Abstract
Clade C is one of the most prevalent genetic subtypes of human immunodeficiency virus type 1 (HIV-1) in the world today and one of the least studied with respect to neutralizing antibodies. Most information on HIV-1 serology as it relates to neutralization is derived from clade B. Clade C primary isolates of HIV-1 from South Africa and Malawi were shown here to resemble clade B isolates in their resistance to inhibition by soluble CD4 and their sensitivity to neutralization by human monoclonal antibody immunoglobulin G1b12 and, to a lesser extent, 2F5. Unlike clade B isolates, however, all 16 clade C isolates examined resisted neutralization by 2G12. Infection with clade C HIV-1 in a cohort of female sex workers in South Africa generated antibodies that neutralized the autologous clade C isolate and T-cell-line-adapted (TCLA) strains of clade B. Neutralization of clade B TCLA strains was much more sensitive to the presence of autologous gp120 V3 loop peptides compared to the neutralization of clade C isolates in most cases. Thus, the native structure of gp120 on primary isolates of clade C will likely pose a challenge for neutralizing antibody induction by candidate HIV-1 vaccines much the same as it has for clade B. The autologous neutralizing antibody response following primary infection with clade C HIV-1 in South Africa matured slowly, requiring at least 4 to 5 months to become detectable. Once detectable, extensive cross-neutralization of heterologous clade C isolates from South Africa was observed, suggesting an unusual degree of shared neutralization determinants at a regional level. This high frequency of cross-neutralization differed significantly from the ability of South African clade C serum samples to neutralize clade B isolates but did not differ significantly from results of other combinations of clade B and C reagents tested in checkerboard assays. Notably, two clade C serum samples obtained after less than 2 years of infection neutralized a broad spectrum of clade B and C isolates. Other individual serum samples showed a significant clade preference in their neutralizing activity. Our results suggest that clades B and C are each comprised of multiple neutralization serotypes, some of which are more clade specific than others. The clustering of shared neutralization determinants on clade C primary HIV-1 isolates from South Africa suggests that neutralizing antibodies induced by vaccines will have less epitope diversity to overcome at a regional level.
An important goal in the development of an effective human immunodeficiency virus type 1 (HIV-1) vaccine is to overcome the extensive genetic heterogeneity of the virus. Nucleotide sequence comparisons have been used to define three groups of the virus known as group M (main), group O (outlier), and group N (non-M, non-O) (50, 69). Group M is further divided into 10 phylogenically related genetic subtypes (clades A, B, C, D, F1, F2, G, H, J, and K) that, together with a growing number of circulating intersubtype recombinant forms, comprise the majority of HIV-1 variants in the world today. Clade C is emerging as most prevalent, being common in India (15, 16, 31, 41) and the southern African countries of Botswana, Zimbabwe, Malawi, Mozambique, and South Africa (7, 8, 25, 26, 60, 64, 79, 81). Clade B is dominant in North America and Western Europe and has been a major focus for vaccine development (27). It is uncertain whether vaccines that are ultimately effective against clade B will be capable of targeting other genetic subtypes of the virus.
The uncertain relevance of genetic subtype to HIV-1 vaccines is owed in part to a poor understanding of the immunotype diversity of the virus as it relates both to cellular and humoral immunity. The fact that genetic subtypes tend to cluster geographically raises the possibility that distinct immunotypes of the virus have evolved along similar lines and, although a growing body of evidence suggests that this may not be true in a strict sense (4, 12, 29, 38, 56, 61, 82), additional studies seem warranted. For example, with respect to humoral immunity, the sporadic neutralizing activity of sera from HIV-1-infected individuals appears to be independent of genetic subtype (38, 56, 61, 82). That observation has led to a general notion that genetic subtype does not predict the neutralization serotype of the virus. An exception has been noted for clades B and E (E is now known as recombinant subtype A/E [32]), which appear to consist of different neutralization serotypes relative to each other. That conclusion was based on results of checkerboard assessments made with four serum samples and virus isolates from each clade (47) and when serum pools from both clades, selected for high neutralizing antibody titers, were tested with a larger panel of clade B and E isolates (45).
The concept of HIV-1 immunotypes may be particularly relevant to neutralizing antibodies. Neutralizing antibodies target the surface gp120 and transmembrane gp41 envelope glycoproteins of the virus (62, 65) and could be a valuable antiviral immune response to generate with vaccines (44, 49, 52). These glycoproteins exist as a trimolecular complex of gp120-gp41 heterodimers (17, 24, 43, 83) and are essential for virus entry. Antibody-mediated neutralization of HIV-1 may take place either by blocking gp120 from binding its cellular receptor (CD4) or coreceptor (CCR5 and CXCR4) or by preventing gp41 from mediating fusion with the target cell membrane (2, 18, 21, 22, 28, 30, 39, 40, 74, 84). Both glycoproteins display an unusual degree of sequence variation that gives rise to complex epitopes. One manifestation of this variation during infection is the evolution of neutralization escape variants. For example, serum from individuals infected with clade B HIV-1 often fails to neutralize contemporaneous and later virus isolates but neutralizes earlier isolates from the respective individuals quite potently (1, 3, 6). Epitopes responsible for primary isolate neutralization by serum samples from HIV-1-infected individuals remain largely unknown.
Genetic variation also affects the higher-order structure of the native envelope glycoprotein complex, having a profound effect on antigenicity. For example, certain N-glycans and tertiary folds in the gp120 core can render primary HIV-1 isolates resistant to neutralization by soluble CD4 (sCD4) and many antibody specificities compared to T-cell-line-adapted (TCLA) strains of the virus (20, 68, 72, 85). This is very common for epitopes in the third variable cysteine-cysteine loop (V3 loop) of gp120 (5, 73, 77). Antibodies in sera from infected individuals and vaccinated volunteers may have potent neutralizing activity against the matched (autologous) HIV-1 variant but those antibodies have limited neutralizing activity against heterologous variants (14, 34, 55, 58, 63). This is especially true in the case of vaccine-elicited antibodies (9, 48). Some exceptions include a small number of human monoclonal antibodies (e.g., 2G12, immunoglobulin G1b12 [IgG1b12], and 2F5) and sera from a subset of HIV-1-infected long-term nonprogressors, in which cases a broad spectrum of HIV-1 isolates are neutralized (10).
Immunotype diversity could adversely impact the ability to develop a single HIV-1 vaccine that is broadly effective on a global scale. Alternatively, it may be possible to tailor vaccines to match the virus variant(s) circulating in specific regions targeted for vaccination. In either case, the ability to identify and predict the relevant immunotypes of the virus would hasten vaccine efforts. With respect to neutralizing antibodies, very few virus isolates and serum samples belonging to clade C have been characterized, and in fact, very little is known about the neutralization properties of clade C isolates and serum samples from South Africa. The present studies were conducted to define the neutralization properties of these virologic and serologic reagents in preparation for future vaccine clinical trials in South Africa and other areas where clade C is common. Our results identify important similarities and differences between clades B and C of HIV-1. Moreover, we provide evidence that neutralization serotypes of HIV-1 may cluster geographically within a clade. The implications of these findings for vaccine development are discussed.
MATERIALS AND METHODS
Viruses, cells, and serum samples.
Eight clade B and 17 clade C primary HIV-1 isolates were used in these studies. All clade B isolates possessed an R5 phenotype and were obtained during early seroconversion from subjects in the United States, Trinidad, and Italy (9). Nine clade C isolates (which begin with the prefix Du) were isolated in 1998 from female sex workers recruited from four truck stops along the major trucking route between Durban and Johannesburg, South Africa. These women were participating in a multicenter clinical trial of a potential vaginal microbicide (78). Details of the recruitment procedures are documented elsewhere (67). The women were screened monthly for sexually transmitted infections including HIV. HIV-1 seroconversion was assessed by enzyme-linked immunosorbent assay (ELISA) (Abbott, Chicago, Ill.) and confirmed by using the Vironostika HIV Uniform II micro-ELISA 4 system (Omnimed, Madison, Wis.). All HIV-1-positive tests were further confirmed by the Institute of Tropical Medicine, Antwerp, Belgium--the central laboratory for the multicenter trial. At each visit, women were given pre- and posttest HIV-1 counseling and were provided with intensive condom counseling. Sexually transmitted diseases were treated according to the South African syndromic management guidelines. Virus was isolated within 4 months of initial seroconversion from five of these sex workers (Du123, 3 months; Du151, 1.5 months; Du156, 1 month; Du172, 1 month; Du422, 4 months) and at later time points from four additional sex workers (Du174, 19.5 months; Du179, 21 months; Du204, 12 months; Du368, 7.5 months). Sequence analysis of gag, pol, and env confirmed that all South African isolates were clade C and lacked intersubtype recombination (Williamson et al., submitted for publication).
Eight additional clade C isolates (which begin with the prefix S) were obtained from individuals attending the sexually transmitted disease clinic of the Lilongwe Central Hospital in Malawi (64). These isolates were shown to be clade C by serologic and sequence analysis of the V3 loop of gp120 (64). All clade C isolates from Africa had an R5 phenotype except for Du179, which was R5X4 and induced syncytium formation in MT-2 cells. Coreceptor usage of clade C isolates from South Africa was assessed in U87.CD4 cells transfected to express either CCR5 or CXCR4 as described (57). The coreceptor usage of the remaining isolates was reported previously (9, 64).
