Abstract
A time course analysis of SL3-3 murine leukemia virus (SL3) infection in thymus and bone marrow of NIH/Swiss mice was performed to assess changes that occur during the early stages of progression to lymphoma. Virus was detectable in thymocytes, bone marrow, and spleen as early as 1 to 2 weeks postinoculation (p.i.). In bone marrow, virus infection was detected predominantly in immature myeloid or granulocytic cells. Flow cytometry revealed significant reductions of the Ter-119+ and Mac-1+ populations, and significant expansions of the Gr-1+ and CD34+ populations, between 2 and 4 weeks p.i. Analysis of colony-forming potential confirmed these findings. In the thymus, SL3 replication was associated with significant disruption in thymocyte subpopulation distribution between 4 and 7 weeks p.i. A significant thymic regression was observed just prior to the clonal outgrowth of tumor cells. Proviral long terminal repeats (LTRs) with increasing numbers of enhancer repeats were observed to accumulate exclusively in the thymus during the first 8 weeks p.i. Observations were compared to the early stages of infection with a virtually nonpathogenic SL3 mutant, termed SL3ΔMyb5, which was shown by real-time PCR to be replication competent. Comparison of SL3 with SL3ΔMyb5 implicated certain premalignant changes in tumorigenesis, including (i) increased proportions of Gr-1+ and CD34+ bone marrow progenitors, (ii) a significant increase in the proportion of CD4− CD8− thymocytes, (iii) thymic regression prior to tumor outgrowth, and (iv) accumulation of LTR enhancer variants. A model in which disrupted bone marrow hematopoiesis and thymopoiesis contribute to the development of lymphoma in the SL3-infected animal is discussed.
The murine leukemia virus (MuLV)-mediated induction of lymphoma involves pathophysiologic changes that occur early in infection and function to establish a preleukemic state favorable for tumor development. These preleukemic changes involve the target organ for tumor formation but are not limited to that tissue. The early stages of MuLV-mediated lymphoma are perhaps best understood in the case of infection with Moloney MuLV (M-MuLV). When inoculated into neonatal mice, M-MuLV reliably induces lymphomas of T-cell origin, most of which originate in the thymus and exhibit the surface phenotype of immature T cells (CD4− CD8− or CD4+ CD8+) (8, 9). Some of the tumors exhibit a more mature surface phenotype (CD4+ CD8− or CD4− CD8+), an observation which has been interpreted to suggest that the virus transforms an immature T-cell target that continues to differentiate to various degrees thereafter (29). During the premalignant stage of M-MuLV-mediated lymphomagenesis, a distinctive hyperplasia of the spleen occurs. The hyperplasia leads to a predictable splenomegaly characterized by two- to threefold enlargement by 4 to 8 weeks after infection (12). Early infection of the bone marrow is also essential for efficient lymphomagenesis mediated by M-MuLV (4). In fact, hyperplasia of the spleen is thought to result secondarily from an inhibition of bone marrow hematopoiesis at early times after viral infection; thus, the splenic hyperplasia is apparently a compensatory extramedullary hematopoiesis that plays an integral role in the malignant process (15, 32). A hallmark of M-MuLV-mediated disease is the appearance of polytropic env gene recombinants with the ability to utilize a cellular receptor different from that of the ecotropic parent virus. These polytropic recombinants are known as mink cell focus-inducing (MCF) viruses owing to their altered host range (15). Although MCF viruses are not absolutely required for M-MuLV to induce disease (49, 53), efficient lymphomagenesis in mice is clearly associated with their generation. Finally, the long terminal repeat (LTR) sequence of M-MuLV has been implicated as a primary determinant of the T-cell specificity and potency of disease induction. The role of the LTR in disease induction may be to increase virus replication in the target tissue for transformation, thus increasing the opportunity for insertional mutagenesis of a relevant proto-oncogene. Approximately 80% of T-cell lymphomas induced by M-MuLV in NIH/Swiss mice show proviral insertions near the proto-oncogene pim-1, c-myc, or the linked pvt-1 region (15).
Preleukemia or prelymphoma has also been studied in the AKR mouse. Early disease development during spontaneous tumorigenesis or following exogenous infection has received considerable attention (see, e.g., references 11, 19, 39, 40, 56, and 57). SL3-3 (SL3) is an oncogenic, ecotropic MuLV derived from a spontaneous lymphoma of an AKR mouse. SL3 infection reliably induces T-cell lymphoma, typically of thymic origin, approximately 70 days following inoculation of neonatal mice (31, 41). Although less is known about the mechanisms of disease induction in SL3-infected animals, some similarities to M-MuLV infection are apparent. For example, SL3-induced tumors are invariably of T-cell origin and exhibit a range of surface phenotypes represented most prominently by the features of immature cells (21). Equally clear are distinct differences in SL3-induced disease compared to M-MuLV, suggesting that the mechanisms of lymphomagenesis may be different. For example, splenic hyperplasia is not characteristic of the early phase of SL3-induced disease. In our recent time course analysis of infection with SL3 or with SL3 LTR variants, spleen weights were observed to increase normally with mouse development until the appearance of frank lymphomas. Only at that time were significantly enlarged spleens evident (35). Thus, the compensatory extramedullary hematopoiesis and consequent splenic hyperplasia characteristic of early M-MuLV infection (15) apparently do not occur during SL3 infection. Further, our studies and others show that viruses containing polytropic recombinant genomes arise during SL3 infection (44, 51, 52); however, no infectious viruses can be isolated from tumor cells that exhibit a polytropic host range (25, 51, 52). Finally, although the pattern of proto-oncogene activation in SL3-induced tumors has not yet been thoroughly elucidated, it is clearly distinct from that of M-MuLV. Insertions at c-myc and pim-1 account for only about 20% of SL3-induced tumors (1, 22, 37, 47). Novel genes have been identified as targets in some tumors (48), and other, yet-unidentified proto-oncogenes may be activated by proviral insertion in the majority of the tumors. This difference in proto-oncogene involvement may be reflective of a distinct pathogenic mechanism.
A major determinant of the malignant potential and T-cell specificity of SL3 is encoded in the LTR. Evidence implicates the transcriptional enhancers within the SL3 LTR as determinants of disease progression, likely through their ability to drive expression of viral and cellular genes at the sites of proviral integration (7, 14, 18, 30, 33-35, 38). A three-nucleotide substitution that eliminates the c-Myb binding site in the SL3 enhancer was previously shown to render the virus virtually nonpathogenic. This enhancer mutant, termed SL3ΔMyb5, was shown to induce lymphoma in only 3 of 11 animals after prolonged latency of 230 days. No lymphomas were detected within the 100-day period that is typically examined. In addition, SL3ΔMyb5 was shown to be a very poor activator of the c-myc promoter in T cells (38). SL3ΔMyb5 was subsequently shown to be replication competent in NIH/Swiss mice, as evidenced by the detection of proviral DNA in bone marrow, spleen, and thymus collected from 1 to 8 weeks postinoculation (p.i.) (44). As in SL3 infection, polytropic recombinant viral genomes were detected during SL3ΔMyb5 infection of NIH/Swiss mice, although recombinants were generated less frequently and with delayed timing of appearance (44). In the present study, SL3ΔMyb5 was used as a tool to define molecular and cellular alterations during the premalignant period of infection that may be linked to oncogenesis.
During the premalignant period, SL3 replication occurs in bone marrow and spleen as well as in thymus, although thymus is the ultimate site of tumor formation (43). The pathophysiologic impact of SL3 replication on bone marrow hematopoiesis and/or thymopoiesis is not well understood. Bone marrow may represent the site of the initial transforming event, since transplantable potential leukemia cells (PLC) are consistently found in bone marrow at 3 to 4 weeks following SL3 inoculation. Transplanted PLC can give rise to lymphoma in the recipient animal, in a manner dependent on the presence of the thymus (20, 50). In the thymus, SL3 infection in AKR mice has been associated with alterations in thymocyte subset distribution at 6 to 8 weeks p.i. and with the appearance of immature thymocytes that fail to mature normally upon transplantation (23, 24). In the present study, the impact of SL3 replication on hematopoiesis and thymopoiesis was examined during the early preleukemic period, defined as 1 to 8 weeks p.i., and was compared to SL3ΔMyb5 infection in age-matched animals. The results show that hematopoiesis and thymopoiesis are significantly disrupted by virus replication. Comparison of SL3 and SL3ΔMyb5 infection during the early weeks after inoculation implicates certain premalignant changes as being important to the tumorigenic process. Those changes include (i) significant increases in the proportions of Gr-1+ and CD34+ progenitors in the bone marrow early in infection, (ii) a significant increase in the proportion of CD4− CD8− thymocytes, (iii) thymic regression prior to tumor outgrowth, and (iv) accumulation of LTR enhancer variants.
MATERIALS AND METHODS
Virus stocks, animal inoculations, and tissue collection.
Stocks of infectious SL3 or SL3ΔMyb5 were developed by concatamerization of a molecularly cloned, single-LTR SL3 or SL3ΔMyb provirus and introduction into NIH 3T3 cells by lipid-mediated transfection (Lipofectamine Plus reagent; Life Technologies, Inc.). The culture supernatant of infected cells was collected, filtered through a low-protein-binding cellulose acetate filter (0.2-μm pore size; Costar), aliquoted, and frozen at −80°C for storage. The titer of the virus stock was determined by end point dilution on SC-1 cells. Newborn NIH/Swiss mice (<24 h of age) were inoculated intraperitoneally with 106 infectious units (0.1 ml) of virus. Mice were sacrificed by carbon dioxide asphyxiation at weekly intervals following inoculation. Age-matched uninfected control mice were included in parallel. Thymus, bone marrow, spleen, liver, and kidney were removed at timed collections as indicated in Results. For preparation of a thymocyte suspension, the thymus was weighed and minced in cold phosphate-buffered saline (PBS) (containing, per liter, 6.12 g of NaCl, 1.5 g of Na2HPO4, and 0.4 g of KH2PO4) by using surgical scissors. A single-cell thymocyte suspension was prepared by repeated passage of the minced tissue through an 18-gauge needle. Stromal components were removed by filtration of the suspension through a sterile 0.6-μm nylon mesh (Spectrum). Viable thymocytes were enumerated by trypan blue exclusion.
Preparation of genomic DNA and PCR amplification for proviral DNA.
