increase or decrease of the 48S and 80S peaks (Fig. 5A). In contrast to eIF4B, only trace amounts of the 39-, 50-, 57-, and 68-kDa proteins were detected in the ribosomal complexes. In conclusion, the IRES-bound eIF4B had been selectively incorporated into the ribosomal initiation complexes, whereas other proteins were largely excluded from interaction with the viral RNA in the ribosomal complexes. These results indicate that eIF4B plays an important role in the formation of the initiation complexes with the poliovirus IRES RNA.
Binding of eIF4B to the poliovirus IRES is strong compared with its binding to cellular poly(A)+ RNA.
Initiation factors eIF2 and eIF3 dissociate from the ribosomal 48S preinitiation complex when the 60S subunit joins to form a complete 80S ribosome (5, 14). However, the fate of eIF4B in this process has not been elucidated in detail, perhaps due to its weak interaction with cellular mRNAs. Accordingly, the association of eIF4B with a picornavirus IRES in complete 80S ribosomes may be viewed as controversial. Therefore, we directly compared the strength of the interaction of eIF4B with the poliovirus IRES with its binding to cellular mRNAs in competition experiments, and we investigated the fate of eIF4B in initiation complexes formed with cellular mRNAs.
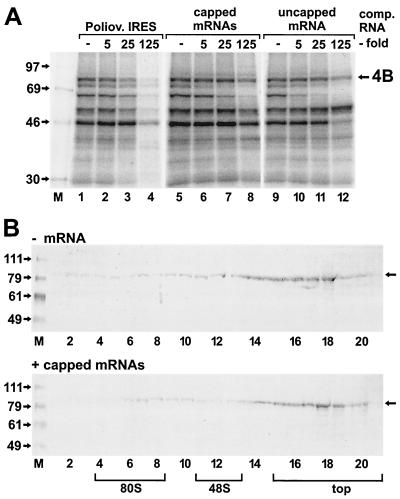
The binding of eIF4B to the radiolabeled poliovirus IRES was strongly competed with by unlabeled poliovirus IRES RNA (Fig. 6A, lanes 1 to 4). In contrast, equivalent molar amounts of total cellular poly(A)+ RNAs competed with the binding of eIF4B to the poliovirus IRES to an extent about 1 order of magnitude weaker (lanes 5 to 8). Also, an in vitro-transcribed uncapped eIF2β mRNA (nucleotides 1 to 417) competed only weakly with the binding of eIF4B to the poliovirus IRES, paralleling data obtained previously for the FMDV IRES (29).
FIG. 6.
Weak interaction of eIF4B with cellular mRNAs. (A) Competition reactions. [α-32P]CTP-labeled poliovirus IRES RNA 1-745 was used in all lanes. For competition, either the poliovirus IRES RNA 1-745 (Poliov. IRES), mouse liver poly(A)+ RNA (capped mRNAs), or eIF2β mRNA 1-417 (uncapped mRNA) was used as an unlabeled competitor RNA in the molar concentration indicated on the top. 4B, eIF4B. (B) Immunoblots of eIF4B from sucrose gradients. Either no RNA (− mRNA) or 10 μg of mouse liver poly(A)+ RNA (+ capped mRNAs) was added to the binding reaction mixture containing 100 μl of RRL. Reactions were incubated at 30°C for 10 min and separated on sucrose gradients as in Fig. 5. Proteins were resolved on SDS-polyacrylamide gels, transferred to nitrocellulose, and probed with an antibody directed against recombinant eIF4B. Molecular masses of marker proteins (M) are given in kilodaltons. The position of eIF4B is indicated by arrows on the right.
We also investigated the interaction of eIF4B with cellular poly(A)+ RNAs in ribosomal initiation complexes (Fig. 6B). When no RNA was added to the nuclease-treated RRL, eIF4B was detected largely in the upper fractions of the gradient that do not contain ribosomal subunits (Fig. 6B, upper panel). Only minute amounts of eIF4B were detected in the lower fractions of the gradient, possibly due to the ability of eIF4B to weakly interact even with purified ribosomes in the absence of mRNA (15). When 10 μg of cellular poly(A)+ RNA was added, no change in the detection of eIF4B throughout the gradient fractions was observed (Fig. 6B, lower panel), indicating that no eIF4B had been incorporated into the 80S ribosomes by a possible interaction of eIF4B with the added mRNA.