Viruses were isolated by peripheral blood mononuclear cell (PBMC) coculture as described (53, 57). All primary isolates were of a low passage number (three passages or fewer) in PBMC exclusively. Stocks of the TCLA strains, HIV-1IIIB, HIV-1MN, and HIV-1SF-2, were generated in H9 cells (54). All virus stocks were made cell free by filtration (filter pore size, 0.45-μm) and stored in aliquots at −80°C until use. Two additional cell lines used in neutralization assays, MT-2 and CEMx174, have been described previously (35, 70). PBMC were maintained in RPMI 1640 containing 20% heat-inactivated fetal bovine serum and supplemented with gentamicin (50 μg/ml) and human interleukin-2 (4%). Growth medium for the H9, MT-2, and CEMx174 cells consisted of RPMI 1640 containing 12% heat-inactivated fetal bovine serum supplemented with gentamicin (50 μg/ml).
Serum samples presumed to be clade B were obtained from HIV-1-infected individuals attending the Duke University Medical Center Infectious Diseases Clinic in Durham, N.C. The individuals had been infected with HIV-1 for ≥2 years and donated serum between 1998 and 2000. A previous study assessed these serum samples for neutralizing activity against the eight clade B primary isolates used here (9). Additional serum samples were collected at multiple time points from each of the nine sex workers in South Africa. Due to personal preference, participation in a structured treatment-interruption protocol, or nonavailability, no subjects were on antiretroviral therapy at the time of serum collection; this was done to avoid possible in vitro antiviral activity of the drugs that might be misinterpreted as neutralizing antibody activity at low serum dilutions. All sera were heat inactivated at 56°C for 45 min.
Monoclonal antibodies and sCD4.
IgG1b12, 2G12, and 2F5 are human monoclonal antibodies that bind conserved epitopes and cross-neutralize a variety of TCLA strains and primary isolates (11, 23, 36, 66, 75, 76). The epitope for IgG1b12 is located in the CD4-binding domain of gp120 and is sensitive to mutations in V2 and C3 (51). 2G12 recognizes an epitope in the C2-V4 region of gp120 that involves sites of N glycosylation (76). 2F5 recognizes a linear epitope in the ectodomain of gp41 having the amino acid sequence ELDKWA (59). sCD4 comprising the full-length extracellular domain of human CD4 and produced in Chinese hamster ovary cells was obtained from Progenics Pharmaceuticals, Inc. (Tarrytown, N.Y.).
DNA sequencing.
The DNA sequence of the V3 loop of gp120 was determined by direct sequencing of amplified products generated by RT-PCR. PCR primers, with the reverse outer primer used as the reverse transcriptase primer in the cDNA synthesis step, were o-env, 6201 to 6227 and 9067 to 9095; i-env, 6815 to 6838 and 7322 to 7349 (numbered using the HIV-1 HXBr sequence, Los Alamos HIV sequence database). Amplified DNA fragments were purified using the QIAQUICK PCR Purification Kit (Qiagen, Valencia, Calif.). Sequencing was done using the Sanger dideoxyterminator strategy with fluorescence dyes attached to the dideoxynucleotides, and the sequence determination was made by electrophoresis using an ABI 377 sequencer. A portion of the gag (939 bases), pol (834 bases), and gp120-gp41 junction (340 bases) was also sequenced and shown to be clade C with no evidence of recombination (Williamson et al., submitted).
gp120-V3 peptides.
Peptides corresponding to amino acid sequences in the V3 loop of clade C isolates were synthesized, purified, and analyzed by SynPep Corporation (Dublin, Calif.). The HIV-1IIIB V3 peptide was purchased from Sigma (Saint Louis, Mo.), whereas the HIV-1MN V3 peptide was purchased from American BioTechnologies, Inc. (Cambridge, Mass.). All peptides were >90% pure as judged by high-pressure liquid chromatography and mass spectrometry.
Neutralizing antibody assays.
Neutralization of primary isolates was measured in PBMC by using a reduction in p24 Gag antigen synthesis as described previously (9). Briefly, 500 50% tissue culture infective doses of virus were incubated with various dilutions of test samples (serum, monoclonal antibodies, and sCD4) in triplicate for 1 h at 37°C in 96-well U-bottom culture plates. PHA-PBMC were added and incubated for one day. The cells were then washed three times with growth medium and resuspended in 200 μl of fresh growth medium. Culture supernatants (25 μl) were collected twice daily thereafter and mixed with 225 μl of 0.5% Triton X-100. The 25 μl of culture fluid removed each day was replaced with an equal volume of fresh growth medium. Concentrations of p24 Gag antigen in Triton X-100 lysates were measured in an antigen capture ELISA as described by the supplier (DuPont/NEN Life Sciences, Boston, Mass.). Concentrations of p24 in virus control wells (virus plus cells but no test serum) were determined for each harvest day. Concentrations in all remaining wells were determined for a harvest day that corresponded to a time when p24 production in virus control wells was in an early linear phase of increase that exceeded 3 ng/ml, which is when optimum sensitivity is achieved in this assay (87). The limit of detection in the p24 ELISA was 0.1 ng of p24/ml. Neutralization titers are given as the reciprocal of the minimum serum dilution (calculated prior to the addition of cells) that reduced p24 synthesis by 80% relative to a negative control serum sample from a healthy, HIV-1-negative individual.
Neutralization assay for TCLA strains were performed in either MT-2 cells (HIV-1IIIB and HIV-1MN) or CEMx174 cells (HIV-1SF2) by using neutral red to quantify the percentage of cells that survived virus-induced killing (54). Briefly, 500 50% tissue culture infective doses of virus were incubated with multiple dilutions of serum samples in triplicate for 1 h at 37°C in 96-well flat-bottom culture plates. Cells were added and the incubation continued until most but not all of the cells in virus control wells (cells plus virus but no serum sample) were involved in syncytium formation (usually 4 to 6 days). Cell viability was quantified by neutral red uptake as described (54). Neutralization titers are defined as the reciprocal serum dilution (before the addition of cells) at which 50% of cells were protected from virus-induced killing. A 50% reduction in cell killing corresponds to an approximate 90% reduction in p24 Gag antigen synthesis in this assay (9). Each set of assays included a positive control serum that had been assayed multiple times and had a known average titer.
V3-specific neutralizing antibodies were assessed by incubating diluted serum samples (diluted with an equal volume of phosphate-buffered saline, pH 7.4) for 1 h at 37°C in the presence and absence of V3 peptide (50 μg/ml). Titers of neutralizing antibodies were then determined in either the PBMC assay (in the case of primary isolates) or the MT-2 cell assay (in the case of HIV-1IIIB and HIV-1MN) as described above.
ELISA.
V3 peptide-specific binding antibodies were assessed by ELISA in Nunc (Roskilde, Denmark) Immuno plates (MaxiSorb F96) using alkaline phosphatase-conjugated, goat anti-monkey IgG as described (19). Plasma samples were assayed in duplicate at a 1:50 dilution, and values are given as the average absorbance at a wavelength of 405 nm.
Statistical analyses.
Differences in positive response rates (i.e., positive neutralization at any serum dilution) between genetic subtypes of the virus for a given serum sample were tested for statistical significance by using the two-sided Fisher's exact test. Differences in mean log neutralizing antibody titers between genetic subtypes of the virus for a given serum sample were tested by using the two-sided Wilcoxon rank sum test. For overall differences in either the positive response rates or mean log neutralizing antibody titers between genetic subtypes of the virus, pairwise results were pooled as a sample and tested for significance by using the nonparametric Wilcoxon sign rank test. All differences were considered significant if P was ≤0.05. Analyses of overall differences included only heterologous virus-serum pairs. This was done to eliminate bias, since only the Du samples contained autologous pairs.
RESULTS
Neutralization of clade C primary HIV-1 isolates with sCD4 and monoclonal antibodies.
Primary isolates from eight subjects in South Africa and eight subjects in Malawi were characterized in neutralization assays with sCD4 and three broadly neutralizing human monoclonal antibodies. These clade C isolates all exhibited a high level of resistance to inhibition by sCD4 (Table 1). Of the three monoclonal antibodies tested, IgG1b12 was most effective, neutralizing five of eight isolates from South Africa and four of eight isolates from Malawi at doses in the range of <1.8 to 50 μg/ml. Monoclonal antibody 2F5 neutralized two isolates from Malawi but none from South Africa. A third monoclonal antibody, 2G12, failed to neutralize all 16 clade C isolates tested. The broad resistance of clade C primary isolates to neutralization by 2G12 was in striking contrast to the ability of this same preparation of 2G12 to neutralize six of eight clade B isolates tested previously (9). Control assays performed with TCLA strains and primary isolates belonging to clade B confirmed no loss of activity of the sCD4 and monoclonal antibodies used in these experiments (data not shown).
TABLE 1.
Neutralization sensitivity of HIV-1 clade C isolates as measured with sCD4 and human monoclonal antibodies
| Reagent | ID80 (μg/ml) for isolatea
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| South African
|
Malawian
|
||||||||||||||||
| Du123 | Du151 | Du156 | Du172 | Du174 | Du179 | Du368 | Du422 | S080 | S021 | S009 | S018 | S007 | S180 | S017 | S103 | ||
| rsCD4 | >50 | >50 | >50 | >50 | >50 | 41 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | |
| 2G12 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | |
| IgG1b12 | 16 | 50 | >50 | 29 | >50 | >50 | 30 | 23 | 33 | >50 | 50 | 2 | >50 | >50 | >50 | <2 | |
| 2F5 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 | 49 | >50 | >50 | >50 | >50 | >50 | 7 | |
Values below each virus strain are the minimum concentration of sCD4 and monoclonal antibodies 2G12, IgG1b12, and 2F5 required to achieve an 80% reduction in p24 synthesis in PBMC (ID80). Positive neutralization is shown in boldface type.