Genomic DNA was isolated for PCR amplification using the QiaAMP Tissue Kit (Qiagen) and was quantified spectrophotometrically. DNA (1 μg) was amplified by PCR using primers SL3A (5" TCATAAGGCTTAGCCAGCTAACTGCAG 3") and SL3C (5" AACCTTGAGACAGTTTCTGG 3") to detect the integrated proviral LTR of SL3 or SL3ΔMyb5 as previously described (3). The 346-bp amplification product was resolved by agarose gel electrophoresis and visualized by ethidium bromide staining. As a control for the presence of amplifiable DNA, parallel reactions were conducted using primers for the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene as previously described (3).
Real-time PCR analysis.
Real-time PCR amplifications were performed according to specifications provided by Perkin-Elmer (PE) Applied Biosystems. Each reaction mixture contained 100 ng of genomic DNA, 1× Taqman Universal PCR Master Mix (PE Applied Biosystems), 900 nM (each) SL3for1 and SL3rev1 primers, and 250 nM sequence-specific 6-carboxy-fluorescein (5")/6-carboxy-tetramethyl-rhodamine (3")-labeled probe designed using the Primer Express software (PE Applied Biosystems). The sequences of the primers and probe are as follows: SL3for1, 5"-CTGCAGTAACGCCATTTTGC; SL3rev1, 5"-TGTACTTCCTTGTTCTTGTTTTTCTGA; and probe, 5"-FAM-AACATCAGCTCTGGTATTTTTCCCATGCCT-TAMRA-3". The primers amplify a 79-bp fragment from the SL3 or SL3ΔMyb5 LTR. Amplifications were performed and analyzed in a 96-well plate format using the ABI Prism 7700 DNA Sequence Detector. The data from this analysis are reported as critical threshold (CT) values, which represent the amplification cycle at which a threshold signal is first detected. The CT value was extrapolated to indicate a precise mass amount of target sequence by comparison to parallel amplification of a standard dilution series. The standard curve was generated by adding known amounts of pSL3LTR plasmid to thymus or bone marrow DNA from uninfected NIH/Swiss mice. In the present analysis, the slopes of the standard curves ranged from −3.1 to −3.6, and the threshold was set to 0.4. The measurement of each sample was repeated in triplicate.
Flow cytometry.
Thymocytes (1 × 106 to 2 × 106 cells per assay) or bone marrow cells (0.5 × 106 to 1 × 106 cells per assay) were suspended in 100 μl of standard azide buffer (containing, per liter, 10 g of FA Bacto buffer [Difco], 1 g of sodium azide, and 10 ml of heat-inactivated fetal bovine serum). For staining of surface antigens, antibody was added at a 1/40 dilution of the total reaction volume and allowed to incubate with the cells on ice for 20 min. The cells were then washed and resuspended in 100 μl of standard azide buffer with 100 μl of 2% paraformaldehyde. The antibodies used were as follows: fluorescein isothiocyanate (FITC)-conjugated rat anti-mouse CD4 monoclonal antibody (RM4-4; BD Pharmingen), phycoerythrin (PE)-conjugated rat anti-mouse CD8a monoclonal antibody (53-6.7; BD Pharmingen), FITC-conjugated goat anti-mouse immunoglobulin G (IgG) monoclonal antibody (F-0257; Sigma), PE-conjugated anti-Mac-1 (Caltag), PE-conjugated anti-Gr-1 (Caltag), PE-conjugated anti-Ter-119 (BD Pharmingen), PE-conjugated anti-CD34 (Caltag), and supernatant from the 24-8 hybridoma (a gift of Miles Cloyd). The 24-8 hybridoma produces a monoclonal antibody of the IgG2a isotype specific for gp70-p15E polyprotein precursor of the AKR family of viruses (42). Isotype controls were included for each antibody used. Surface labeling for CD4 and CD8 was analyzed on a Becton Dickinson FacsVantage dual-laser cell sorter and interpreted with Becton Dickinson Cell Quest software. An EPICS ELITE flow cytometer and EPICS ELITE software was used to analyze surface labeling of SL3 SU and for lineage-restricted antigens on bone marrow cells. Forward- and side-scatter characteristics were used to measure cell size and density, respectively, for the purpose of gating out cell debris. Electronic compensation for spectral overlap of the fluorochromes was performed using single-color staining of controls prepared from experimental material.
Immunohistochemical staining for virus infection in bone marrow cells.
Bone marrow was recovered by flushing 500 μl of cold PBS through the femur into a microcentrifuge tube. The bone marrow was mixed with an equal volume of Cytospin Collection Fluid (Shandon) and was deposited onto gelatin-coated glass slides by cytocentrifugation for 5 min at 1,000 rpm (Cytospin; Shandon). Slides were air dried for several hours before fixation in 2% paraformaldehyde-PBS for 15 min at room temperature. Slides were rinsed in PBS for 5 min and stained using a polyclonal goat anti-AKR gp70 antiserum (National Cancer Institute [NCI] repository, no. 80S000008) according to the protocol supplied with the Vectastain Elite ABC kit (Vector Labs). The staining procedure was as follows: 30 min in 0.3% hydrogen peroxide in methanol, 5 min in PBS, 20 min in normal goat serum (NCI repository, no. 79S000703) diluted 1/1,000 in PBS, 5 min in PBS, 30 min in goat anti-AKR gp70 antiserum (NCI repository, no. 80S000008) diluted 1/1,000 in PBS, 5 min in PBS, 30 min in biotinylated anti-goat IgG (Vector Labs) diluted to 5 μg/ml in PBS, 5 min in PBS, 30 min in ABC reagent (avidin-biotinylated horseradish peroxidase complex), 5 min in PBS, and 2 to 10 min in diaminobenzidine tetrahydrochloride. After immunohistochemical staining, the slides were briefly counterstained in Harris' modified hematoxylin solution for 30 s, rinsed in tap water, and treated with 0.1% ammonium acetate for 30 s. The slides were dried and mounted with coverslips using Permount.
Bone marrow colony-forming assays.
Bone marrow cells were flushed from the femur under sterile conditions with Iscove's modified Dulbecco's medium containing 2% fetal bovine serum. Cells were enumerated by trypan blue exclusion and were deposited in 35-mm-diameter petri dishes at a concentration of 2 × 104 cells per 1.1 ml in Methocult 3434 (StemCell Technologies, Inc.) Colonies were enumerated by microscopic examination after 12 days of growth at 37°C in a humidified atmosphere containing 5% CO2. Duplicate plates were from prepared from the bone marrow of each animal, and four or five animals were examined at each timed collection. Age-matched uninfected control animals were examined in parallel. To amplify SL3 proviral DNA, representative colonies of the CFU-granulocyte-macrophage (CFU-GM), CFU-granulocyte-erythrocyte-macrophage-mekaryocyte (CFU-GEMM), and burst-forming unit-erythrocyte (BFU-E) types were aspirated from the plate with a fine-tipped Pasteur pipette. Cells were washed repeatedly in 1× Hanks balanced salt solution and were lysed for 10 min at 65°C in 5 μl of alkaline lysis buffer (50 mM dithiothreitol, 200 mM NaOH). The reaction was neutralized with 5 μl of neutralization buffer (0.8 M Tris-Cl [pH 8.3], 0.3 M KCl, 0.2 M HCl). Proviral LTR and GAPDH sequences were amplified from the resulting DNA as described above.
RESULTS
Virus replication in target hematopoietic tissues and the accumulation of viral enhancer variants.
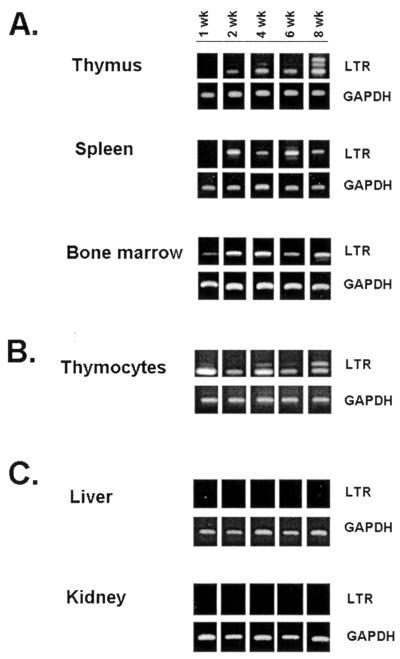
Previous studies have shown that bone marrow, spleen, and thymus are targets of SL3 infection at early times p.i. and that the number of infected cells in the thymus exceeds that in other tissues. In those studies, tissues were collected from SL3-infected NFS mice at 4 to 8 weeks p.i., and the number of infected cells was quantified using a serological focus assay (43). In the present study, neonatal NIH/Swiss mice were inoculated intraperitoneally with 106 infectious units of SL3 or SL3ΔMyb5 and were sacrificed at 1, 2, 4, 6, and 8 weeks p.i. Thymus, spleen, bone marrow, liver, and kidney were retrieved at each collection, and genomic DNA was isolated from the tissues. A single-cell suspension of thymocytes was also prepared at each timed collection. PCR amplification was used to detect integrated proviral DNA as evidence for virus replication in each tissue. The oligonucleotide primers used for amplification (SL3A and SL3C) were specific for the SL3 or SL3ΔMyb5 LTR and have been previously shown not to amplify endogenous viral sequences from NIH/Swiss mice (3). Thus, amplification with these primers is useful as an indicator of viral infection in target tissues. As shown in Fig. 1A, SL3 proviral DNA was readily detectable in the spleen, thymus, and bone marrow of SL3-infected animals as early as 1 to 2 weeks p.i. Isolated thymocytes were also positive for proviral DNA (Fig. 1B). Proviral DNA was not detected in nonhematopoietic tissues such as liver or kidney (Fig. 1C). Thus, SL3 replication is apparently restricted to hematopoietic target tissues, beginning as early as 1 to 2 weeks p.i. A similar pattern was observed in tissues from age-matched SL3ΔMyb5-infected animals (data not shown).
FIG. 1.
PCR amplification of SL3 proviral LTR or GAPDH sequences in tissues from virus-infected NIH/Swiss mice. Thymus, spleen, and bone marrow (A), isolated thymocytes (B), and liver and kidney (C) were collected at 1, 2, 4, 6, and 8 weeks after intraperitoneal inoculation of neonatal animals with SL3. Genomic DNA was isolated from the tissues, and 1 μg of the DNA was amplified with primers SL3A and SL3C to detect the presence of integrated SL3 provirus in the host genome. GAPDH sequences were examined as a control for the presence of amplifiable DNA. Representative results from one animal of four examined at each timed collection are shown.