In this assay using 100 μl of RRL, the molar amount of cellular poly(A)+ RNA added was in the same range as the molar amount of eIF4B which can be estimated from previous studies. Per 100 μl of RRL, about 2 × 10−12 mol of eIF4B was obtained after purification (49). Taking into account that purification yielded only about 20% of the total eIF4B (4), we can assume that 100 μl of RRL can contain 10 × 10−12 mol of eIF4B. The cellular poly(A)+ RNA we used has a mean size of about 2,000 bp (data not shown). Thus, 10 μg of this mRNA corresponds to about 15 × 10−12 mol of mRNA.
Taken together, the binding of eIF4B to cellular mRNAs must be regarded as weak compared with its binding to the poliovirus IRES, and the association of eIF4B with the complete 80S ribosomal initiation complexes appears to be different from the interaction of eIF4B with cellular mRNAs.
The order of binding of eIF4B and ribosomes to the IRES RNA.
To determine the order in which eIF4B and the ribosomes bind to the poliovirus IRES, the ribosomes were separated from the soluble fraction of the reticulocyte lysate. First, the RRL was adjusted to 250 mM K acetate, ensuring that a sufficient amount of initiation factors, particularly eIF4B, dissociated from the ribosomes and was present in the soluble fraction. Then the ribosomes were pelleted. The ribosome-free supernatant was then used to bind the initiation factors to the poliovirus IRES RNA in the absence of ribosomes. To determine the possible temperature sensitivity of this reaction, this step was performed at either 0 or 30°C. After that, UV cross-linking was performed at 0°C to covalently cross-link the bound proteins to the RNA. After UV cross-linking, the ribosomes were added back, and the reaction was incubated again at either 0 or 30°C for the association of the ribosomes. The reaction products were then loaded on sucrose gradients and analyzed as before.
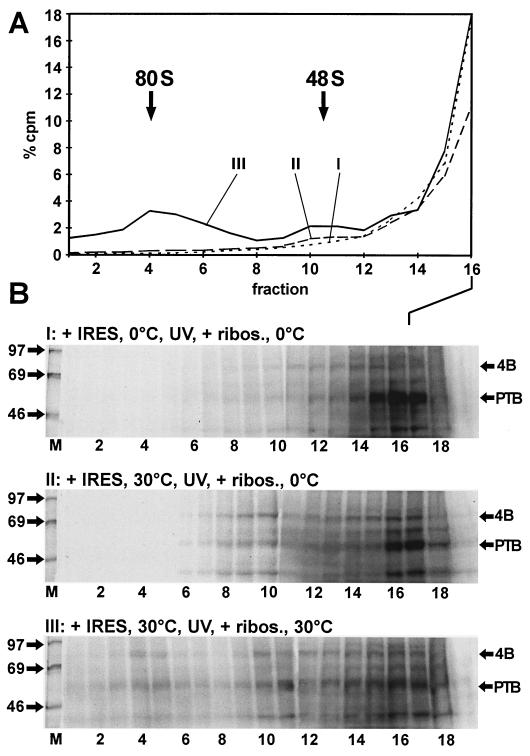
In the first reaction (reaction I), the IRES RNA was incubated with the RRL devoid of ribosomes at 0°C. After UV cross-linking, ribosomes were added back and the reaction mixture was incubated again for 10 min at 0°C. In this reaction, only small amounts of 48S complexes were formed and PTB appeared in the top fractions, but eIF4B bound only marginally to the IRES (Fig. 7). This indicates that (i) the binding of eIF4B to the IRES is largely temperature dependent whereas the binding of PTB is temperature independent (32), and (ii) the small ribosomal subunit is not able to associate with the IRES in the absence of bound initiation factors.