Neutralizing antibody response following infection with clade C HIV-1.
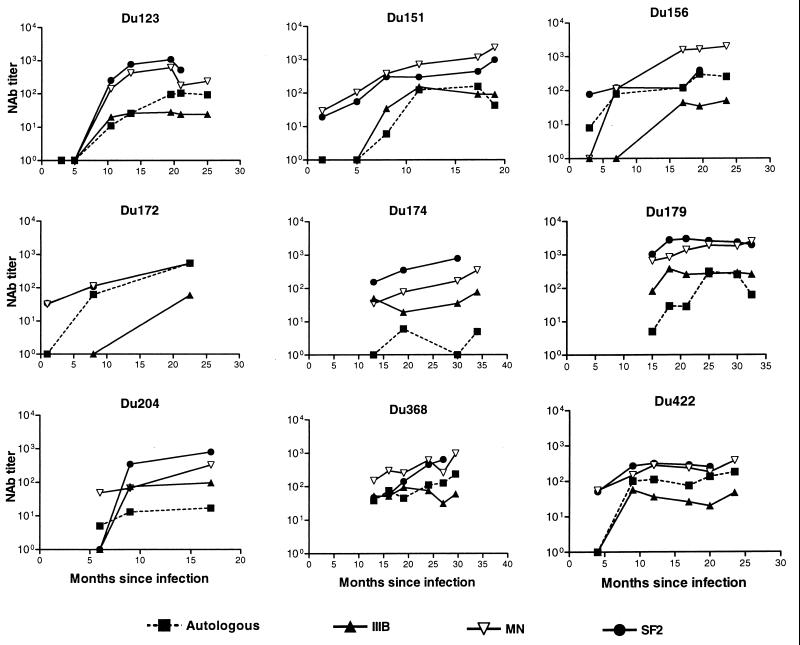
Serum samples collected at multiple time points from nine South African sex workers were assayed for neutralizing activity against the autologous virus isolate and three TCLA strains of clade B HIV-1. All subjects developed a neutralizing antibody response against their own virus (Fig. 1). The magnitude of this response varied between individuals and was relatively potent in most cases (titers of >100 in seven of nine cases). Although the timing of serum collection did not permit a refined assessment of the temporal response, it is accurate to say that autologous neutralizing antibodies were undetectable for at least the first 4 to 5 months of infection in three subjects (Du123, Du151, and Du422). Despite this delay, all three subjects eventually mounted a potent autologous neutralizing antibody response. Autologous neutralizing antibodies were low or undetectable at early time points and later rose in magnitude for subjects Du123, Du151, Du156, Du172, and Du422 as evidence that these five study subjects were in an early stage of seroconversion at the time of their enrollment. Autologous neutralizing antibodies in these five subjects were assessed with virus that was obtained shortly after infection. Autologous neutralizing antibodies in the remaining cases were measured with virus that was obtained either before (Du368) or after (Du174, Du179, and Du204) the time of first serum collection (an estimated 6 to 15 months from initial ELISA positivity). Of these latter cases, virus from subject Du368 was neutralized potently by all serum samples collected 5.5 months later and thereafter. Virus isolated at month 21 from subject Du179 was neutralized weakly by serum samples obtained at month 15 and was neutralized potently by serum samples obtained at month 25 and thereafter. Virus from subject Du174, isolated at month 19.5, was neutralized weakly or not at all by serum collected at months 13, 18, 30, and 34. A second study subject, Du204, had a similar weak autologous neutralizing antibody response.
FIG. 1.
Neutralizing antibody response over time in infected individuals in South Africa. Neutralizing antibodies were measured in PBMC with the early autologous isolate and in either MT-2 or CEMx174 cells with the clade B TCLA strains IIIB (MT-2), MN (MT-2), and SF2 (CEMx174). Results that were negative (neutralization titers of <4 for autologous isolates and <20 for IIIB, MN, and SF2) were assigned a value of 1 for representation.
TCLA strains belonging to clade B HIV-1 were highly sensitive to neutralization by the clade C serum samples. Overall neutralization-sensitivity with clade C serum samples was HIV-1MN ≅ HIV-1SF2 > HIV-1IIIB (Fig. 1). Neutralizing antibodies were sometimes detected with clade B TCLA strains before being detected with the autologous clade C isolate. Moreover, the potency of neutralization detected with TCLA strains usually exceeded the potency detected with the autologous isolate.
Two additional observations are worth noting. First, as mentioned above, sera from 5 subjects were capable of neutralizing the autologous isolate obtained prior to serum collection (Du123, 10.5 months; Du151, 8 months; Du156, 3 months; Du172, 8 months; Du422, 9 months). Autologous isolates in these cases were obtained 1 to 3 months after initial seroconversion and, as such, could closely resemble the transmitted variant that drove the initial antibody response, leading to antibodies that neutralized the early variant. Two subjects enrolled later in infection (Du179 and Du368) had detectable autologous neutralizing antibodies that increased in magnitude over the observation period. Autologous isolates in these two cases were obtained 21 and 7.5 months after infection and therefore could be variants that escaped an earlier neutralizing antibody response. In this event, the neutralizing antibodies we detected might have arisen as a de novo antibody response to escape epitopes. Although earlier isolates were not available to test this possibility, a later isolate from subject Du179 (36.5 months) was sensitive to neutralization (titer >108) by serum samples collected at months 25, 30, and 32.5, suggesting that the neutralization determinants were not evolving at a fast pace.
Antibody response to the V3 loop of gp120.
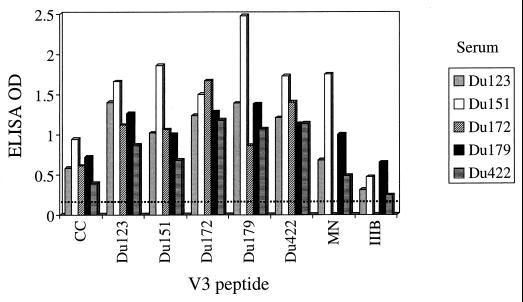
V3-specific antibodies were assessed by ELISA and in competitive neutralization assays with peptides from five different clade C isolates from South Africa (Du123, Du151, Du172, Du179, and Du422), the clade C consensus sequence (37), and the clade B TCLA strains HIV-1MN and HIV-1IIIB. The amino acid sequence of each V3 peptide is shown in Table 2. Sera from South African subjects reacted strongly with the autologous peptide and exhibited broad cross-reactivity with heterologous clade C peptides (Fig. 2). Reactivity was somewhat diminished in the case of the consensus clade C peptide and more so in the case of the IIIB peptide. Reactivity to the HIV-1MN peptide was variable, being relatively strong for subjects Du151 and Du179 and low to moderate for subjects Du123 and Du422.
TABLE 2.
Amino acid sequences of V3 peptides
| HIV-1 isolate | Amino acid sequencea |
|---|---|
| CC | TRPNNNTRKSIRI--GPGQTFYATGDIIGDIRQ |
| Du123 | IRPNNNTRKSIRI--GPGQTFYATNDIIGDIRQ |
| Du151 | TRPNNNTRKSIRI--GPGQTFYATDAIIGNIRE |
| Du172 | TRPSNNTRKSVRI--GPGQTFFATGDIIGDIRQ |
| Du179 | TRPGNNTRKSIRI--GPGQAFY-TNHIIGDIRQ |
| Du422 | TRPNNNTRKSVRI--GPGQTFYATGAIIGDIRE |
| MN | NKRKRIHI--GPGRAFYTTKNIIGTI |
| IIIB | NNNTRKSIRIQRGPGRAFVTIGK-IG |
Amino acids that differ from the consensus clade C sequence (CC) are shown in boldface type
FIG. 2.
ELISA reactivity to V3 peptides. Serum samples (1:50 dilution) were evaluated by ELISA for the presence of antibodies that could bind individual peptides derived from the V3 loop of the indicated virus strains. The amino acid sequence of each peptide is shown in Table 2. The dotted line represents the cutoff value for positive reactivity (twice the optical density [OD] of a control serum sample from a healthy, HIV-1-negative individual). The following serum samples were obtained after the estimated number of months of infection: Du123 (13.5 months), Du151 (11.3 months), Du172 (8 months), Du179 (25 months), Du422 (17 months). CC, clade C consensus sequence.