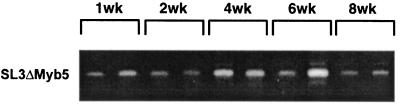
Previous studies of SL3 proviruses retrieved from end-stage tumors demonstrated frequent variation in the number of 72-bp enhancer repeats within the LTR. The formation of these enhancer variants, attributed to template misalignment errors during reverse transcription, may confer a selective replicative advantage to the virus in tumor cells. Alternatively, the accumulation of proviruses containing additional enhancer repeats may confer a selective growth advantage on the host cell, perhaps through optimal transcriptional activation of an adjacent oncogene (35, 37). PCR analysis of the premalignant thymus in SL3-infected animals demonstrated LTRs containing predominantly the predicted two enhancer repeats, as evidenced by the presence of a single 346-bp amplification product at 2, 4, and 6 weeks p.i. By 8 weeks p.i., larger amplification products were clearly evident in the thymus (Fig. 1A) and within thymocytes (Fig. 1B). Sequence analysis demonstrated these larger products to contain up to four copies of the enhancer repeat (data not shown). In an amplification product containing four enhancer repeats, the newly acquired sequences were identical except for a deletion of the core II binding site (data not shown). Interestingly, this site has been previously shown to be of minimal importance for pathogenicity of SL3 (18). Analysis of premalignant tissues further demonstrated the accumulation of enhancer repeats uniquely in the thymus and not in spleen or bone marrow (Fig. 1A). Thus, the selective advantage conferred by the accumulation of enhancer repeats may be effective only in thymocytes. Analysis of SL3ΔMyb5-infected animals showed that proviruses with increased numbers of enhancer repeats did not accumulate in the thymus during infection (Fig. 2), nor were they detectable in spleen or bone marrow of the same animals (data not shown). Thus, the accumulation of additional enhancer repeats is unique to SL3-infected mice and occurs exclusively in the thymuses of those animals.
FIG. 2.
PCR amplification of SL3ΔMyb5 proviral LTR from thymuses of virus-infected NIH/Swiss mice. Thymuses were collected at 1, 2, 4, 6, and 8 weeks after intraperitoneal inoculation of neonatal animals with SL3ΔMyb5. Genomic DNA was isolated from the tissues, and 1 μg of the DNA was amplified with primers SL3A and SL3C to detect the presence of integrated SL3ΔMyb5 provirus in the host genome. Representative results for two animals of four examined at each timed collection are shown.
Real-time PCR quantitation of SL3 and SL3ΔMyb5 replication during premalignancy.
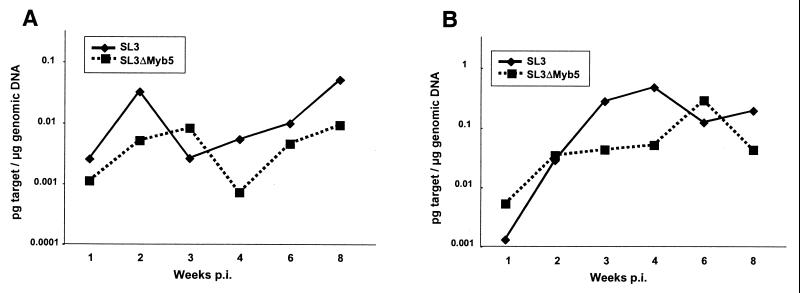
While the replication competence of SL3ΔMyb5 was demonstrated by PCR analysis of proviral DNA throughout the premalignant period in the thymus, bone marrow, and spleen of infected animals (Fig. 2) (44), the relative levels of SL3 and SL3ΔMyb5 replication were not quantified. We quantified the proviral load in thymus and bone marrow during the premalignant time period by using real-time PCR. Neonatal NIH/Swiss mice were inoculated intraperitoneally with 106 infectious units of wild-type SL3 or SL3ΔMyb5 and were sacrificed at 1, 2, 3, 4, 6, and 8 weeks p.i. Thymus and bone marrow were retrieved at each collection, and genomic DNA was prepared. Genomic DNA was amplified with primers SL3for1 and SL3rev1 in the presence of a sequence-specific Taqman FAM(5")/TAMRA(3")-labeled probe, using the ABI 7700 DNA Sequence Detector for real-time detection and analysis. Analysis of each sample was repeated in triplicate. A standard dilution series in which known amounts of SL3 plasmid DNA were added to DNA from uninfected tissues was amplified in parallel. By comparison to the standard curve thus generated, the CT of each amplification reaction was extrapolated to indicate the precise mass, expressed as picograms of 79-bp proviral LTR target sequence per microgram of total genomic DNA (Fig. 3; Table 1). The results showed that proviral loads in both SL3- and SL3ΔMyb5-infected mice increased during the first few weeks after inoculation, especially in the thymus. Proviral loads of SL3ΔMyb5 were generally lower than those of SL3 and increased more slowly. For example, when the data were transposed to indicate provirus copy number per cell equivalent (Table 1), the SL3-infected thymus was observed to contain 0.38, 6.5, and 1.7 copies per cell equivalent at 2, 4, and 6 weeks p.i., respectively. At the same time points, the SL3ΔMyb5-infected thymus was observed to contain 0.5, 0.7, and 3.9 copies per cell equivalent.
FIG. 3.
Quantification of SL3 or SL3ΔMyb5 viral load by real-time PCR analysis. A 79-bp LTR sequence was amplified from the bone marrow (A) or thymuses (B) of NIH/Swiss mice infected with wild-type SL3 or the SLΔMyb5 mutant. Genomic DNA was isolated from infected tissues at periodic intervals as indicated after neonatal inoculation. Genomic DNA (100 ng) from each tissue was amplified with primers SL3for1 and SL3rev1 in the presence of a sequence-specific FAM(5")/TAMRA(3")-labeled probe by using Taqman reagents (PE Applied Biosystems). A standard dilution series in which known amounts of SL3 plasmid DNA were added to DNA from uninfected tissues was amplified in parallel. By comparison to the standard curve thus generated, the CT of the amplification reaction from each test sample was extrapolated to indicate a precise mass amount of 79-bp proviral LTR target sequence. The data are expressed as picograms of 79-bp LTR target sequence per microgram of genomic DNA. Shown are the means of triplicate measurements from three or four animals at each timed collection. Raw data, including standard errors of the means, are shown in Table 1.
TABLE.
1. Proviral loads in tissues collected from SL3-infected or SL3ΔMyb5-infected mice during the premalignant time course
| Tissue | Virus | Wk p.i. | Proviral loada mean ± SEM | Proviruses per cell equivalentb |
|---|---|---|---|---|
| Bone marrow | SL3 | 1 | 2.54 × 10−3 ± 1.37 × 10−3 | 0.06 |
| 2 | 3.27 × 10−2 ± 3.23 × 10−2 | 0.65 | ||
| 3 | 2.60 × 10−3 ± 1.87 × 10−3 | 0.05 | ||
| 4 | 5.42 × 10−3 ± 2.34 × 10−3 | 0.1 | ||
| 6 | 9.98 × 10−3 ± 6.47 × 10−3 | 0.2 | ||
| 8 | 5.12 × 10−2 ± 2.49 × 10−2 | 1.0 | ||
| SL3ΔMyb5 | 1 | 1.12 × 10−3 ± 6.50 × 10−4 | 0.02 | |
| 2 | 5.13 × 10−3 ± 3.78 × 10−3 | 0.1 | ||
| 3 | 8.30 × 10−3 ± 2.48 × 10−3 | 0.15 | ||
| 4 | 7.23 × 10−4 ± 1.75 × 10−4 | 0.02 | ||
| 6 | 4.61 × 10−3 ± 3.95 × 10−3 | 0.09 | ||
| 8 | 9.21 × 10−3 ± 7.69 × 10−3 | 0.17 | ||
| Thymus | SL3 | 1 | 9.23 × 10−4 ± 4.31 × 10−4 | 0.02 |
| 2 | 1.99 × 10−2 ± 7.99 × 10−3 | 0.38 | ||
| 3 | 1.99 × 10−1 ± 6.63 × 10−2 | 3.8 | ||
| 4 | 3.40 × 10−1 ± 1.62 × 10−1 | 6.5 | ||
| 6 | 8.72 × 10−2 ± 3.68 × 10−2 | 1.7 | ||
| 8 | 1.39 × 10−1 ± 4.57 × 10−2 | 2.7 | ||
| SL3ΔMyb5 | 1 | 3.76 × 10−3 ± 2.75 × 10−3 | 0.07 | |
| 2 | 2.46 × 10−2 ± 1.66 × 10−2 | 0.5 | ||
| 3 | 3.06 × 10−2 ± 5.98 × 10−3 | 0.6 | ||
| 4 | 3.62 × 10−2 ± 2.24 × 10−2 | 0.7 | ||
| 6 | 2.05 × 10−1 ± 1.95 × 10−1 | 3.9 | ||
| 8 | 3.07 × 10−2 ± 3.07 × 10−2 | 0.6 |
Three or four animals were examined at each timed collection. Each sample was subjected to real-time PCR analysis in triplicate. For calculation of the mass amount of amplification target, a standard curve in which known amounts of SL3 plasmid DNA were added to DNA from uninfected NIH/Swiss bone marrow or thymus was amplified in parallel. Values are reported as picograms of 79-bp LTR target sequence per microgram of genomic DNA. Proviral loads in SL3-infected animals were statistically indistinguishable from those in SL3ΔMyb5-infected animals at each timed collection as determined by Student's t test, except at 4 weeks p.i. At that time, the SL3ΔMyb5 proviral load was significantly lower than the SL3 proviral load in the thymus (P < 0.05).
Calculated number of copies of proviral DNA per cell equivalent, based on the quantified mass amount of 79-bp LTR amplification target per microgram of genomic DNA and assuming a genome size of 3 × 109 bp and two LTRs per provirus.
Proportion of SU+ (gp70+) thymocytes in SL3- or SL3ΔMyb5-infected mice.