FIG. 7.
Order and temperature dependency of binding events. Binding reactions and gradient runs were performed with a reconstituted RRL system. The ribosomes had been removed from the RRL by centrifugation before this experiment. Binding of initiation factors from the ribosomal supernatant, UV cross-linking, and the subsequent binding of reconstituted ribosomes were performed separately and at the temperatures indicated. (A) Gradient profiles of fractions 1 to 16 of reactions I, II, and III. RRL devoid of ribosomes was incubated with RNA 1-745 (+ IRES) for 10 min at 30 or 0°C as indicated. The reactions were then irradiated with UV at 0°C to covalently cross-link the RNA to the initiation factors. The ribosomes were then added (+ ribos.), and the reaction product was again incubated for 10 min at 30 or 0°C as indicated. The reaction products were then loaded onto sucrose gradients and analyzed as for Fig. 5. (B) UV cross-linked proteins from the gradients shown in panel A. Molecular masses of marker proteins (M) are given in kilodaltons. The positions of eIF4B (4B) and PTB are indicated.
In reaction II, IRES RNA was incubated with RRL devoid of ribosomes at 30°C for 10 min, and after UV cross-linking, ribosomes were added back and the reaction mixture was incubated for another 10 min at 0°C (Fig. 7B). In this reaction, eIF4B bound to the IRES. Moreover, a large amount of eIF4B was present in the 48S peak. From these results we conclude that (i) binding of eIF4B and possibly of other initiation factors to the IRES occurs in an energy-dependent manner in the absence of ribosomes, (ii) binding of the initiation factors (most likely including eIF4B) may be a prerequisite for the binding of the ribosomal 40S subunit to the IRES, (iii) this binding of the small ribosomal subunit to an IRES preloaded with initiation factors is not energy dependent, and (iv) joining of the 60S subunit again involves energy-dependent reactions, since no 80S complexes were formed at 0°C after the addition of ribosomal subunits.
In reaction III, incubation with ribosomes was performed at 30°C. In this case, not only 48S but also 80S complexes were formed, confirming that the joining of the 60S subunit requires energy-dependent reactions.
These experiments show that eIF4B binds in an energy-dependent manner to the poliovirus IRES, and this binding of initiation factors (most likely including eIF4B) may be a prerequisite for the entry of the small ribosomal subunit into the poliovirus IRES, a step which is energy independent per se. The subsequent association of the 60S subunit again occurs in an energy-dependent manner due to GTP hydrolysis by eIF2 and eIF5B (40).
DISCUSSION
In this study, we show that eIF4B interacts directly with the poliovirus IRES. Binding of the initiation factors eIF4B, eIF4G, and eIF4A to picornaviral IRES elements had been reported only for type II IRES elements, namely, those of FMDV (23, 29, 45, 46) and EMCV (19, 38). In contrast, for the picornavirus type I IRES elements, e.g., those of poliovirus or the rhinoviruses, as well as for the type III IRES of HAV, no such interaction of initiation factors has been described so far. In cell-free reconstitution experiments, the more distantly related IRES elements of hepatitis C virus and pestiviruses, two members of the genera Hepacivirus and Pestivirus, respectively, of the Flaviviridae family, bind the small ribosomal subunit completely independently of any initiation factors, even without eIF2 and eIF3 (41).
The binding site for eIF4B in the poliovirus IRES resides mainly in the IRES domain V, whereas domain VI sequences, including the silent AUG at nucleotide 586, contribute to the binding of eIF4B to a lesser extent, which is a finding consistent with the observation that domain VI is not absolutely essential for poliovirus translation (21, 25). In domain V, several linker scanning mutations seriously impaired the binding of eIF4B (see Fig. 3). These mutations also seriously impaired poliovirus translation and growth, leading to either lethal or temperature-sensitive phenotypes (11). This correlation suggests that eIF4B may play an important role in the initiation of translation from the poliovirus IRES.