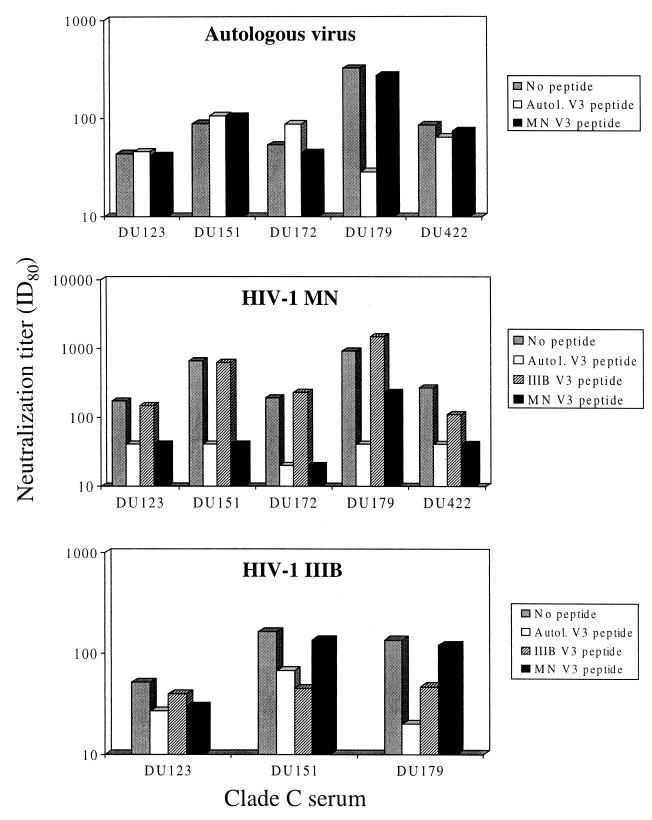
To determine whether these V3-specific antibodies had neutralizing activity, serum samples were preincubated with V3 peptides and then assayed for neutralizing activity against the autologous virus and two TCLA strains (Fig. 3). A major fraction of autologous neutralizing activity in serum Du179 was blocked by the matched Du179 V3 peptide, suggesting the presence of neutralizing antibodies directed against the V3 loop of this virus. As a control, the HIV-1MN peptide had no effect on the autologous neutralizing activity of this serum sample. Matched V3 peptides and the HIV-1MN V3 peptide had no effect on the autologous neutralizing activity of four other clade C serum samples. By comparison, the HIV-1MN-specific neutralizing activity of all five serum samples was dramatically reduced in the presence of matched V3 peptides and the HIV-1MN V3 peptide but not the HIV-1IIIB V3 peptide. In addition, the HIV-1IIIB-specific neutralizing activity was reduced in the presence of matching V3 peptide and the HIV-1IIIB V3 peptide but not the HIV-1MN V3 peptide. We note that the reduction in HIV-1IIIB-specific neutralization was minor in one of three cases examined (Du123). Due to sample availability and low neutralization titers against HIV-1IIIB, the remaining two samples could not be tested in these competition assays with HIV-1IIIB.
FIG. 3.
Ability of V3 peptides to out-compete the neutralizing activity of clade C serum samples from South Africa. Serum samples shown in Fig. 1 were evaluated for neutralizing activity in the presence and absence of gp120 V3 peptides (50 μg/ml) as described in Materials and Methods. Autologous (Autol.) V3 peptide refers to a peptide bearing the amino acid sequence of the virus from the subject who was the source of serum listed on the x axis. The top panel shows autologous virus-serum combinations, the middle panel shows clade C serum samples assayed with the HIV-1MN virus, and the bottom panel shows clade C serum samples assayed with the HIV-1IIIB virus. ID80, 80% inhibitory dose.
Analysis of intra- and interclade neutralization.
A checkerboard analysis was performed with clade B and C virus isolates and serum samples. Clade C serum samples were selected from the South African sex workers at a time when neutralizing activity was detected with the autologous virus and TCLA strains. Clade B serum samples were obtained from individuals in North Carolina who had documented HIV-1 infection for ≥2 years. These same clade B serum samples were assayed recently against the clade B viruses in our panel (9), and therefore, those results were used in our comparative analyses. We caution that a potential source of error in our checkerboard assays is the lack of sequence information to confirm the genetic subtype of virus from our presumed clade B serum donors from North Carolina. These subjects were presumed to be infected with clade B based on the high incidence of clade B in North America. We also note that our partial genetic and serologic characterization of the clade C isolates from Malawi does not entirely eliminate the possibility that these are recombinant viruses.
Cross-neutralization within and between genetic subtypes was readily apparent (Table 3). The clade C isolates appeared to be more sensitive to neutralization overall compared to clade B isolates. Specifically, the Du isolates were neutralized 70% of the time (excluding autologous virus-serum combinations) by Du serum samples, and 54% of the time by clade B serum samples. Moreover, the Malawi isolates were neutralized 48% of the time by Du serum samples. By comparison, clade B isolates were neutralized 25% of the time by Du serum samples (Table 3) and 27% of the time by clade B serum samples (9). Overall trends were analyzed statistically based on positive neutralization frequencies and mean neutralization titers. Only the positive response rates between Du and clade B isolates tested with Du serum samples were found to be statistically significant (Table 4). We note that no significant differences were identified based on mean neutralization titers (Table 4).
TABLE 3.
Checkerboard analysis of clade B and C neutralization
| ID80 fora:
|
||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| South African clade C isolates
|
Clade B isolates
|
Malawian clade C isolates
|
||||||||||||||||||||||||
| Serum | Cladeb | Du123 | Du151 | Du156 | Du172 | Du174 | Du179 | Du368 | Du422 | P15 | P27 | QH0515 | QH0692 | 1168 | 1196 | PVO | TRO | S080 | S021 | S009 | S018 | S007 | S180 | S017 | S103 | |
| T88580 | B | <4 | <4 | <4 | 7 | 15 | <4 | <4 | <4 | |||||||||||||||||
| F22934 | B | <4 | 4 | <4 | <4 | <4 | 60 | <4 | <4 | |||||||||||||||||
| JAF-01 | B | 4 | 4 | <4 | 7 | <4 | 7 | <4 | 5 | |||||||||||||||||
| NLS-02 | B | 8 | 5 | 4 | <4 | <4 | 9 | 5 | 8 | |||||||||||||||||
| LEH-03 | B | 16 | 16 | 12 | 4 | <4 | 21 | 20 | 6 | |||||||||||||||||
| SCE-06 | B | 4 | 5 | <4 | <4 | <4 | <4 | 6 | 7 | |||||||||||||||||
| Du123 | C | (26) | 4 | <4 | 5 | 4 | <4 | 4 | 4 | <4 | 7 | <4 | <4 | <4 | 4 | <4 | <4 | <4 | 5 | <4 | 4 | <4 | <4 | <4 | 12 | |
| Du151 | C | 15 | (123) | 7 | 6 | 64 | 6 | 11 | 11 | <4 | 27 | 5 | <4 | 20 | <4 | <4 | <4 | <4 | <4 | <4 | 10 | <4 | <4 | <4 | 46 | |
| Du156 | C | <4 | <4 | <4 | 7 | <4 | <4 | <4 | 17 | |||||||||||||||||
| Du172 | C | <4 | <4 | 6 | (62) | 55 | <4 | 4 | <4 | |||||||||||||||||
| Du179 | C | 22 | 7 | 20 | 20 | 44 | (310) | 11 | 14 | 18 | 42 | <4 | <4 | 6 | 108 | 6 | 26 | 5 | 12 | 6 | 18 | 5 | 4 | 5 | 55 | |
| Du204 | C | 5 | 6 | <4 | <4 | 64 | <4 | 8 | 6 | <4 | <4 | <4 | <4 | <4 | <4 | <4 | <4 | |||||||||
| Du368 | C | 17 | <4 | <4 | 6 | 4 | 4 | (45) | 4 | <4 | <4 | <4 | <4 | <4 | 16 | <4 | <4 | 6 | 5 | <4 | 5 | <4 | <4 | <4 | 37 | |
| Du422 | C | <4 | 12 | <4 | <4 | 8 | <4 | 8 | (112) | <4 | <4 | <4 | <4 | <4 | <4 | <4 | <4 | 6 | <4 | <4 | 7 | 5 | <4 | <4 | 39 | |
Values below each virus isolate are the reciprocal serum dilution at which p24 synthesis was reduced 80% relative to a control serum sample from an HIV-1-negative individual (ID80). Positive neutralization is shown in boldface type. Values in parentheses are matched virus-serum combinations.
Clade B serum samples were from HIV-1-infected individuals in North Carolina. Clade C serum samples were from HIV-1-infected subjects in South Africa. These serum samples were obtained after the estimated number of months of infection: Du123 (13.5 months), Du151 (11.3 months), Du172 (8 months), Du179 (25 months), Du204 (9 months), Du368 (19 months), Du422 (12 months).
TABLE 4.
Statistical analysis of checkerboard results
| Seruma | Virusesa |
P derived fromb:
|
|
|---|---|---|---|
| Positive response rate | Mean NAb titer | ||
| Du | Du vs B | 0.03 | 0.31 |
| B | Du vs B | 0.13 | 0.13 |
| Du | Mw vs B | 0.19 | 0.50 |
| Du | Du vs Mw | 0.25 | 0.13 |
Du, clade C from South Africa; B, clade B; Mw, clade C from Malawi.
Overall differences in either the positive neutralization response rate or mean log neutralizing antibody (NAb) titers between genetic subtypes of the virus were tested for significance by using the nonparametric Wilcoxon sign rank test. Differences were considered significant if P was ≤0.05. To eliminate bias, results of autologous serum-virus pairs in the Du samples were excluded from analysis, since this was the only group where autologous pairs were present.
A number of clade-specific reactivities were observed within the overall analysis: (i) isolate Du174 was neutralized preferentially by clade C sera (P < 0.01 based on response rate), (ii) serum Du151 exhibited a preferential neutralization of clade C isolates from South Africa (P = 0.01 based on response rate; P = 0.02 based on mean neutralization titer), (iii) serum NLS-02 exhibited a preferential neutralization of clade C isolates from South Africa (P = 0.01 by both criteria), and (iv) serum Du204 exhibited a preferential neutralization of clade C isolated from South Africa (P ≤ 0.05 based on mean neutralization titer). Notably, one serum sample from the Du cohort (Du179) neutralized 16 of 16 clade C isolates and six of eight clade B isolates (Table 3).