As another measure of relative replication rate, the proportion of SU+ (gp70+) thymocytes in SL3- or SL3ΔMyb5-infected mice was quantified throughout the premalignant time course by flow cytometry. Neonatal NIH/Swiss mice were inoculated with 106 infectious units of SL3 or SL3ΔMyb5, and the thymus was recovered at 2, 4, 6, and 8 weeks p.i. (from four to six animals at each timed collection). The thymus was minced, and a single-cell thymocyte suspension was prepared. The percentage of SL3 SU+ thymocytes was determined by flow cytometry using as a probe the supernatant of the 24-8 hybridoma. 24-8 produces a monoclonal antibody specific for the envelope precursor polyprotein of the AKR family of viruses (42). An FITC-conjugated secondary antibody, goat anti-mouse IgG2a, was used to detect the signal. By this measure, the percentage of SU+ cells in the thymuses of uninfected age-matched control mice was low or undetectable (average, 3.7% [data not shown]). At the earliest time point examined (2 weeks p.i.), SU was detectable on 14 to 23% of thymocytes in SL3- or SL3ΔMyb5-infected animals, increasing to ≈35% by 4 weeks p.i. (Table 2). By 8 weeks p.i. in most SL3-infected animals examined, the majority of thymocytes (51 to 82%) were SU+. Noteworthy among the infected animals examined at 8 weeks p.i. were two in which <10% of thymocytes expressed SU. Interestingly, the spleens of these animals were grossly enlarged at the time of sacrifice at 8 weeks. Southern blot analysis of genomic DNA from the spleen samples revealed clonal rearrangement of the T-cell receptor beta locus, evidence of T-cell tumor formation in that tissue (data not shown). In SL3ΔMyb5-infected mice, in contrast, the proportion of SU+ cells in the thymus declined to ≈17% by 8 weeks p.i. (Table 2). Taken together with the results of real-time PCR, these studies show that SL3ΔMyb5 is replication competent, although it replicates to lower levels than does SL3 in both bone marrow and thymus.
TABLE.
2. Percentage of thymocytes staining positive for viral SU protein (gp70) by flow cytometry
| Infecting virus | % Positive at wk p.i.a:
|
|||
|---|---|---|---|---|
| 2 | 4 | 6 | 8 | |
| SL3 | 6.5 | 63.3 | 50.2 | 82.2 |
| 14.6 | 57.7 | 19.4 | 77.6 | |
| 64.6 | 13 | 5.8 | 74.6 | |
| 17.8 | 27.2 | 15.5 | 6.9 | |
| 17.9 | 28.3 | 20.7 | 50.9 | |
| 16.9 | 20.6 | 49.7 | 3.6 | |
| Mean | 23.1 | 35 | 26.9 | 49.3 |
| SL3ΔMyb5 | 6.3 | 59.7 | 15.5 | 1 |
| 18.2 | 57.1 | 16.3 | 18 | |
| 15.9 | 2 | 14.6 | 19.9 | |
| 14.9 | 10.4 | 5.2 | 27.8 | |
| 15.9 | 19.2 | |||
| Mean | 14.2 | 32.3 | 14.2 | 16.7 |
Percentage of thymocytes expressing SL3 SU protein (gp70) in neonatally inoculated NIH/Swiss mice infected with SL3 or SL3ΔMyb5 and collected at 2, 4, 6, and 8 weeks p.i. Surface SU expression was assessed by flow cytometry using the 24-8 monoclonal antibody and an FITC-conjugated anti-mouse IgG2a secondary antibody. Four to six animals were examined at each timed collection. The percentage of SU+ thymocytes from each individual animal is shown, as is the mean for each timed collection. The percentage of SU+ cells in uninfected age-matched control mice was low or undetectable (average = 3.7%) (data not shown).
Virus replication in bone marrow progenitor cells.
Several lines of evidence indicate that early infection of the bone marrow plays an important role in SL3 pathogenesis. Our findings (Fig. 1) and those of others (43) show that SL3 replicates in bone marrow cells at early times after inoculation. Bone marrow may be the site of the initial transforming event, inasmuch as transplantation assays using SL3-infected AKR mice demonstrated cells with leukemic potential in bone marrow at 3 to 4 weeks p.i. (50). The target cells for virus replication in the bone marrow have not yet been identified, nor is the role of bone marrow in SL3 pathogenesis clearly understood. In the present study, we used an immunohistochemical approach to analyze replication of SL3 and SL3ΔMyb5 in bone marrow during the premalignant period. Neonatal NIH/Swiss mice were inoculated intraperitoneally with either virus, and bone marrow was collected from the femur at weekly intervals thereafter (from five to nine animals per timed collection). Immunohistochemical analysis was performed with the Vectastain Elite ABC Kit (Vector Labs), using as primary antibody a goat polyclonal antiserum directed against the SU protein of AKR-derived MuLV including SL3 (obtained from the NCI Repository). Following reaction with a biotinylated anti-goat IgG antiserum as secondary antibody and with avidin-peroxidase (Elite ABC reagent), the addition of diaminobenzidine tetrahydrochloride as a peroxidase substrate was used to produce a reddish-brown precipitate indicative of SU expression. The bone marrow preparation was then counterstained with hematoxylin to permit visualization of the cells.
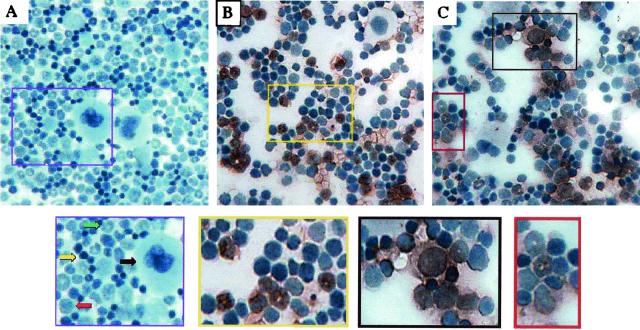
Although cells cannot be positively identified on the basis of morphology in these preparations, four populations are clearly evident in the bone marrow of uninfected animals (Fig. 4A): (i) very large cells with abundant cytoplasm and irregular nuclei, characteristic of megakaryocytes; (ii) medium-sized cells with large, often eccentric nuclei and relatively abundant cytoplasm, a morphology consistent with immature myeloid precursors; (iii) medium-sized cells with crescent- or ring-shaped nuclei typical of maturing cells of the granulocyte lineage; and (iv) small cells with scant cytoplasm and condensed nuclei. This morphology is characteristic of a heterogeneous population including myeloid precursors, erythroid precursors, and mature and maturing lymphocytes. In the bone marrow of SL3-infected animals, virus infection was localized by 3 weeks p.i. predominantly in maturing cells of the myeloid lineage. Brown staining was clearly evident over cells with distinctive ring-shaped nuclei characteristic of maturing granulocytes (Fig. 4B). At 5 weeks p.i., granulocytic cells remained infected, but infection was also evident in less mature cells of the myeloid lineage (data not shown). As in SL3 infection, SL3ΔMyb5 replication was detected predominantly in mature and maturing granulocytes and in less mature cells of the myeloid lineage (Fig. 4C).
FIG. 4.
Immunohistochemical staining for SL3 SU protein in bone marrow cells from uninfected NIH/Swiss mice (A) or from mice infected with SL3 (B) or SL3ΔMyb5 (C). Bone marrow was collected from the femur at 3 weeks p.i., deposited onto glass slides by cytocentrifugation, and stained immunohistochemically for SL3 SU (reddish-brown precipitate). Cells were then counterstained with hematoxylin. The enlarged image from panel A (purple box) shows four morphologically distinct types of cells in uninfected marrow: (i) very large cells with abundant cytoplasm and irregular nuclei, characteristic of megakaryocytes (black arrow); (ii) medium-sized cells with large, often eccentric nuclei and relatively abundant cytoplasm, a morphology consistent with immature myeloid precursors (red arrow); (iii) medium-sized cells with crescent- or ring-shaped nuclei typical of maturing cells of the granulocyte lineage (green arrow); and (iv) small cells with scant cytoplasm and condensed nuclei, characteristic of a heterogeneous population including myeloid precursors, erythroid precursors, and mature and maturing lymphocytes (yellow arrow). The enlarged image from panel B (yellow box) shows SL3 infection predominantly in immature and maturing granulocytes with ring-shaped nuclei. The enlarged images from panel C show SL3ΔMyb5 infection predominantly in immature myeloid precursors (black box) and maturing granulocytes (red box).
Alterations in bone marrow hematopoiesis in the early, premalignant stages of SL3 or SL3ΔMyb5 infection.
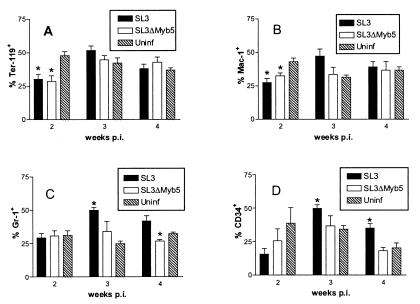
To determine the impact of virus replication on bone marrow hematopoiesis at early times after infection, bone marrow was recovered from neonatally inoculated NIH/Swiss mice at 2, 3, and 4 weeks p.i. Cells were stained with antibodies to quantify surface expression of the lineage-restricted markers Ter-119, Mac-1, Gr-1, and CD34 and were analyzed by flow cytometry. Ter-119 is expressed on immature erythroid progenitors (28). Mac-1 (CD11b) is expressed on monocytes, macrophages, granulocytes, and natural killer cells, increasing with cell maturation (55). Gr-1 (Ly 6G) is present on granulocytes, increasing in expression as differentiation progresses, and is expressed transiently on monocytes and neutrophils (16, 27). CD34 is expressed on a heterogenous population of immature bone marrow cells including multipotent progenitors and committed myelomonocytic progenitors (2). The results revealed a statistically significant alteration in each of the bone marrow subpopulations examined during the early premalignant period of infection with SL3 (Fig. 5). Compared to that from age-matched uninfected controls, bone marrow from SL3-infected mice was significantly depleted of cells expressing Ter-119 or Mac-1 by 2 weeks p.i. In contrast, the proportion of Gr-1+ cells was significantly increased at 3 weeks p.i. Similarly, the proportion of CD34+ cells was significantly increased at 3 and 4 weeks p.i. Infection with SL3ΔMyb5 also resulted in a significant decrease in the proportion of bone marrow cells positive for Ter-119 or Mac-1 markers at 2 weeks p.i. (Fig. 5A and B). In contrast to the case for SL3-infected mice, however, no significant expansion of the Gr-1+ population (Fig. 5C) or the CD34+ population (Fig. 5D) was observed in SL3ΔMyb5-infected mice. In fact, the proportion of Gr-1+ cells was significantly decreased by 4 weeks p.i. in SL3ΔMyb5-infected animals.
FIG. 5.