Even if these linker scanning mutations may alter the actual structure of domain V, we can conclude that separate determinants located both in its lower part and in its upper part are required for eIF4B binding and for translation. Secondary structure calculations (reference 24 and data not shown) for the lower part of domain V and for the bulge predict a variety of different possible secondary structures with similar energy minima. Accordingly, the RNA may easily change its conformation, possibly to meet different requirements for the binding of various cellular proteins. All linker scanning mutations in this region seriously impaired binding of eIF4B (this study) as well as the viability of the corresponding virus (11). In contrast, all secondary structures for the upper part of domain V are predicted to be similar, regardless of whether they are in the context of unmutated domain V or of variants with altered predicted secondary structures of the lower part. Thus, the serious effect seen of the X482 mutation implies that the upper part of domain V must bear a determinant for the binding of eIF4B (this study) and for virus growth (11) that is separate from those located in the lower part.
In the relative position of its eIF4B binding site within the arrangement of secondary structures, the poliovirus IRES resembles type II IRES elements like those of FMDV and EMCV (3, 8, 30). Also, in FMDV, the determinant for the binding of eIF4B resides in the IRES 3"-region, mainly including the Y-shaped domain 4 (29, 45). A common feature of the eIF4B binding regions in both types of IRES elements is that they are located downstream of the large central domain structure, which is referred to as domain 3 in FMDV and domain IV in poliovirus, respectively.
At a first glance, this arrangement of cis elements in the picornaviral IRES elements may also reflect a segmentation of the IRES at a functional level. The 3" part of the IRES appears to be the area for binding the canonical translation initiation factors that probably guide the ribosome to the starting window at the IRES 3" border. This process may also involve interactions between the IRES-bound eIF4B and the 170-kDa subunit of the ribosome-bound eIF3 (28). In addition, the oligopyrimidine tract in the 3" region of the IRES may interact with the ribosomal 18S RNA (44, 47), perhaps facilitated by the RNA annealing activity of eIF4B (1) and the RNA helicase activity of eIF4A (35). In contrast, the upstream parts of the IRES may bind regulatory proteins, like the 39-kDa poly(rC) binding protein 2 (6), which has been speculated to be involved in a switch from translation to minus-strand synthesis later in the poliovirus replication cycle (9).
However, this simple view of a clear segmentation of the IRES elements is complicated by the finding that the apical parts of the poliovirus IRES domains V and VI bear determinants for neuropathogenicity (10). Moreover, PTB appears to bind to multiple sites of the poliovirus IRES, including domain V (reference 12 and this study). Thus, the determinants for the binding of cellular proteins which may be involved in regulatory phenomena appear to overlap with the binding site for at least one of the canonical initiation factors, eIF4B. However, although binding of these factors appears to depend on closely neighboring determinants in domain V, binding of PTB does not interfere with the binding of eIF4B. Further studies will be required to investigate the possible interactions of known or yet unknown cellular factors with the assembled core of canonical initiation factors in the 3" region of the poliovirus IRES.
The events during the assembly of the complete 80S ribosome on the poliovirus RNA can be clearly subdivided into different steps that occur sequentially. The first step is the binding of soluble initiation factors, including eIF4B, to the IRES. This step is energy dependent, consistent with the observation that binding of eIF4B to the FMDV IRES is ATP dependent (29). This binding of initiation factors occurs independently of ribosomes, since eIF4B was UV cross-linked to the IRES in the complete absence of ribosomes. Only after this binding does the small ribosomal subunit enter this RNA-protein complex. This second step is energy independent per se but requires the previous binding of the initiation factors. The last step in the assembly of complete 80S ribosomes, the joining of the ribosomal 60S subunit, is again energy dependent due to the hydrolysis of the eIF2-bound GTP prior to subunit joining.
eIF4B remains bound to the poliovirus IRES not only after the association of the small ribosomal subunit. In contrast to the weak interaction of eIF4B with capped cellular mRNAs, eIF4B remains still tightly associated with the poliovirus IRES even after the formation of the complete 80S ribosome. This observation parallels results obtained with the type II FMDV IRES (33) and points to a possible functional difference between the initiation of translation mediated by picornaviral IRES elements and cap-dependent initiation. Probably eIF4B and also other initiation factors remain continuously bound to the IRES 3" region after joining of the ribosomal 60S subunit and perhaps even during repeated events of ribosome entry.