A broader assessment of cross-neutralizing activity with 50 heterologous primary HIV-1 isolates was performed on a small subset of serum samples for which an adequate supply of serum was available. These particular serum samples had potent neutralizing activity against the early autologous clade C isolate (titer > 100) and were able to neutralize TCLA strains belonging to clade B. They were assayed at a 1:20 dilution with the 50 primary isolates to be selective for relatively potent heterologous neutralization specificities. The heterologous primary isolates consisted of the 8 clade B and 16 clade C isolates listed in Table 3 plus an additional 26 clade B isolates. The results are summarized in Table 5 and do not include autologous virus-serum combinations. Serum Du151 neutralized 40% of the isolates as evidence that this serum sample contains broadly cross-reactive neutralizing antibodies. Serum samples Du156 and Du368 were much less cross-reactive but nonetheless neutralized 7 to 17% of isolates. No clade-specific preferences were observed with these two serum samples (P = 1.00).
TABLE 5.
Cross-neutralizing activity of serum samples from three subjects infected with clade C HIV-1
| Subject | No. (%) of HIV-1 isolates neutralized by 1:20-diluted seruma
|
||
|---|---|---|---|
| Clade B (n = 35) | Clade C (n = 15) | Totalb (n = 50) | |
| Du151 | 12 (34) | 8 (53) | 20 (40) |
| Du156 | 6 (17) | 2 (13) | 8 (16) |
| Du368 | 3 (9) | 1 (7) | 4 (8) |
Serum samples from subjects Du151 (17.3 months), Du156 (17 months), and Du368 (27 months) were assayed at a 1:20 dilution with 35 clade B and 15 heterologous clade C primary HIV-1 isolates. Neutralization was considered positive if p24 synthesis was reduced ≥80% relative to a negative control (serum from a healthy, HIV-1-negative individual). Values do not include autologous virus-serum combinations.
This is the fraction and percent of isolates neutralized for the 50 isolates total.
DISCUSSION
The neutralization properties of primary isolates and serum samples from clade C HIV-1-infected individuals were characterized and compared to clade B. Our goal was to develop standard virologic reagents and to acquire information that would benefit vaccine design. A major focus was placed on South Africa because of the high rates of HIV-1 transmission and the lack of information on neutralizing antibodies in that country. A particular emphasis was placed on a group of female sex workers located at multiple stops along a major trucking route between Durban and Johannesburg. These women were participating in HIV-1 prevention programs as part of a microbicide trial and were screened monthly for HIV-1 seroconversion. Virus isolates and serum samples were available in some cases very soon after infection, which made it possible to examine the autologous and heterologous neutralizing activity of the earliest antibodies generated.
All but one clade C isolate possessed the R5 biologic phenotype that is typical of transmitted variants. These isolates (eight from South Africa and eight from Malawi) resembled R5 clade B isolates in their sensitivity to neutralization by some but not all serologic reagents. Shared properties included a high level of resistance to inhibition by sCD4 and the frequent neutralization by monoclonal antibody IgG1b12. Resistance to sCD4 is a general property that distinguishes primary isolates from TCLA strains (20) and is thought to reflect the exposure of critical epitopes in and around the CD4 binding site of gp120 as affected by N-linked glycans and tertiary folds in the native gp120 molecule (85). It may be presumed that we preserved the sCD4-resistant phenotype of these clade C isolates by passing them a minimum number of times in PBMC exclusively (71). Many sCD4-resistant variants of HIV-1 are difficult to neutralize with monoclonal antibodies and sera from infected individuals. The IgG1b12 epitope, which overlaps the CD4 binding domain of gp120 (51), is an exception in that neutralization by this monoclonal antibody is not predicted by sCD4 sensitivity. For example, the 89.6 and 89.6P variants of simian-human immunodeficiency virus are both moderately sensitive to inhibition by sCD4 yet exhibit a striking dichotomy in their sensitivity to neutralization by IgG1b12 (19). The ability of IgG1b12 to neutralize many sCD4-resistant variants of clade C HIV-1 is additional evidence that this antibody specificity would be highly beneficial for broadly effective vaccination.
Another monoclonal antibody, 2F5, that is known to be effective against primary isolates was only able to neutralize 2 of 16 clade C isolates here. Since this low frequency was no different statistically from the three of eight clade B isolates neutralized by this same batch of 2F5 (9), the neutralizing activity of 2F5, like that of IgG1b12, does not appear to distinguish clade B from clade C. A much different outcome occurred with the 2G12 monoclonal antibody in that it failed to neutralize all 16 clade C isolates from South Africa and Malawi up to the highest dose tested (50 μg/ml). 2G12 has been shown to neutralize primary HIV-1 isolates from multiple clades (9, 23, 46, 75, 76), but very high doses were needed to neutralize the small number of clade C isolates tested in those studies (75, 86). We conclude that 2G12 has very poor neutralizing activity against clade C isolates relative to other clades. It seems unlikely that the neutralizing specificity of 2G12 will have an impact in regions where clade C is prevalent, except in cases where it is synergistic with other neutralizing antibody specificities (86).
Our analysis of sequential serum samples from a group of South African sex workers infected with clade C HIV-1 showed a delay in autologous neutralizing antibody production following primary infection of at least 4 to 5 months. That delay was similar to clade B infection (55, 63) and could be due to either early immunologic dysfunction, the poor immunogenicity of the envelope glycoproteins, or a combination of these factors. Following the delay, a potent autologous neutralizing antibody response was detected in most cases (titers > 100) as evidence that the B-cell response to critical epitopes eventually matured. Despite potent neutralizing antibodies, one individual (Du151) never gained control of her viremia and progressed to AIDS rapidly. We do not know the status of this person's HIV-1-specific cellular immune response, nor were we able to determine whether a neutralization escape variant arose that might explain the rapid progression.
In addition to neutralizing the autologous isolate, clade C serum samples from South Africa neutralized TCLA strains of clade B HIV-1 with a level of potency similar to clade B serum samples. This is evidence that the antibody specificities for TCLA HIV-1 neutralization are conserved between clades B and C. In support of this notion, antibodies in clade C serum samples bound peptides corresponding to the V3 loop of clade B and C gp120, which is a major target for the neutralization of clade B TCLA strains (5, 73, 77). Although the amino acid sequence of the V3 loop of clades B and C can be quite different (37, 64), they appear to share antigenic features recognized by monoclonal antibodies (33). This may explain why the neutralizing activity of clade C serum samples against clade B TCLA strains was sensitive to the presence of V3 loop peptides. The inability of V3 loop peptides to block autologous neutralization of clade C primary isolates in four of five cases examined, this despite the presence of antibodies to the autologous V3 peptide, suggests that certain epitopes in the V3 loop of clade C primary isolates are poorly antigenic relative to TCLA strains. A similar phenomenon has been described for clade B (5, 73, 77). This dichotomy in antigenicity is an indication that epitope exposure on the native envelope glycoprotein complex of clade C primary isolates will pose a challenge for neutralizing antibody induction by candidate HIV-1 vaccines much the same as it has for clade B (52, 85).
Primary isolates of clade B and C HIV-1 were further compared in checkerboard neutralization assays that differed from those in previous reports in several ways: (i) a large number of clade C isolates and serum samples were included, (ii) virus isolates were obtained early in infection, and (iii) the clade C serum samples corresponded to an early stage of neutralizing antibody production. In general, the earliest neutralizing antibodies to be detected in an HIV-1-infected individual have a limited range of specificity (14, 55). It was therefore unexpected to find 70% of heterologous clade C virus-serum combinations testing positive from South Africa. This high frequency is unusual and, with the exception of sera from some long-term nonprogressors (13, 63), has not be documented previously.
The extensive cross-neutralization within the South African serum samples and virus isolates may be an indication of shared neutralization determinants. This would be encouraging for vaccine efforts, as it suggests that neutralizing antibodies will have less epitope diversity to overcome at a regional level. The broad implications of this concept depend in part on the geographic range of the shared determinant that defines the corresponding neutralization serotypes. Less epitope diversity would be expected if multiple subjects in our cohort were infected by the same individual. Shared determinants might also arise as a consequence of serial transmission of selected isolates in these sex workers. Early evidence in South Africa suggested multiple introductions of the virus into the country, with the diversity observed being greater than one would expect from founder-type effects such as seen in Thailand (8, 79). Similarly, within this sex worker cohort, the analysis of the relationships between gag, pol, and env sequences showed relatively high diversity, and little phylogenetic clustering of sequences was detected (Williamson et al., submitted). Single- donor and serial transmission of selected isolates are therefore not obvious explanations for our results. However, full-length sequencing does show reliable clustering of two of the isolates (Du151 and Du422), suggesting that at least for these two isolates there was a common source in the not-too-distant past (80). Moreover, we cannot be certain that these female sex workers were not infected multiple times to give rise to a cross-reactive polyvalent neutralizing antibody response. Along this same line, it has been reported that initial virus populations are more heterogeneous in women than in men immediately following transmission (42). Additional studies will be needed to clarify the events that gave rise to the cross-neutralizing antibody responses in the South African subjects and how those events might impact vaccine development.
The 70% positive neutralization rate within the set of South African clade C viruses and serum samples differed significantly only from the overall ability of these same serum samples to neutralize clade B isolates. This case of clade-specific neutralization did not extend to other combinations in our checkerboard assays with clade B and C reagents, suggesting that clades B and C are comprised in part of overlapping neutralization serotypes. Evidence that the overlap was only partial comes from a number of individual cases of clade-specific neutralization. One example is a clade C isolate (Du174) that was neutralized preferentially by clade C serum samples. In addition, two South African clade C serum samples neutralized South African clade C isolates in a specific manner. Taken together with the selective neutralizing activity of 2G12, these examples reflect subtle associations between genetic subtype and neutralization serotype that have gone unnoticed in previous studies, owing in part to the complexity of the neutralization determinants on the virus and the limited number of clade C reagents used in the past. One unexpected finding was the preferential neutralization of clade C isolates by a presumed clade B serum sample. Unfortunately, a lack of sequence information on the virus from this individual makes it possible that infection was with a non-clade B virus. Nonetheless, this is an additional case of a serum that recognized a neutralization serotype found in clade C that was rarely detected in clade B. Our combined results are an indication that within each genetic subtype are multiple neutralization serotypes, some of which are more clade-specific than others.