Flow cytometric analysis of surface antigen expression on bone marrow cells from NIH/Swiss mice infected with SL3 or SL3ΔMyb5 or from age-matched uninfected (Uninf) control animals. Bone marrow was collected at 2, 3, and 4 weeks p.i. from three to five animals at each timed collection. The proportions of Ter-119+ (A), Mac-1+ (B), Gr-1+ (C), and CD34+ (D) bone marrow cells were measured. Because of overlapping patterns of expression of the markers, the total number of surface-positive cells may represent >100%. Asterisks represent significant differences between values from infected and uninfected mice as determined by Student's t test (P < 0.05).
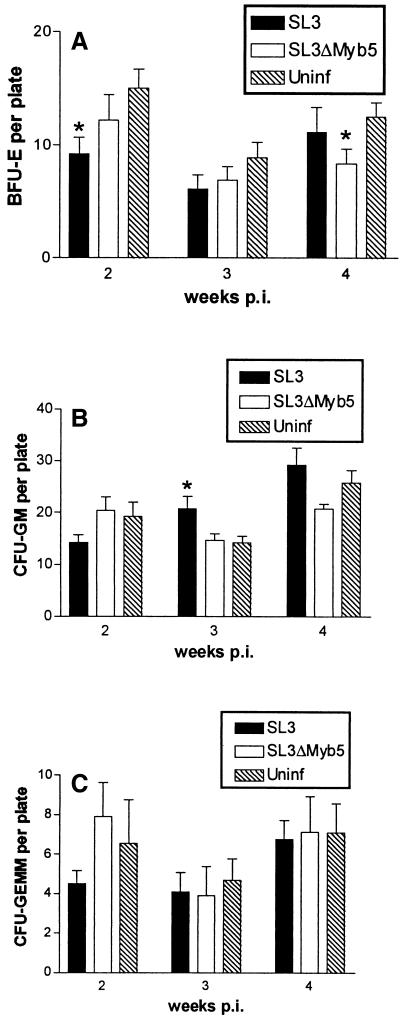
The potential impact of virus infection on bone marrow hematopoiesis was further evaluated by measuring the colony-forming potential of bone marrow progenitor cells in vitro. Bone marrow was retrieved from SL3- or SL3ΔMyb5-infected mice at 2, 3, and 4 weeks p.i. and from age-matched uninfected animals. Bone marrow cells were counted and deposited in semisolid medium supplemented with growth factors. After 12 days, the number of colonies of the BFU-E, CFU-GM, and CFU-GEMM types were enumerated (Fig. 6). The results demonstrated a statistically significant reduction in the number of BFU-E progenitors in bone marrow from SL3-infected mice at 2 weeks p.i. These findings are consistent with the observed reduction in Ter-119+ cells at the same time point (Fig. 5A) and suggest that erythropoiesis is significantly diminished at early times after SL3 infection. A statistically significant increase in the number of CFU-GM progenitors was detected in bone marrow from infected mice at 3 weeks p.i. This finding is consistent with the observed expansion of the Gr-1+ subpopulation at the same time point (Fig. 5C), since Gr-1+ cells are a downstream differentiation product of CFU-GM. In SL3ΔMyb5-infected mice, a significantly reduced number of colonies of the BFU-E type was detected at 4 weeks p.i.; however, no change in the ability of progenitors to form CFU-GM or CFU-GEMM colonies compared to uninfected controls was observed (Fig. 6). SL3 (or SL3ΔMyb5) proviral LTR sequences were detectable by PCR in all types of colonies from infected animals, indicating that infection was not restricted from any lineage (data not shown).
FIG. 6.
Enumeration of BFU-E (A), CFU-GM (B), and CFU-GEMM (C) in bone marrow from NIH/Swiss mice infected with SL3 or SL3ΔMyb5 or from age-matched uninfected (Uninf) control animals, as determined by assays for colony-forming potential. Bone marrow was collected at 2, 3, and 4 weeks p.i. from four or five animals at each time point and deposited in semisolid medium. Colonies were counted after 12 days of incubation at 37°C. Asterisks represent significant differences between values from infected and uninfected animals as determined by Student's t test (P < 0.05).
Alterations in thymocyte subpopulation distribution in the early, premalignant stages of infection.
In normal thymopoiesis, T-cell precursors originate in the bone marrow and travel to the thymus for maturation. The phenotype of the bone marrow-derived precursor is not precisely known, but it lacks both the CD4 and CD8 cell surface antigens and thus is termed double negative. As T cells mature in the thymus, they travel from the cortex to the medulla, interacting with important stromal components that mediate developmental changes. Transition to the CD4+ CD8+ (double-positive) T-cell stage then occurs, during which time the high degree of expansion and proliferation among double-negative T cells arrests. In the double-positive stage, thymocytes undergo positive and negative selection processes, upregulate T-cell receptor expression to high levels, lose either CD4 or CD8 expression to become mature single-positive T cells, and emerge to the periphery (17, 45). To examine the potential pathophysiologic impact of SL3 or SL3ΔMyb5 infection on thymopoiesis, a study was performed to assess and quantify the CD4 CD8 thymocyte subpopulation distribution throughout the premalignant time course. Neonatal NIH/Swiss mice were inoculated intraperitoneally with 106 infectious units of either virus and were sacrificed at weekly intervals between 1 and 8 weeks p.i. (four to seven animals at each timed collection). The thymus was collected from each animal, and a single-cell thymocyte suspension was prepared. Two-color flow cytometry was then used to determine the relative proportions of double-negative (CD4− CD8−), double-positive (CD4+ CD8+), and single-positive (CD4+ CD8− and CD4− CD8+) thymocytes. Age-matched uninfected control animals were examined in parallel to establish baseline values. The results showed that the thymocyte subpopulation distribution was significantly disrupted early in SL3 infection, beginning at 4 weeks p.i. At that time, the proportion of CD4− CD8− thymocytes was observed to increase 3.5-fold over that in uninfected animals (Table 3). In parallel, a decrease in the proportion of CD4+ CD8+ thymocytes was also detected, but little change in the single-positive subpopulations was observed. The observed CD4− CD8− expansion represented an increase not only in the proportion but also in the absolute number of cells. Cell counts showed that CD4− CD8− thymocytes increased in number in the first 2 to 4 weeks after SL3 inoculation, doubling by 4 weeks p.i. (from 2.1 × 107 at 2 weeks to 4 × 107 at 4 weeks; average of four to seven animals at each timed collection). In uninfected control animals, by comparison, the number of CD4− CD8− thymocytes did not increase between 2 to 4 weeks of age (data not shown). In SL3ΔMyb5-infected animals, expansion of the CD4− CD8− thymocyte subpopulation was also detected beginning at 4 weeks p.i. but became statistically significant (P < 0.05) only after 6 weeks of infection (Table 3). Thus, SL3ΔMyb5 infection was associated with a qualitatively similar, although blunted, effect on the thymocyte subpopulation distribution early after inoculation.
TABLE.
3. Thymocyte subpopulation distribution in weeks 1 to 8 following inoculation of neonatal NIH/Swiss mice with SL3 or SL3ΔMyb5a
| Subpopulation | Wk p.i. | % (mean ± SEM)b in:
|
||
|---|---|---|---|---|
| SL3-inoculated mice | SL3ΔMyb5-inoculated mice | Uninfected controls | ||
| CD4− CD8− (DN) | 1 | 15.8 ± 2.1 | 7.7∗ ± 3.7 | 16.3 ± 2.5 |
| 2 | 11.6 ± 1.9 | 10.3 ± 1.8 | 10.5 ± 1.8 | |
| 3 | 19.0 ± 5.1 | 12.6 ± 1.9 | 17.9 ± 3.1 | |
| 4 | 19.0∗ ± 2.9 | 12.2 ± 3.6 | 5.4 ± 0.6 | |
| 5 | 19.0∗ ± 3.3 | 15.5 ± 3.5 | 10.7 ± 1.7 | |
| 6 | 22.6 ± 4.2 | 27.2∗ ± 6.3 | 16.4 ± 0.7 | |
| 7 | 19.2∗ ± 1.0 | 15.5 ± 3.9 | 13.8 ± 1.9 | |
| 8 | 22.0 ± 4.1 | 16.5 ± 3.4 | 23.1 ± 1.8 | |
| CD4+ CD8+ (DP) | 1 | 65.2 ± 4.5 | 70.3 ± 4.4 | 70.0 ± 2.1 |
| 2 | 65.3 ± 6.1 | 75.7 ± 2.4 | 73.4 ± 0.9 | |
| 3 | 59.5 ± 5.1 | 64.5 ± 6.5 | 58.2 ± 3.2 | |
| 4 | 58.3∗ ± 2.1 | 67.1∗ ± 3.9 | 77.6 ± 2.6 | |
| 5 | 61.9 ± 3.8 | 64.2 ± 2.2 | 70.1 ± 2.8 | |
| 6 | 55.6∗ ± 3.5 | 50.1∗ ± 4.3 | 63.5 ± 3.0 | |
| 7 | 48.9∗ ± 1.9 | 62.6 ± 3.7 | 58.4 ± 4.4 | |
| 8 | 57.3 ± 4.8 | 56.2 ± 2.6 | 48.7 ± 4.7 | |
| CD4+ CD8− (CD4) | 1 | 17.3 ± 5.7 | 16.7 ± 5.7 | 11.1 ± 2.5 |
| 2 | 20.1 ± 4.7 | 11.6 ± 1.4 | 13.9 ± 2.1 | |
| 3 | 17.5 ± 2.5 | 19.0 ± 4.9 | 19.7 ± 2.1 | |
| 4 | 21.5∗ ± 0.8 | 18.1 ± 1.0 | 15.0 ± 2.3 | |
| 5 | 14.6 ± 1.9 | 16.7 ± 2.6 | 13.6 ± 3.2 | |
| 6 | 16.9 ± 2.0 | 18.1 ± 3.4 | 16.5 ± 2.9 | |
| 7 | 23.7 ± 1.4 | 18.6 ± 1.3 | 21.2 ± 3.0 | |
| 8 | 14.7 ± 5.0 | 19.6 ± 2.5 | 22.5 ± 1.8 | |
| CD4− CD8+ (CD8) | 1 | 1.6 ± 0.4 | 2.6 ± 1.7 | 2.5 ± 1.2 |
| 2 | 2.0 ± 0.6 | 2.1 ± 0.7 | 1.9 ± 0.2 | |
| 3 | 3.9 ± 0.4 | 3.7 ± 0.4 | 4.3 ± 0.7 | |
| 4 | 1.5 ± 0.3 | 2.6 ± 0.5 | 2.1 ± 0.4 | |
| 5 | 4.6 ± 1.2 | 3.7* ± 0.3 | 5.6 ± 0.7 | |
| 6 | 5.0 ± 0.9 | 3.9 ± 0.5 | 3.6 ± 0.9 | |
| 7 | 8.2 ± 1.0 | 3.3 ± 1.2 | 6.6 ± 1.6 | |
| 8 | 6.0 ± 2.8 | 5.4 ± 1.9 | 5.7 ± 1.3 | |
Relative proportions of the thymocyte subpopulations CD4− CD8− (double negative [DN]), CD4+ CD8+ (double positive [DP]), CD4− CD8+ (single positive [CD8]) and CD4+ CD8− (single positive [CD4]) were examined at weekly intervals following inoculation of neonatal NIH/Swiss mice with SL3 MuLV or SL3ΔMyb5. At each timed collection, single-cell thymocyte suspensions were prepared and dually stained with FITC-conjugated anti-CD4 and PE-conjugated anti-CD8. The percentage of thymocytes staining for one or both markers was assessed by flow cytometry and compared to values from age-matched uninfected control animals examined in parallel. Four to seven animals were examined at each timed collection.