Acknowledgments
We thank B. L. Semler for kindly providing the poliovirus domain V and VI mutants 5"-NC-X482, -X515, -X521, -X529, -X539, -X543, and -X585 (11); C. U. T. Hellen for plasmid pT7XLmyr(−); M. Garçia-Blanco for plasmid pGEX-2TKhuPTB; M. Bachmann for anti-La antibodies 4B6 and 3B9; M. Bachmann and T. Heise for recombinant La protein; and E. Beck for helpful discussions.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 535 and GK 370).
REFERENCES
- 1.Altmann, M., B. Wittmer, N. Méthot, N. Sonenberg, and H. Trachsel. 1995. The Saccharomyces cerevisiae translation initiation factor Tif3 and its mammalian homologue, eIF-4B, have RNA annealing activity. EMBO J. 14:3820-3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthony, D. D., and W. C. Merrick. 1991. Eukaryotic initiation factor (eIF)-4F. Implications for a role in internal initiation of translation. J. Biol. Chem. 266:10218-10226. [PubMed] [Google Scholar]
- 3.Belsham, G. J., and R. J. Jackson. 2000. Translation initiation on picornavirus RNA, p. 869-900. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 4.Benne, R., M. L. Brown Luedi, and J. W. Hershey. 1979. Protein synthesis initiation factors from rabbit reticulocytes: purification, characterization, and radiochemical labeling. Methods Enzymol. 60:15-35. [DOI] [PubMed] [Google Scholar]
- 5.Benne, R., and J. W. Hershey. 1978. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J. Biol. Chem. 253:3078-3087. [PubMed] [Google Scholar]
- 6.Blyn, L. B., J. S. Towner, B. L. Semler, and E. Ehrenfeld. 1997. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 71:6243-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dildine, S. L., K. R. Stark, A. A. Haller, and B. L. Semler. 1991. Poliovirus translation initiation: differential effects of directed and selected mutations in the 5" noncoding region of viral RNAs. Virology 182:742-752. [DOI] [PubMed] [Google Scholar]
- 8.Ehrenfeld, E., and B. L. Semler. 1995. Anatomy of the poliovirus internal ribosome entry site. Curr. Top. Microbiol. Immunol. 203:65-83. [DOI] [PubMed] [Google Scholar]
- 9.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gromeier, M., B. Bossert, M. Arita, A. Nomoto, and E. Wimmer. 1999. Dual stem loops within the poliovirus internal ribosomal entry site control neurovirulence. J. Virol. 73:958-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haller, A. A., and B. L. Semler. 1992. Linker scanning mutagenesis of the internal ribosome entry site of poliovirus RNA. J. Virol. 66:5075-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hellen, C. U., T. V. Pestova, M. Litterst, and E. Wimmer. 1994. The cellular polypeptide p57 (pyrimidine tract-binding protein) binds to multiple sites in the poliovirus 5" nontranslated region. J. Virol. 68:941-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hellen, C. U., G. W. Witherell, M. Schmid, S. H. Shin, T. V. Pestova, A. Gil, and E. Wimmer. 1993. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl. Acad. Sci. USA 90:7642-7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hershey, J. W. B., and W. C. Merrick. 2000. Pathway and mechanism of initiation of protein synthesis, p. 33-88. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Hughes, D. L., T. E. Dever, and W. C. Merrick. 1993. Further biochemical characterization of rabbit reticulocyte eIF-4B. Arch. Biochem. Biophys. 301:311-319. [DOI] [PubMed] [Google Scholar]
- 16.Hühn, P., G. J. Pruijn, W. J. van Venrooij, and M. Bachmann. 1997. Characterization of the autoantigen La (SS-B) as a dsRNA unwinding enzyme. Nucleic Acids Res. 25:410-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jang, S. K., H. G. Kräusslich, M. J. Nicklin, G. M. Duke, A. C. Palmenberg, and E. Wimmer. 1988. A segment of the 5" nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaminski, A., S. L. Hunt, J. G. Patton, and R. J. Jackson. 1995. Direct evidence that polypyrimidine tract binding protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA. RNA 1:924-938. [PMC free article] [PubMed] [Google Scholar]