During the course of our studies we discovered two individuals in South Africa (Du151 and Du179) whose serum after less than 2 years of infection neutralized a large number of clade B and C primary isolates. This cross-neutralizing activity was not unlike that of sera from some HIV-1-infected long-term nonprogressors (13, 63) and appears to be relatively unique for recent seroconverters. The cross-reactive neutralizing activity could be due to host factors, viral factors, or a combination of the two. It also serves to confirm that clades B and C consist in part of overlapping neutralization serotypes. The viral envelope glycoproteins from these two individuals are interesting candidates for vaccine development.
It seems possible that some neutralization serotypes track more with geography than with genetic subtype, as was suggested by our observations in a cohort of sex workers in South Africa infected with clade C HIV-1. If generally true, targeted immunogens might prove beneficial for vaccines in this and other parts of the world. The selection of immunogens for such tailored vaccines will require extensive surveys of virus isolates and serum samples. Alternatively, new approaches for cross-reactive neutralizing antibody induction are under investigation that may make tailoring efforts unnecessary (52). With few exceptions (e.g., 2G12 epitope), an immunogen that generates antibodies capable of neutralizing many clade B isolates may be expected to neutralize many clade C isolates and vice versa. Until such immunogens become available, efforts to induce neutralizing antibodies with candidate HIV-1 vaccines might benefit the most by including envelope glycoproteins from one or more strains of the virus that predominate in the specific regions targeted for vaccination. We add as a cautionary note that it is not clear at this time what level of neutralizing antibody induction and clinical benefit will be achieved with those envelope glycoproteins.
Acknowledgments
We thank Mary Phoswa and Melissa Kerkau for virus isolation and Tonie Cilliers for virus phenotyping. We also thank Myron S. Cohen for providing access to samples from Malawi, Dennis Burton for IgG1b12, and Herman Katinger and John Mascola for 2G12 and 2F5.
This work was supported by the U.S. National Institutes of Health (grants AI46705, P30-HD37260, and DK49381), the South African AIDS Vaccine Initiative, the Poliomyelitis Foundation, and the National Research Foundation of South Africa.
REFERENCES
- 1.Albert, J., B. Abrahamsson, K. Nagy, E. Aurelius, H. Gaines, G. Nystrom, and E. M. Fenyö. 1990. Rapid development of isolate-specific neutralizing antibodies after primary HIV-1 infection and consequent emergence of virus variants which resist neutralization by autologous sera. AIDS 4:107-112. [DOI] [PubMed] [Google Scholar]
- 2.Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955-1958. [DOI] [PubMed] [Google Scholar]
- 3.Arendrup, M., C. Nielsen, J.-E. S. Hanson, C. Pedersen, L. Mathiesen, and J. O. Nielsen. 1992. Autologous HIV-1 neutralizing antibodies: emergence of neutralization-resistant escape virus and subsequent development of escape virus neutralizing antibodies. J. Acquir. Immune Defic. Syndr. 5:303-307. [PubMed] [Google Scholar]
- 4.Betts, M. R., J. Krowka, C. Santamaria, K. Balsamo, F. Gao, G. Mulundu, C. Luo, N. N’Gandu, H. Sheppard, B. H. Hahn, S. Allen, and J. A. Frelinger. 1997. Cross-clade human immunodeficiency virus (HIV)-specific cytotoxic T-lymphocyte responses in HIV-infected Zambians. J. Virol. 71:8908-8911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bou-Habib, D. C., G. Roderiquez, T. Oravecz, P. W. Berman, P. Lusso, and M. A. Norcross. 1994. Cryptic nature of envelope V3 region epitopes protects primary monocytotropic human immunodeficiency virus type 1 from antibody neutralization. J. Virol. 68:6006-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradney, A. P., S. Scheer, J. M. Crawford, S. P. Buchbinder, and D. C. Montefiori. 1999. Neutralization escape in human immunodeficiency virus type 1-infected long-term nonprogressors. J. Infect. Dis. 179:1264-1267. [DOI] [PubMed] [Google Scholar]
- 7.Bredell, H., G. Hunt, B. Morgan, C. T. Tiemessen, D. J. Martin, and L. Morris. 2000. Identification of HIV-1 inter-subtype recombinants in South Africa using env and gag heteroduplex mobility assays. AIDS Res. Hum. Retrovir. 16:493-497. [DOI] [PubMed] [Google Scholar]
- 8.Bredell, H., C. Williamson, P. Sonnenberg, D. J. Martin, and L. Morris. 1998. Genetic characterization of HIV type 1 from migrant workers in three South African gold mines. AIDS Res. Hum. Retrovir. 14:677-684. [DOI] [PubMed] [Google Scholar]
- 9.Bures, R., A. Gaitan, T. Zhu, C. Graziosi, K. McGrath, J. Tartaglia, P. Caudrelier, R. E. L. Habib, M. Klein, A. Lazzarin, D. Stablein, M. Deers, L. Corey, M. L. Greenberg, D. H. Schwartz, and D. C. Montefiori. 2000. Immunization with recombinant canarypox vectors expressing membrane-anchored gp120 followed by gp160 protein boosting fails to generate antibodies that neutralize R5 primary isolates of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 16:2019-2035. [DOI] [PubMed] [Google Scholar]
- 10.Burton, D. R., and D. C. Montefiori. 1997. The antibody response in HIV-1 infection. AIDS 11(Suppl. A):S87-S98. [PubMed] [Google Scholar]
- 11.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. H. I. Parren, L. S. W. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, M. Lamacchia, E. Garratty, E. R. Stiehm, Y. J. Bryson, Y. Cao, J. P. Moore, D. D. Ho, and C. F. Barbas III. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 12.Cao, H., P. Kanki, J.-L. Sankalé, A. Dieng-Sarr, G. P. Mazzara, S. A. Kalams, B. Korber, S. Mboup, and B. D. Walker. 1997. Cytotoxic T-lymphocyte cross-reactivity among different human immunodeficiency virus type 1 clades: implications for vaccine development. J. Virol. 71:8615-8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao, Y., L. Qin, L. Zhang, J. Safrit, and D. D. Ho. 1995. Virologic and immunologic characterization of long-term survivors of human immunodeficiency virus type 1 infection. N. Engl. J. Med. 332:201-208. [DOI] [PubMed] [Google Scholar]
- 14.Carotenuto, P., D. Looij, L. Keldermans, F. de Wolf, and J. Goudsmit. 1998. Neutralizing antibodies are positively associated with CD4+ T-cell counts and T-cell function in long-term AIDS-free infection. AIDS 12:1591-1600. [DOI] [PubMed] [Google Scholar]
- 15.Cassol, S., B. G. Weniger, P. G. Babu, M. O. Salminen, X. Zheng, M. T. Htoon, A. Delaney, M. O'Shaughnessy, and C.-Y. Ou. 1996. Detection of HIV type 1 env ubtypes A, B, C, and E in Asia using dried blood spots: a new surveillance tool for molecular epidemiology. AIDS Res. Hum. Retrovir. 12:1435-1441. [DOI] [PubMed] [Google Scholar]
- 16.Cecilia, D., S. S. Kulkarni, S. P. Tripathy, R. R. Gangakhedkar, R. S. Paranjape, and D. A. Gadkari. 2000. Absence of coreceptor switch with disease progression in human immunodeficiency virus infections in India. Virology 271:253-258. [DOI] [PubMed] [Google Scholar]
- 17.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 18.Choe, H., M. Farzan, Y. Sun, N. Sullivan, B. Rollins, P. D. Ponath, L. Wu, C. R. Mackay, G. LaRosa, W. Newman, N. Gerard, C. Gerard, and J. Sodroski. 1996. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135-1148. [DOI] [PubMed] [Google Scholar]
- 19.Crawford, J. M., P. L. Earl, B. Moss, K. A. Reimann, M. S. Wyand, K. H. Manson, M. Bilska, J. T. Zhou, C. D. Pauza, P. W. H. I. Parren, D. R. Burton, J. G. Sodroski, N. L. Letvin, and D. C. Montefiori. 1999. Characterization of primary isolate-like variants of simian-human immunodeficiency virus. J. Virol. 73:10199-10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daar, E. S., X. L. Li, T. Moudgil, and D. D. Ho. 1990. High concentrations of recombinant soluble CD4 are required to neutralize primary human immunodeficiency virus type 1 isolates. Proc. Natl. Acad. Sci. USA 87:6574-6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 22.Dragic, T., V. Litwin, G. P. Allaway, S. R. Martin, Y. Huang, K. A. Nagashima, C. Cayanan, P. J. Maddon, R. A. Koup, J. P. Moore, and W. A. Paxton. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667-673. [DOI] [PubMed] [Google Scholar]
- 23.D'Souza, M. P., D. Livnat, J. A. Bradac, and S. H. Bridges. 1997. Evaluation of monoclonal antibodies to human immunodeficiency virus type 1 primary isolates by neutralization assays: performance criteria for selecting candidate antibodies for clinical trials. J. Infect. Dis. 175:1056-1062. [DOI] [PubMed] [Google Scholar]