Asterisks indicate values from SL3- or SL3ΔMyb5-infected mice that are statistically distinct from those from age-matched uninfected controls as determined by Student's t test (P < 0.05).
Thymic atrophy immediately preceding tumor outgrowth.
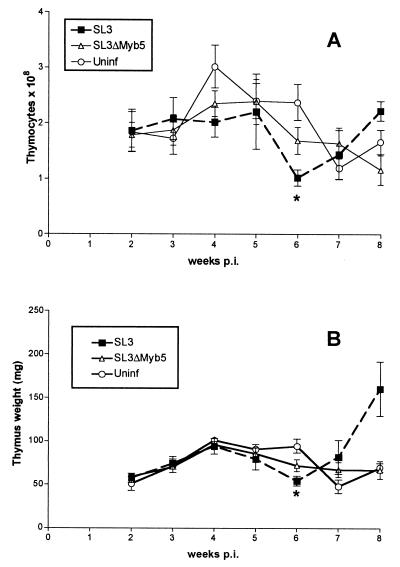
The weight of the thymus and the total number of thymocytes were monitored at weekly intervals throughout the premalignant time course in infected animals and were compared to results for age-matched uninfected controls (Fig. 7). In uninfected NIH/Swiss mice, thymus weights and thymocyte counts were observed to increase in the first 4 weeks after birth and then to decline gradually through the 8-week time point. By contrast, the pronounced increase in cellularity of the thymus characteristic of the first few weeks of life was not seen in SL3-infected animals. By 6 weeks p.i., a statistically significant reduction in both thymus weight and thymocyte count was apparent in SL3-infected animals. Shortly thereafter, by 8 weeks p.i., SL3-infected mice displayed a marked increase in thymus weight and thymocyte content consistent with the outgrowth of a tumor. In fact, Southern blot analysis of genomic DNA from the 8-week thymus demonstrated clonal outgrowth of a T-cell population with distinctive T-cell receptor beta gene rearrangement (data not shown). In SL3ΔMyb5-infected animals, no significant reduction in thymocyte cell count or thymus weight was observed during the premalignant period (Fig. 7).
FIG. 7.
Thymocyte cell count (A) and average thymus weight (B) in NIH/Swiss mice infected with SL3 or SL3ΔMyb5 and collected at weekly intervals after inoculation or in age-matched uninfected (Uninf) control animals. Four to seven animals in each group were examined at each timed collection. Statistically significant differences between values from infected and infected mice were determined by Student's t test (P < 0.05) and are represented by asterisks.
DISCUSSION
The MuLV-mediated induction of lymphoma is thought to represent a multistep process that begins early in infection, long before clinical evidence of disease. Pathophysiologic changes occur in the target organs for virus replication, including the eventual target for tumor development and other tissues as well. In the case of SL3, the thymus is thought to be the primary site of tumor formation, because the tumor typically displays the phenotype of an immature T cell (21). In addition to occurring in the thymus, however, virus replication also occurs in the spleen and bone marrow at early times after inoculation and may be required at these sites to establish a preleukemic state favorable for development of the thymic tumor. Previous studies using biological assays showed that SL3 can be detected in thymus, spleen, and bone marrow as early as 4 to 8 weeks following intraperitoneal inoculation (43). The present study extends those findings by demonstrating virus replication in all three tissues as early as 1 to 2 weeks after inoculation (Fig. 1A). SL3 proviral DNA was demonstrated in isolated thymocytes by 1 week p.i. (Fig. 1B), a finding confirmed by flow cytometry for expression of SL3 SU (Table 2). These findings are generally consistent with a recent analysis demonstrating SL3 replication in the thymus as early as 2 weeks p.i. (54). In that study, however, infection did not occur in thymocytes and was apparently restricted to cells identified as stromal components. Why SL3 replication was detected within thymocytes in the present study but not in the previous study is unclear, but it may be related to the increased sensitivity of PCR amplification compared to the infectious-cell center assay used in that study (54). A potential problem with the use of PCR amplification to identify proviral DNA is that the tissues examined are contaminated with circulating blood cells that may be infected, thereby yielding an artifactually positive amplification signal. The amplification of SL3 proviral DNA from the tissues and cells examined here apparently does not represent blood contamination, however, since no amplification signal was detected from liver or kidney (Fig. 1C). PCR amplification also demonstrated the accumulation of SL3 LTRs containing multiple enhancer repeats, which was evident in thymocytes by 8 weeks p.i. It is noteworthy that these enhancer variants were readily detected in the thymus but not in infected spleen or bone marrow (Fig. 1A and B). The accumulation of enhancer repeat variants uniquely in the thymus may reflect a selective replicative advantage to the virus in tumor cells. Alternatively, the variants may confer a selective growth advantage on the host cell, perhaps through optimal transcriptional activation of an adjacent oncogene (35, 37).
Although the thymus is the target tissue for tumor formation, preleukemic alterations were initially detected in the bone marrow. Immunohistochemical analysis of bone marrow localized SL3 infection predominantly to immature myeloid cells and maturing granulocytes at early times after inoculation (Fig. 4). These findings are consistent with significant increases observed in the number of colony-forming progenitors of the CFU-GM type (Fig. 6B) and of Gr-1+ cells (Fig. 5C), a downstream differentiation product of CFU-GM. The data indicate that SL3 infection may be directly involved in expansion of the myeloid progenitor population in infected bone marrow, perhaps to increase the number of available target cells for virus replication. Flow cytometric analysis further revealed expansion of the primitive CD34+ population in the early preleukemic period (3 to 4 weeks) (Fig. 5D). This is an intriguing observation, since an expanding CD34+ population may be vulnerable to transforming events such as those giving rise to the PLC identified in the bone marrow of SL3-infected mice about 4 weeks p.i. (50). The mechanism is reminiscent of that described for M-MuLV-induced disease in which a compensatory extramedullary hematopoiesis in the spleen is thought to provide hyperplastic progenitor cells that may represent the transformation targets (15). Both SL3 and M-MuLV apparently act by stimulating the proliferation of primitive hematopoietic progenitor cells, either in the bone marrow or at an extramedullary site. In the case of SL3, polytropic recombinant viruses are unlikely to be involved in the process, since they are not detected in bone marrow until 6 weeks p.i. (44).
During the early weeks of infection, SL3 replication in the thymus increased rapidly as determined by measurement of proviral load (Fig. 3; Table 1) or surface SU expression (Table 2). As was detected previously by serological focus assay (43), the viral load varied widely among animals examined at the same timed collection. Despite the observed variation in viral load, SL3 replication in the thymus was associated with a statistically significant disruption of the thymocyte subpopulation distribution (Table 3). Infected animals demonstrated a statistically significant increase, up to 3.5-fold, in the proportion of immature CD4− CD8− thymocytes between 4 and 7 weeks p.i. The observed increase in the proportion of the most immature thymocyte subset is consistent with previous studies of SL3 infection in AKR mice, in which infected thymic lobes displayed increased percentages of CD4− CD8− thymocytes between 6 and 8 weeks p.i. (24). It was further shown that CD4− CD8− thymocytes from SL3-infected AKR mice did not mature normally when introduced intrathymically into irradiated syngeneic hosts (23). Similarly, in the preleukemic M-MuLV-infected thymus, the most immature cells were also detected at an increased proportion (13). These findings implicate physiologic alteration of thymocyte development as an important element in the premalignant process mediated by MuLV. In addition to disrupted thymopoiesis, a significant thymic regression was detected in SL3-infected animals at 6 weeks p.i. as measured by thymus weight or thymocyte count (Fig. 7). A similar thymic atrophy preceding tumor outgrowth was previously described for AKR mice infected with MCF13 MuLV (58) and in NIH/Swiss mice infected with M-MuLV (5). In the former case, thymic atrophy was attributed to increased apoptosis in the CD4+ CD8+ thymocyte subpopulation at 6 to 8 weeks p.i. Apoptotic cells occurred predominantly in the gp70+ population, suggesting that apoptosis was a result of virus infection (58). In M-MuLV-infected mice, preleukemic thymic atrophy was also attributed to a higher apoptotic rate in the infected thymus (5). In fact, a high rate of apoptosis was shown to persist in the transformed cell and was characteristic of the end-stage tumor (6). In SL3-infected animals, the mechanism responsible for thymic regression has not yet been evaluated. Polytropic recombinant viruses may have a role, since they are present in the thymus as early as 2 weeks p.i. (44). In M-MuLV-infected mice, however, MCF virus recombinants are not required for preleukemic thymic atrophy (5).