- 19.Kolupaeva, V. G., T. V. Pestova, C. U. Hellen, and I. N. Shatsky. 1998. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J. Biol. Chem. 273:18599-18604. [DOI] [PubMed] [Google Scholar]
- 20.Kuge, S., N. Kawamura, and A. Nomoto. 1989. Genetic variation occurring on the genome of an in vitro insertion mutant of poliovirus type 1. J. Virol. 63:1069-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuge, S., and A. Nomoto. 1987. Construction of viable deletion and insertion mutants of the Sabin strain type 1 poliovirus: function of the 5" noncoding sequence in viral replication. J. Virol. 61:1478-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.La Monica, N., and V. R. Racaniello. 1989. Differences in replication of attenuated and neurovirulent polioviruses in human neuroblastoma cell line SH-SY5Y. J. Virol. 63:2357-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez de Quinto, S., and E. Martinez-Salas. 2000. Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA 6:1380-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 25.Meerovitch, K., R. Nicholson, and N. Sonenberg. 1991. In vitro mutational analysis of cis-acting RNA translational elements within the poliovirus type 2 5" untranslated region. J. Virol. 65:5895-5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meerovitch, K., Y. V. Svitkin, H. S. Lee, F. Lejbkowicz, D. J. Kenan, E. K. Chan, V. I. Agol, J. D. Keene, and N. Sonenberg. 1993. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 67:3798-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Méthot, N., G. Pickett, J. D. Keene, and N. Sonenberg. 1996. In vitro RNA selection identifies RNA ligands that specifically bind to eukaryotic translation initiation factor 4B: the role of the RNA recognition motif. RNA 2:38-50. [PMC free article] [PubMed] [Google Scholar]
- 28.Méthot, N., M. S. Song, and N. Sonenberg. 1996. A region rich in aspartic acid, arginine, tyrosine, and glycine (DRYG) mediates eukaryotic initiation factor 4B (eIF4B) self-association and interaction with eIF3. Mol. Cell. Biol. 16:5328-5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer, K., A. Petersen, M. Niepmann, and E. Beck. 1995. Interaction of eukaryotic initiation factor eIF-4B with a picornavirus internal translation initiation site. J. Virol. 69:2819-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niepmann, M. 1999. Internal initiation of translation of picornaviruses, hepatitis C virus and pestiviruses. Recent Res. Dev. Virol. 1:229-250. [Google Scholar]
- 31.Niepmann, M. 1996. Porcine polypyrimidine tract-binding protein stimulates translation initiation at the internal ribosome entry site of foot-and-mouth-disease virus. FEBS Lett. 388:39-42. [DOI] [PubMed] [Google Scholar]
- 32.Niepmann, M., A. Petersen, K. Meyer, and E. Beck. 1997. Functional involvement of polypyrimidine tract-binding protein in translation initiation complexes with the internal ribosome entry site of foot-and-mouth disease virus. J. Virol. 71:8330-8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ochs, K., R. C. Rust, and M. Niepmann. 1999. Translation initiation factor eIF4B interacts with a picornavirus internal ribosome entry site in both 48S and 80S initiation complexes independently of initiator AUG location. J. Virol. 73:7505-7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pathak, V. K., P. J. Nielsen, H. Trachsel, and J. W. Hershey. 1988. Structure of the beta subunit of translational initiation factor eIF-2. Cell 54:633-639. [DOI] [PubMed] [Google Scholar]