- 24.Earl, P. L., R. W. Doms, and B. Moss. 1990. Oligomeric structure of the human immunodeficiency virus type 1 envelope glycoprotein. Proc. Natl. Acad. Sci. USA 87:648-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engelbrecht, S., I. Koulinska, T. L. Smith, J. Barreto, and E. J. van Rensburg. 1998. Variation in HIV type 1 V3 region env sequences from Mozambique. AIDS Res. Hum. Retrovir. 14:803-805. [DOI] [PubMed] [Google Scholar]
- 26.Engelbrecht, S., T. L. Smith, P. Kasper, E. Faatz, M. Zeier, D. Moodley, C. G. Clay, and E. J. van Rensburg. 1999. HIV type 1 V3 domain serotyping and genotyping in Gauteng, Mpumalanga, KwaZulu-Natal, and Western Cape Provinces of South Africa. AIDS Res. Hum. Retrovir. 15:325-328. [DOI] [PubMed] [Google Scholar]
- 27.Esparza, J., and N. Bhamarapravati. 2000. Accelerating the development and future availability of HIV-1 vaccines: why, when, where and how? Lancet 355:2061-2066. [DOI] [PubMed] [Google Scholar]
- 28.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 29.Ferrari, G., W. Humphrey, M. J. McElrath, J.-L. Excler, A.-M. Duliege, M. L. Clements, L. C. Corey, Dani P. Bolognesi, and K. J. Weinhold. 1997. Clade B-based HIV-1 vaccines elicit cross-clade cytotoxic T lymphocyte reactivities in uninfected volunteers. Proc. Natl. Acad. Sci. USA 94:1396-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freed, E. O., E. L. Delwart, G. L. Buchschacher, Jr., and A. T. Panganiban. 1992. A mutation in the human immunodeficiency virus type 1 transmembrane glycoprotein gp41 dominantly interferes with fusion and infectivity. Proc. Natl. Acad. Sci. USA 89:70-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gadkari, D. A., D. Moore, H. W. Sheppard, S. S. Kulkarni, S. M. Mehendale, and R. C. Bollinger. 1998. Transmission of genetically diverse strains of HIV-1 in Puna, India. Indian J. Med. Res. 107:1-9. [PubMed] [Google Scholar]
- 32.Gao, F., D. L. Robertson, S. G. Morrison, H. Hui, S. Craig, J. Decker, P. N. Fultz, M. Girard, G. M. Shaw, B. H. Hahn, and P. M. Sharp. 1996. The heterosexual human immunodeficiency virus type 1 epidemic in Thailand is caused by an intersubtype (A/E) recombinant of African origin. J. Virol. 70:7013-7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorny, M. K., T. C. VanCott, C. Hioe, Z. R. Israel, N. L. Michael, A. J. Conley, C. Williams, J. A. Kessler, I. I., P. Chigurupati, S. Burda, and S. Zolla-Pazner. 1997. Human monoclonal antibodies to the V3 loop of HIV-1 with intra- and interclade cross-reactivity. J. Immunol. 159:5114-5122. [PubMed] [Google Scholar]
- 34.Graham, B. S. 1994. Serologic responses to candidate AIDS vaccines. AIDS Res. Hum. Retrovir. 10:S145-S148. [PubMed] [Google Scholar]
- 35.Harada, S., Y. Koyanagi, and N. Yamamoto. 1985. Infection of HTLV-III/LAV in HTLV-I-carrying cells MT-2 and MT-4 and application in a plaque assay. Science 229:563-566. [DOI] [PubMed] [Google Scholar]
- 36.Kessler, J. A., II, P. M. McKenna, E. A. Emini, C. P. Chan, M. D. Patel, S. K. Gupta, G. E. Mark III, C. F. Barbas III, D. R. Burton, and A. J. Conley. 1997. The recombinant human monoclonal antibody IgG1b12 neutralizes diverse human immunodeficiency virus type 1 primary isolates. AIDS Res. Hum. Retrovir. 13:575-582. [DOI] [PubMed] [Google Scholar]
- 37.Korber, B., B. Hahn, B. Foley, J. W. Mellors, T. Leitner, G. Meyers, F. McCutchen, and C. L. Kuiken. 1997. Human Retroviruses and AIDS 1997: a compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 38.Kostrikis, L. G., Y. Cao, H. Ngai, J. P. Moore, and D. D. Ho. 1996. Quantitative analysis of serum neutralization of human immunodeficiency virus type 1 from subtypes A, B, C, D, E, F, and I: lack of direct correlation between neutralization serotypes and genetic subtypes and evidence for prevalent serum-dependent infectivity enhancement. J. Virol. 70:445-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kowalski, M., J. Potz, L. Basiripour, T. Dorfman, W. C. Goh, E. Terwilliger, A. Dayton, C. Rosen, W. Haseltine, and J. Sodroski. 1987. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science 237:1351-1355. [DOI] [PubMed] [Google Scholar]
- 40.Lasky, L. A., G. Nakamura, D. H. Smith, C. Fennie, C. Shimasaki, E. Patzer, P. Berman, T. Gregory, and D. J. Capon. 1987. Delineation of a region of the human immunodeficiency virus gp120 glycoprotein critical for interaction with the CD4 receptor. Cell 50:975-985. [DOI] [PubMed] [Google Scholar]
- 41.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Long, E. M., H. L. Martin, Jr., J. K. Kreiss, S. M. J. Rainwater, L. Lavreys, D. J. Jackson, J. Rakwar, K. Mandaliya, and J. Overbaugh. 2000. Gender differences in HIV-1 diversity at the time of infection. Nat. Med. 6:71-75. [DOI] [PubMed] [Google Scholar]
- 43.Lu, M., S. C. Blacklow, and P. S. Kim. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 2:1075-1082. [DOI] [PubMed] [Google Scholar]
- 44.Mascola, J. R., M. G. Lewis, G. Stiegler, D. Harris, T. C. VanCott, D. Hayes, M. K. Louder, C. R. Brown, C. V. Sapan, S. S. Frankel, Y. Lu, M. L. Robb, H. Katinger, and D. L. Birx. 1999. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J. Virol. 73:4009-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mascola, J. R., M. K. Louder, S. R. Surman, T. C. VanCott, X. F. Yu, J. Bradac, K. R. Porter, K. E. Nelson, M. Girard, J. G. McNeil, F. E. McCutchan, D. L. Birx, and D. S. Burke. 1996. Human immunodeficiency virus type 1 neutralizing antibody serotyping using serum pools and an infectivity reduction assay. AIDS Res. Hum. Retrovir. 12:1319-1328. [DOI] [PubMed] [Google Scholar]
- 46.Mascola, J. R., M. K. Louder, T. C. VanCott, C. V. Sapan, J. S. Lambert, L. R. Muenz, B. Bunow, D. L. Birx, and M. L. Robb. 1997. Potent and synergistic neutralization of human immunodeficiency virus (HIV) type 1 primary isolates by hyperimmune anti-HIV immunoglobulin combined with monoclonal antibodies 2F5 and 2G12. J. Virol. 71:7198-7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mascola, J. R., J. Louwagie, F. E. McCutchan, C. L. Fischer, P. A. Hegerich, K. F. Wagner, A. K. Fowler, J. G. McNeil, and D. S. Burke. 1994. Two antigenically distinct subtypes of human immunodeficiency virus type 1: viral genotype predicts neutralization serotype. J. Infect. Dis. 169:48-54. [DOI] [PubMed] [Google Scholar]
- 48.Mascola, J. R., S. W. Snyder, O. S. Weislow, S. M. Belay, R. B. Belshe, D. H. Schwartz, M. L. Clements, R. Dolin, B. S. Graham, G. J. Gorse, M. C. Keefer, M. J. McElrath, M. C. Walker, K. F. Wagner, J. G. McNeil, F. E. McCutchan, and D. S. Burke. 1996. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. J. Infect. Dis. 173:340-348. [DOI] [PubMed] [Google Scholar]
- 49.Mascola, J. R., G. Stiegler, T. C. VanCott, H. Katinger, C. B. Carpenter, C. E. Hanson, H. Beary, D. Hayes, S. S. Frankel, D. L. Birx, and M. G. Lewis. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 6:207-210. [DOI] [PubMed] [Google Scholar]
- 50.McCutchan, F. E., M. O. Salminen, J. K. Carr, and D. S. Burke. 1996. HIV genetic diversity. AIDS 10(Suppl. 3):S13-S20. [PubMed] [Google Scholar]
- 51.Mo, H., L. Stamatatos, J. E. Ip, C. F. Barbas, P. W. H. I. Parren, D. R. Burton, J. P. Moore, and D. D. Ho. 1997. Human immunodeficiency virus type 1 mutants that escape neutralization by human monoclonal antibody IgG1b12. J. Virol. 71:6869-6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Montefiori, D. C., and T. G. Evans. 1999. Toward an HIV-1 vaccine that generates potent, broadly cross-reactive neutralizing antibodies. AIDS Res. Hum. Retrovir. 15:689-698. [DOI] [PubMed] [Google Scholar]
- 53.Montefiori, D. C., G. Pantaleo, L. M. Fink, J. T. Zhou, J. Y. Zhou, M. Bilska, G. D. Miralles, and A. S. Fauci. 1996. Neutralizing and infection-enhancing antibody responses to human immunodeficiency virus type 1 in long-term nonprogressors. J. Infect. Dis. 173:60-67. [DOI] [PubMed] [Google Scholar]