These and other studies clearly demonstrate that pathophysiologic changes occur in the target tissue for tumor formation, and in other infected tissues, long before clinical evidence of SL3-mediated disease (23, 24, 44, 50). The relationship of those early changes to the subsequent outgrowth of lymphoma remains incompletely understood. In the present study, we used the SL3 mutant SL3ΔMyb5 as a tool to distinguish early changes associated with chronic retroviral infection from those that may be specifically linked to oncogenesis. We considered that SL3ΔMyb5 would be a useful tool for this purpose because it replicates in thymus, spleen, and bone marrow throughout the preleukemic period (44) but is essentially nonpathogenic (38). Like for SL3, SL3ΔMyb5 proviral DNA was demonstrated in target tissues as early as 1 week p.i. (Fig. 2) (44). Quantification of proviral load (Fig. 3; Table 1) and surface SU expression (Table 2) showed that infection levels varied widely, consistent with previous measurements made using biological assays of viral load (43). Our results showed a rapid increase in proviral load during weeks 1 to 3 p.i., especially in the thymus (Fig. 3B). By 4 weeks p.i. and thereafter, however, SL3ΔMyb5 replication steadily declined, while SL3 replication increased (Tables 1 and 2 ). By 8 weeks p.i., mean SL3 proviral loads in bone marrow and thymus exceeded SL3ΔMyb5 proviral loads by fivefold (Table 1). It is noteworthy that, although SL3ΔMyb5 replication declined during the preleukemic period, some similar changes were induced by SL3 and SL3ΔMyb5 in target tissues. Both SL3 and SL3ΔMyb5 disrupted the thymocyte subpopulation distribution (Table 3), both viruses were localized predominantly in mature and maturing myeloid cells in the bone marrow (Fig. 4), and both viruses disrupted bone marrow hematopoiesis (Fig. 5 and 6). Despite the similarities observed, differences were apparent during the early weeks of SL3 and SL3ΔMyb5 infection that may be informative as to the tumorigenic mechanism. At 2 to 4 weeks p.i., hematopoiesis in the bone marrow was disrupted by infection with either virus as detected by a decline in the proportion of Ter-119+ cells, a corresponding decrease in the number of BFU-E colony-forming progenitors, and a decline in the proportion of Mac-1+ cells. Not detected in the SL3ΔMyb5-infected animals, however, were the significant increases in Gr-1+ cells, in the corresponding CFU-GM colony-forming potential, and in CD34+ progenitors at 3 to 4 weeks p.i. (Fig. 5 and 6). The mechanism by which SL3, but not SL3ΔMyb5, induces these changes remains unknown. A possibility to consider is that polytropic recombinant viruses arise in SL3ΔMyb5-infected bone marrow in considerably fewer animals than during SL3 infection (25 versus 100% at 6 weeks p.i. [44]). However, since polytropic recombinant viruses were not detected in bone marrow until 6 weeks p.i., this difference is unlikely to be involved in the effects on hematopoiesis detected at 2 to 4 weeks p.i. More likely, the reduced level of replication of SL3ΔMyb5 in bone marrow compared to SL3 may account for the differences in pathophysiologic impact.
By 4 weeks after inoculation, the normal distribution of thymocyte subpopulations was disrupted in animals infected with either virus (Table 3). In both SL3- and SL3ΔMyb5-infected animals, a significant increase in the proportion of the most immature thymocyte subset (CD4− CD8− double negative) was observed, although the magnitude and duration of the increase were blunted in SL3ΔMyb5 infection. Again, this difference in pathophysiologic impact may result from reduced replication of SL3ΔMyb5 in the thymus (Fig. 3; Table 1). Of note is that the disruption of the thymocyte subset distribution in SL3ΔMyb5-infected animals was not accompanied by the significant thymic atrophy detected in SL3 infection (Fig. 7), thus implicating thymic regression in the malignant process. Further, SL3ΔMyb5 replication in the thymus was not accompanied by an accumulation of proviruses bearing increased numbers of enhancer repeats (Fig. 2). If the accumulation of enhancer repeats confers a selective growth advantage on the host cell, through optimal transcriptional activation of an adjacent oncogene as previously suggested (35, 37), the diminished pathogenicity of SL3ΔMyb5 might be due in part to its reduced ability to participate in insertional activation. Not only might the failure to accumulate enhancer repeats reduce the potential to activate adjacent gene sequences, but so might the elimination of c-Myb binding. Indeed, many cellular genes have been identified that are activated synergistically by the cooperative binding of c-Myb and Ets-1 or core binding factor, transcription factors whose binding sites lie in close proximity on the SL3 LTR (10, 26, 36, 46). Elimination of the c-Myb binding site in SL3ΔMyb5 might disrupt the formation of specific transcription factor complexes required for synergistic activation, thereby restricting the number of potential enhancer activation events mediated by the mutant virus.
Taken together, the results presented here and previously (44) implicate certain premalignant alterations in the malignant process, including (i) significant increases in the proportions of Gr-1+ or CD34+ progenitors in the bone marrow early in infection, (ii) a significant increase in the proportion of CD4− CD8− thymocytes, (iii) significant thymic atrophy just prior to tumor outgrowth, and (iv) accumulation of proviral LTRs bearing increased numbers of enhancer repeats. The data support a model of SL3-mediated lymphomagenesis in which virus inoculated into the peritoneum is ferried rapidly to the bone marrow, where it replicates in actively dividing hematopoietic progenitors. Bone marrow hematopoiesis is thereby altered, particularly with the generation and maturation of increasing numbers of CD34+ progenitors, immature and maturing myeloid cells. The expanding CD34+ population is likely to include precursors to prothymocytes, perhaps the previously described PLC (19, 50), which exit the bone marrow and migrate to the thymus for further development. The newly arrived CD4− CD8− thymocytes, at least in part from virally induced CD34+ precursors, now find a thymic environment more suitable for their continued expansion. As a consequence of SL3 replication since the first week after inoculation, thymopoiesis has been disrupted such that the immature CD4− CD8− subpopulation is increased in size and proportion. The mechanism of this increase is not yet known, but it may be the result of increased proliferative capacity and/or prolonged survival in the CD4− CD8− compartment. Within weeks of the CD4− CD8− expansion, a marked thymic atrophy occurs, which is followed shortly by tumor expansion. In an environment of thymic regression, an immature CD4− CD8− cell with increased proliferative or survival capacity would have a strong selective advantage. The selection of an infected, immature thymocyte, perhaps the PLC, may render it vulnerable to the late-occurring genetic events that lead to frank malignancy, e.g., insertional mutagenesis. Proviral LTRs whose structure optimally accomplishes the enhancer activation of adjacent cellular proto-oncogenes might then be selected, leading eventually to the clonal expansion of fully malignant tumor cells.
Acknowledgments
This work was supported by Public Health Service grant CA83823, by development funds of the Tulane Cancer Center, and by a grant from the W. M. William Keck Foundation of Los Angeles.
We are grateful to Miles Cloyd for providing the 24-8 hybridoma and to Donald Phinney for helpful discussions and advice.
REFERENCES
- 1.Amtoft, H. W., A. B. Sorensen, C. Bareil, J. Schmidt, A. Luz, and F. S. Pedersen. 1997. Stability of AML1 (core) site enhancer mutations in T lymphomas induced by attenuated SL3-3 murine leukemia virus mutants. J. Virol. 71:5080-5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews, R. G., J. W. Singer, and I. D. Bernstein. 1986. Monoclonal antibody 12-8 recognizes a 115-kd molecule present on both unipotent and multipotent hematopoietic colony-forming cells and their precursors. Blood 67:842-845. [PubMed] [Google Scholar]
- 3.Beaty, R. M., K. Rulli, K. L. Bost, J. Pantginis, J. Lenz, and L. S. Levy. 1999. High levels of IL-4 and IL-10 mRNA and low levels of IL-2, IL-9 and IFN-gamma mRNA in MuLV-induced lymphomas. Virology 261:253-262. [DOI] [PubMed] [Google Scholar]
- 4.Belli, B., and H. Fan. 1994. The leukemogenic potential of an enhancer variant of Moloney murine leukemia virus varies with the route of inoculation. J. Virol. 68:6883-6889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonzon, C., and H. Fan. 1999. Moloney murine leukemia virus-induced preleukemic thymic atrophy and enhanced thymocyte apoptosis correlate with disease pathogenicity. J. Virol. 73:2434-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonzon, C., and H. Fan. 2000. Moloney murine leukemia virus-induced tumors show altered levels of proapoptotic and antiapoptotic proteins. J. Virol. 74:8151-8158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boral, A. L., S. A. Okenquist, and J. Lenz. 1989. Identification of the SL3-3 virus enhancer core as a T-lymphoma cell-specific element. J. Virol. 63:76-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brightman, B. K., K. G. Chandy, R. H. Spencer, S. Gupta, P. K. Pattengale, and H. Fan. 1988. Characterization of lymphoid tumors induced by a recombinant murine retrovirus carrying the avian v-myc oncogene. Identification of novel (B-lymphoid) tumors in the thymus. J. Immunol. 141:2844-2854. [PubMed] [Google Scholar]
- 9.Brightman, B. K., B. R. Davis, and H. Fan. 1990. Preleukemic hematopoietic hyperplasia induced by Moloney murine leukemia virus is an indirect consequence of viral infection. J. Virol. 64:4582-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burk, O., and K. H. Klempnauer. 1999. Myb and Ets transcription factors cooperate at the myb-inducible promoter of the tom-1 gene. Biochim. Biophys. Acta 1446:243-252. [DOI] [PubMed] [Google Scholar]