- 35.Pause, A., N. Méthot, Y. Svitkin, W. C. Merrick, and N. Sonenberg. 1994. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 13:1205-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pelletier, J., G. Kaplan, V. R. Racaniello, and N. Sonenberg. 1988. Cap-independent translation of poliovirus mRNA is conferred by sequence elements within the 5" noncoding region. Mol. Cell. Biol. 8:1103-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pelletier, J., and N. Sonenberg. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320-325. [DOI] [PubMed] [Google Scholar]
- 38.Pestova, T. V., C. U. Hellen, and I. N. Shatsky. 1996. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16:6859-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pestova, T. V., C. U. Hellen, and E. Wimmer. 1994. A conserved AUG triplet in the 5" nontranslated region of poliovirus can function as an initiation codon in vitro and in vivo. Virology 204:729-737. [DOI] [PubMed] [Google Scholar]
- 40.Pestova, T. V., I. B. Lomakin, J. H. Lee, S. K. Choi, T. E. Dever, and C. U. Hellen. 2000. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 403:332-335. [DOI] [PubMed] [Google Scholar]
- 41.Pestova, T. V., I. N. Shatsky, S. P. Fletcher, R. J. Jackson, and C. U. Hellen. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12:67-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pilipenko, E. V., V. M. Blinov, B. K. Chernov, T. M. Dmitrieva, and V. I. Agol. 1989. Conservation of the secondary structure elements of the 5"-untranslated region of cardio- and aphthovirus RNAs. Nucleic Acids Res. 17:5701-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pilipenko, E. V., A. P. Gmyl, S. V. Maslova, G. A. Belov, A. N. Sinyakov, M. Huang, T. D. Brown, and V. I. Agol. 1994. Starting window, a distinct element in the cap-independent internal initiation of translation on picornaviral RNA. J. Mol. Biol. 241:398-414. [DOI] [PubMed] [Google Scholar]
- 44.Pilipenko, E. V., A. P. Gmyl, S. V. Maslova, Y. V. Svitkin, A. N. Sinyakov, and V. I. Agol. 1992. Prokaryotic-like cis elements in the cap-independent internal initiation of translation on picornavirus RNA. Cell 68:119-131. [DOI] [PubMed] [Google Scholar]
- 45.Rust, R. C., K. Ochs, K. Meyer, E. Beck, and M. Niepmann. 1999. Interaction of eukaryotic initiation factor eIF4B with the internal ribosome entry site of foot-and-mouth disease virus is independent of the polypyrimidine tract-binding protein. J. Virol. 73:6111-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saleh, L., R. C. Rust, R. Füllkrug, E. Beck, G. Bassili, K. Ochs, and M. Niepmann. 2001. Functional interaction of translation initiation factor eIF4G with the foot-and-mouth-disease virus internal ribosome entry site. J. Gen. Virol. 82:757-763. [DOI] [PubMed] [Google Scholar]
- 47.Scheper, G. C., H. O. Voorma, and A. A. Thomas. 1994. Basepairing with 18S ribosomal RNA in internal initiation of translation. FEBS Lett. 352:271-275. [DOI] [PubMed] [Google Scholar]
- 48.Svitkin, Y. V., K. Meerovitch, H. S. Lee, J. N. Dholakia, D. J. Kenan, V. I. Agol, and N. Sonenberg. 1994. Internal translation initiation on poliovirus RNA: further characterization of La function in poliovirus translation in vitro. J. Virol. 68:1544-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas, A., H. Goumans, H. Amesz, R. Benne, and H. O. Voorma. 1979. A comparison of the initiation factors of eukaryotic protein synthesis from ribosomes and from the postribosomal supernatant. Eur. J. Biochem. 98:329-337. [DOI] [PubMed] [Google Scholar]
- 50.Trono, D., R. Andino, and D. Baltimore. 1988. An RNA sequence of hundreds of nucleotides at the 5" end of poliovirus RNA is involved in allowing viral protein synthesis. J. Virol. 62:2291-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trono, D., J. Pelletier, N. Sonenberg, and D. Baltimore. 1988. Translation in mammalian cells of a gene linked to the poliovirus 5" noncoding region. Science 241:445-448. [DOI] [PubMed] [Google Scholar]