- 54.Montefiori, D. C., W. E. Robinson, Jr., S. S. Schuffman, and W. M. Mitchell. 1988. Evaluation of antiviral drugs and neutralizing antibodies to human immunodeficiency virus by a rapid and sensitive microtiter infection assay. J. Clin. Microbiol. 26:231-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moog, C., H. J. A. Fleury, I. Pellegrin, A. Kirn, and A. M. Aubertin. 1997. Autologous and heterologous neutralizing antibody responses following initial seroconversion in human immunodeficiency virus type 1-infected individuals. J. Virol. 71:3734-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moore, J. P., Y. Cao, J. Leu, L. Qin, B. Korber, and D. D. Ho. 1996. Inter- and intraclade neutralization of human immunodeficiency virus type 1: genetic clades do not correspond to neutralization serotypes but partially correspond to gp120 antigenic serotypes. J. Virol. 70:427-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morris, L., T. Cilliers, M. Phoswa, and D. J. Martin. 2001. CCR5 is the major coreceptor used by HIV-1 subtype C isolates from patients with active tuberculosis. AIDS Res. Hum. Retrovir. 17:697-701. [DOI] [PubMed] [Google Scholar]
- 58.Mulligan, M. J., and J. Weber. 1999. Human trials of HIV-1 vaccines. AIDS 13(Suppl. A):S105-S112. [PubMed] [Google Scholar]
- 59.Muster, T. F. Steindl, M. Purtscher, A. Trkola, A. Klima, G. Himmler, F. Rüker, and H. Katinger. 1993. A conserved neutralization epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67:6642-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Novitsky, V. A., M. A. Montano, M. F. McLane, B. Renjifo, F. Vannberg, B. T. Foley, T. P. Ndung'u, M. Rahman, M. J. Makhema, R. Marlink, and M. Essex. 1999. Molecular cloning and phylogenetic analysis of human immunodeficiency virus type 1 subtype C: a set of 23 full-length clones from Botswana. J. Virol. 73:4427-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nyambi, P. N., J. Nkengasong, P. Lewi, K. Andries, W. Janssens, K. Fransen, L. Heyndrickx, P. Piot, and G. van der Groen. 1996. Multivariate analysis of human immunodeficiency virus type 1 neutralization data. J. Virol. 70:6235-6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parren, P. W. H. I., J. P. Moore, D. R. Burton, and Q. J. Sattentau. 1999. The neutralizing antibody response to HIV-1: viral evasion and escape from humoral immunity. AIDS 13(Suppl. A):S137-S162. [PubMed] [Google Scholar]
- 63.Pilgrim, A. K., G. Pantaleo, O. J. Cohen, L. M. Fink, J. Y. Zhou, J. T. Zhou, D. P. Bolognesi, A. S. Fauci, and D. C. Montefiori. 1997. Neutralizing antibody responses to human immunodeficiency virus type 1 in primary infection and long-term non-progressive infection. J. Infect. Dis. 176:924-932. [DOI] [PubMed] [Google Scholar]
- 64.Ping, L.-H., J. A. E. Nelson, I. F. Hoffman, J. Schock, S. L. Lamers, M. Goodman, P. Vernazza, P. Kazembe, M. Maida, D. Zimba, M. M. Goodenow, J. J. Eron, Jr., S. A. Fiscus, M. S. Cohen, and R. Swanstrom. 1999. Characterization of V3 sequence heterogeneity in subtype C human immunodeficiency virus type 1 isolates from Malawi: underrepresentation of X4 variants. J. Virol. 73:6271-6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Poignard, P., P. J. Klasse, and Q. J. Sattentau. 1996. Antibody neutralization of HIV-1. Immunol. Today 17:239-246. [DOI] [PubMed] [Google Scholar]
- 66.Purtscher, M., A. Trkola, A. Grassauer, P. M. Schultz, A. Klima, S. Döpper, G. Gruber, A. Buchacher, T. Muster, and H. Katinger. 1996. Restricted antigenic variability of the epitope recognized by the neutralizing gp41 antibody 2F5. AIDS 10:587-593. [DOI] [PubMed] [Google Scholar]
- 67.Ramjee, G., S. A. Karim, and W. Sturm. 1998. Prevalence of sexually transmitted infections among sex workers in the Kwazulu-Natal midlands, South Africa. Sex. Transm. Dis. 25:346-349. [DOI] [PubMed] [Google Scholar]
- 68.Reitter, J. N., R. E. Means, and R. C. Desrosiers. 1998. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 4:679-684. [DOI] [PubMed] [Google Scholar]
- 69.Robertson, D. L., J. P. Anderson, J. A. Bradac, J. K. Carr, B. Foley, R. K. Funkhouser, F. Gao, B. H. Hahn, M. L. Kalish, C. Kuiken, G. H. Learn, T. Leitner, F. McCutchan, S. Osmanov, M. Peeters, D. Pieniazek, M. Salminen, P. M. Sharp, S. Wolinski, and B. Korber. 1999. HIV-1 nomenclature proposal—a reference guide to HIV-1 classification, p. 492-505. In C. L. Kuiken,B. Foley, B. Hahn, B. Korber, F. McCutchan, P. A. Marx, J. W. Mellors, J. I. Mullins, J. Sodroski, and S. Wolinski (ed.), Human retroviruses and AIDS 1999: a compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 70.Salter, R. D., D. N. Howell, and P. Cresswell. 1985. Gene regulating HLA class I antigen expression in T-B lymphoblast hybrids. Immunogenetics 21:235-246. [DOI] [PubMed] [Google Scholar]
- 71.Sawyer, L. S. W., M. T. Wrin, L. Crawford-Miksza, B. Potts, Y. Wu, P. A. Weber, R. D. Alfonso, and C. V. Hanson. 1994. Neutralization sensitivity of human immunodeficiency virus type 1 is determined in part by the cell in which the virus is propagated. J. Virol. 68:1342-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schønning, K., B. Jansson, S. Olofsson, J. O. Neilsen, and J.-E. S. Hansen. 1996. Resistance to V3-directed neutralization caused by an N-linked oligosaccharide depends on the quaternary structure of the HIV-1 envelope oligomer. Virology 218:134-140. [DOI] [PubMed] [Google Scholar]
- 73.Spenlehauer, C., S. Saragosti, H. J. A. Fleury, A. Kirn, A.-M. Aubertin, and C. Moog. 1998. Study of the V3 loop as a target epitope for antibodies involved in the neutralization of primary isolates versus T-cell-line-adapted strains of human immunodeficiency virus type 1. J. Virol. 72:9855-9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng-Meyer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184-187. [DOI] [PubMed] [Google Scholar]
- 75.Trkola, A., A. B. Pomales, H. Yuan, B. Korber, P. J. Maddon, G. P. Allaway, H. Katinger, C. F. Barbas III, Dennis R. Burton, D. D. Ho, and J. P. Moore. 1995. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J. Virol. 69:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Trkola. A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vancott, T. C., V. R. Polonis, L. D. Loomis, N. L. Michael, P. L. Nara, and D. L. Birx. 1995. Differential role of V3-specific antibodies in neutralization assays involving primary and laboratory-adapted isolates of HIV type 1. AIDS Res. Hum. Retrovir. 11:1379-1390. [DOI] [PubMed] [Google Scholar]
- 78.van Damme, L., C. Chandeying, G. Ramjee, H. Rees, P. Sirivongrangson, M. Laga, and J. Perriens. 2000. Safety of multiple daily applications of COL-1492, a nonoxynol-9 vaginal gel, among female sex workers. AIDS 14:85-88. [DOI] [PubMed] [Google Scholar]
- 79.van Harmelen, J., R. Wood, M. Lambrick, E. P. Rybicki, A. L. Williamson, and C. Williamson. 1997. An association between HIV-1 subtypes and mode of transmission in Cape Town, South Africa. AIDS 11:81-87. [DOI] [PubMed] [Google Scholar]
- 80.van Harmelen, J., C. Williamson, B. Kim, L. Morris, J. Carr, S. Abdool Karim, and F. McCutchan. 2001. Characterization of the full-length HIV type 1 subtype C sequences from South Africa. AIDS Res. Hum. Retrovir. 17:1527-1531. [DOI] [PubMed]
- 81.van Harmelen, J. H., E. van der Ryst, A. S. Loubser, D. York, S. Madurai, S. Lyons, R. Wood, and C. Williamson. 1999. A predominantly HIV type 1 subtype C-restricted epidemic in South African urban populations. AIDS Res. Hum. Retrovir. 15:395-398. [DOI] [PubMed] [Google Scholar]
- 82.Weber, J., E.-M. Fenyö, S. Beddows, P. Kaleebu, Å. Björndal, and the WHO Network for HIV Isolation and Characterization. 1996. Neutralization serotypes of human immunodeficiency virus type 1 field isolates are not predicted by genetic subtype. J. Virol. 70:7827-7832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 84.Wu, L., N. P. Gerard, R. Wyatt, H. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. A. Cardose, E. Desjardins, W. Newman, C. Gerard, and J. Sodroski. 1996. CD4-induced interaction or primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179-183. [DOI] [PubMed] [Google Scholar]
- 85.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 86.Xu, W., B. A. Smith-Franklin, P.-L. Li, C. Wood, J. He, Q. Du, G. J. Bhat, C. Kankasa, H. Katinger, L. A. Cavacini, M. R. Posner, D. R. Burton, T.-C. Chou, and R. M. Ruprecht. 2001. Potent neutralization of primary human immunodeficiency virus clade C isolates with a synergistic combination of human monoclonal antibodies raised against clade B. J. Hum. Virol. 4:55-61. [PubMed] [Google Scholar]
- 87.Zhou, J. Y., and D. C. Montefiori. 1997. Antibody-mediated neutralization of primary isolates of human immunodeficiency virus type 1 in peripheral blood mononuclear cells is not affected by the initial activation state of the cells. J. Virol. 71:2512-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]