- 11.Cloyd, M. W., J. W. Hartley, and W. P. Rowe. 1980. Lymphomagenicity of recombinant mink cell focus-inducing murine leukemia viruses. J. Exp. Med. 151:542-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis, B. R., B. K. Brightman, K. G. Chandy, and H. Fan. 1987. Characterization of a preleukemic state induced by Moloney murine leukemia virus: evidence for two infection events during leukemogenesis. Proc. Natl. Acad. Sci. USA 84:4875-4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis, B. R., K. G. Chandy, B. K. Brightman, S. Gupta, and H. Fan. 1986. Effects of nonleukemogenic and wild-type Moloney murine leukemia virus on lymphoid cells in vivo: identification of a preleukemic shift in thymocyte subpopulations. J. Virol. 60:423-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ethelberg, S., B. D. Tzschaschel, A. Luz, S. J. Diaz-Cano, F. S. Pedersen, and J. Schmidt. 1999. Increased induction of osteopetrosis, but unaltered lymphomagenicity, by murine leukemia virus SL3-3 after mutation of a nuclear factor 1 site in the enhancer. J. Virol. 73:10406-10415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan, H. 1997. Leukemogenesis by Moloney murine leukemia virus: a multistep process. Trends Microbiol. 5:74-82. [DOI] [PubMed] [Google Scholar]
- 16.Fleming, T. J., M. L. Fleming, and, T. R. Malek. 1993. Selective expression of Ly-6G on myeloid lineage cells in mouse bone marrow. RB6-8C5 mAb to granulocyte-differentiation antigen (Gr-1) detects members of the Ly-6 family. J. Immunol. 151:2399-2408. [PubMed] [Google Scholar]
- 17.Fowlkes, B. J., and D. M. Pardoll. 1989. Molecular and cellular events of T cell development. Adv. Immunol. 44:207-264. [DOI] [PubMed] [Google Scholar]
- 18.Hallberg, B., J. Schmidt, A. Luz, F. S. Pedersen, and T. Grundstrom. 1991. SL3-3 enhancer factor 1 transcriptional activators are required for tumor formation by SL3-3 murine leukemia virus. J. Virol. 65:4177-4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haran-Ghera, N. 1980. Potential leukemic cells among bone marrow cells of young AKR/J mice. Proc. Natl. Acad. Sci. USA 77:2923-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haran-Ghera, N., A. Peled, F. Leef, A. D. Hoffman, and J. A. Levy. 1987. Enhanced AKR leukemogenesis by the dual tropic viruses. I. The time and site of origin of potential leukemic cells. Leukemia 1:442-449. [PubMed] [Google Scholar]
- 21.Hays, E. F., and G. Bristol. 1992. Observations on lymphomagenesis and lymphoma in AKR mice. A description of prelymphoma changes in the thymus and phenotypic diversity of lymphomas induced by SL3-3 virus. Thymus 19:219-234. [PubMed] [Google Scholar]
- 22.Hays, E. F., G. Bristol, and S. McDougall. 1990. Mechanisms of thymic lymphomagenesis by the retrovirus SL3-3. Cancer Res. 50:5631.S-5635S. [PubMed]
- 23.Hays, E. F., G. C. Bristol, J. P. Lugo, and X. F. Wang. 1989. Progression to development of lymphoma in the thymus of AKR mice treated neonatally with SL 3-3 virus. Exp. Hematol. 17:1116-1121. [PubMed] [Google Scholar]
- 24.Hays, E. F., G. C. Bristol, S. McDougall, J. L. Klotz, and M. Kronenberg. 1989. Development of lymphoma in the thymus of AKR mice treated with the lymphomagenic virus SL 3-3. Cancer Res. 49:4225-4230. [PubMed] [Google Scholar]
- 25.Hays, E. F., and J. A. Levy. 1984. Differences in lymphomagenic properties of AKR mouse retroviruses. Virology 138:49-57. [DOI] [PubMed] [Google Scholar]
- 26.Hernandez-Munain, C., and M. S. Krangel. 1995. c-Myb and core-binding factor/PEBP2 display functional synergy but bind independently to adjacent sites in the T-cell receptor delta enhancer. Mol. Cell. Biol. 15:3090-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hestdal, K., F. W. Ruscetti, J. N. Ihle, S. E. Jacobsen, C. M. Dubois, W. C. Kopp, D. L. Longo, and J. R. Keller. 1991. Characterization and regulation of RB6-8C5 antigen expression on murine bone marrow cells. J. Immunol. 147:22-28. [PubMed] [Google Scholar]
- 28.Kina, T., K. Ikuta, E. Takayama, K. Wada, A. S. Majumdar, I. L. Weissman, and Y. Katsura. 2000. The monoclonal antibody TER-119 recognizes a molecule associated with glycophorin A and specifically marks the late stages of murine erythroid lineage. Br. J. Haematol. 109:280-287. [DOI] [PubMed] [Google Scholar]
- 29.Lazo, P. A., A. J. Klein-Szanto, and P. N. Tsichlis. 1990. T-cell lymphoma lines derived from rat thymomas induced by Moloney murine leukemia virus: phenotypic diversity and its implications. J. Virol. 64:3948-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenz, J., D. Celander, R. L. Crowther, R. Patarca, D. W. Perkins, and W. A. Haseltine. 1984. Determination of the leukaemogenicity of a murine retrovirus by sequences within the long terminal repeat. Nature 308:467-470. [DOI] [PubMed] [Google Scholar]
- 31.Lenz, J., R. Crowther, S. Klimenko, and W. Haseltine. 1982. Molecular cloning of a highly leukemogenic, ecotropic retrovirus from an AKR mouse. J. Virol. 43:943-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, Q. X., and H. Fan. 1990. Combined infection by Moloney murine leukemia virus and a mink cell focus-forming virus recombinant induces cytopathic effects in fibroblasts or in long-term bone marrow cultures from preleukemic mice. J. Virol. 64:3701-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.LoSardo, J. E., A. L. Boral, and J. Lenz. 1990. Relative importance of elements within the SL3-3 virus enhancer for T-cell specificity. J. Virol. 64:1756-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martiney, M. J., L. S. Levy, and J. Lenz. 1999. Suppressor mutations within the core binding factor (CBF/AML1) binding site of a T-cell lymphomagenic retrovirus. J. Virol. 73:2143-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martiney, M. J., K. Rulli, R. Beaty, L. S. Levy, and J. Lenz. 1999. Selection of reversions and suppressors of a mutation in the CBF binding site of a lymphomagenic retrovirus. J. Virol. 73:7599-7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCracken, S., S. Leung, R. Bosselut, J. Ghysdael, and N. G. Miyamoto. 1994. Myb and Ets related transcription factors are required for activity of the human lck type I promoter. Oncogene 9:3609-3615. [PubMed] [Google Scholar]
- 37.Morrison, H. L., B. Soni, and J. Lenz. 1995. Long terminal repeat enhancer core sequences in proviruses adjacent to c-myc in T-cell lymphomas induced by a murine retrovirus. J. Virol. 69:446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nieves, A., L. S. Levy, and J. Lenz. 1997. Importance of a c-Myb binding site for lymphomagenesis by the retrovirus SL3-3. J. Virol. 71:1213-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Donnell, P. V., E. Fleissner, H. Lonial, C. F. Koehne, and A. Reicin. 1985. Early clonality and high-frequency proviral integration into the c-myc locus in AKR leukemias. J. Virol. 55:500-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Donnell, P. V., R. Woller, and A. Chu. 1984. Stages in development of mink cell focus-inducing (MCF) virus-accelerated leukemia in AKR mice. J. Exp. Med. 160:914-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pedersen, F. S., R. L. Crowther, D. Y. Tenney, A. M. Reimold, and W. A. Haseltine. 1981. Novel leukaemogenic retroviruses isolated from cell line derived from spontaneous AKR tumour. Nature 292:167-170. [DOI] [PubMed] [Google Scholar]
- 42.Portis, J. L., F. J. McAtee, and M. W. Cloyd. 1982. Monoclonal antibodies to xenotropic and MCF murine leukemia viruses derived during the graft-versus-host reaction. Virology 118:181-190. [DOI] [PubMed] [Google Scholar]
- 43.Rosen, C. A., W. A. Haseltine, J. Lenz, R. Ruprecht, and M. W. Cloyd. 1985. Tissue selectivity of murine leukemia virus infection is determined by long terminal repeat sequences. J. Virol. 55:862-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rulli, K., P. A. Lobelle-Rich, A. Trubetskoy, J. Lenz, and L. S. Levy. 2001. Tissue distribution and timing of appearance of polytropic envelope recombinants during infection with SL3-3 murine leukemia virus or its weakly pathogenic SL3ΔMyb5 mutant. J. Virol. 75:522-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scollay, R. 1991. T-cell subset relationships in thymocyte development. Curr. Opin. Immunol. 3:204-209. [DOI] [PubMed] [Google Scholar]
- 46.Shapiro, L. H. 1995. Myb and Ets proteins cooperate to transactivate an early myeloid gene. J. Biol. Chem. 270:8763-8771. [DOI] [PubMed] [Google Scholar]
- 47.Sorensen, A. B., M. Duch, H. W. Amtoft, P. Jorgensen, and F. S. Pedersen. 1996. Sequence tags of provirus integration sites in DNAs of tumors induced by the murine retrovirus SL3-3. J. Virol. 70:4063-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sorensen, A. B., A. H. Lund, S. Ethelberg, N. G. Copeland, N. A. Jenkins, and F. S. Pedersen. 2000. Sint1, a common integration site in SL3-3-induced T-cell lymphomas, harbors a putative proto-oncogene with homology to the septin gene family. J. Virol. 74:2161-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steffen, D. 1984. Proviruses are adjacent to c-myc in some murine leukemia virus-induced lymphomas. Proc. Natl. Acad. Sci. USA 81:2097-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takeuchi, H., A. Kato, and E. F. Hays. 1984. Presence of prelymphoma cells in the bone marrow of the lymphomagenic virus-treated AKR mouse. Cancer Res. 44:1008-1011. [PubMed] [Google Scholar]
- 51.Thomas, C. Y. 1986. AKR ecotropic murine leukemia virus SL3-3 forms envelope gene recombinants in vivo. J. Virol. 59:23-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomas, C. Y., J. D. Nuckols, C. Murphy, and D. Innes. 1993. Generation and pathogenicity of an NB-tropic SL3-3 murine leukemia virus. Virology 193:1013-1017. [DOI] [PubMed] [Google Scholar]
- 53.Tsichlis, P. N. 1987. Oncogenesis by Moloney murine leukemia virus. Anticancer Res. 7:171-180. [PubMed] [Google Scholar]
- 54.Uittenbogaart, C. H., W. Law, P. J. Leenen, G. Bristol, W. van Ewijk, and E. F. Hays. 1998. Thymic dendritic cells are primary targets for the oncogenic virus SL3-3. J. Virol. 72:10118-10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Voura, E. B., F. Billia, N. N. Iscove, and R. G. Hawley. 1997. Expression mapping of adhesion receptor genes during differentiation of individual hematopoietic precursors. Exp. Hematol. 25:1172-1179. [PubMed] [Google Scholar]
- 56.Yoshimura, F. K., M. Cankovic, R. Smeltz, and S. Ibrahim. 1997. Identification of nucleotide sequences that regulate transcription of the MCF13 murine leukemia virus long terminal repeat in activated T cells. J. Virol. 71:2572-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoshimura, F. K., T. Wang, and M. Cankovic. 1999. Sequences between the enhancer and promoter in the long terminal repeat affect murine leukemia virus pathogenicity and replication in the thymus. J. Virol. 73:4890-4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoshimura, F. K., T. Wang, F. Yu, H. R. Kim, and J. R. Turner. 2000. Mink cell focus-forming murine leukemia virus infection induces apoptosis of thymic lymphocytes. J. Virol. 74:8119-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]