Abstract
We demonstrate that the S4 genome segment of baboon reovirus (BRV) contains two sequential partially overlapping open reading frames (ORFs), both of which are functional in vitro and in virus-infected cells. The 15-kDa gene product (p15) of the 5"-proximal ORF induces efficient cell-cell fusion when expressed by itself in transfected cells, suggesting that p15 is the only viral protein required for induction of syncytium formation by BRV. The p15 protein is a small, hydrophobic, basic, integral membrane protein, properties shared with the p10 fusion-associated small transmembrane (FAST) proteins encoded by avian reovirus and Nelson Bay reovirus. As with p10, the BRV p15 protein is also a nonstructural protein and, therefore, is not involved in virus entry. Sequence analysis indicates that p15 shares no significant sequence similarity with the p10 FAST proteins and contains a unique repertoire and arrangement of sequence-predicted structural and functional motifs. These motifs include a functional N-terminal myristylation consensus sequence, an N-proximal proline-rich motif, two potential transmembrane domains, and an intervening polybasic region. The unique structural properties of p15 suggest that this protein is a novel member of the new family of FAST proteins.
Membrane fusion is an essential cellular event, the precise mechanism of which remains poorly understood. Much of our understanding of membrane fusion is derived from studies of enveloped virus fusion proteins. Structure-function analysis of numerous enveloped virus fusion proteins has led to a generalized model of protein-mediated membrane fusion. This model suggests that triggered conformational changes in the metastable viral fusion proteins expose a previously concealed hydrophobic fusion peptide for insertion into, and destabilization of, the target lipid bilayer (16, 43, 50, 51). Further extensive conformational changes are predicted to be involved in supplying the energy required to pull the donor and target bilayers together and allow the fusion reaction to proceed 1 (6, 50, 51). Based on the nature of the structural rearrangements, the enveloped virus fusion proteins fall into two general classes. Class I proteins, such as the influenza virus hemagglutinin, human immunodeficiency virus gp41, and Ebola virus GP2, use coiled-coil rearrangements to drive the fusion process while class II proteins, such as those of the alphaviruses, flaviviruses, and rhabdoviruses, undergo extensive multimer rearrangements in the absence of apparent coiled-coil interactions (19, 34, 42, 43, 44).
Our recent characterization of a fusion-associated small transmembrane (FAST) protein encoded by certain fusogenic reoviruses (39) suggests that extensive conformational changes in large, complex, multimeric viral fusion proteins may not be a universal requirement for protein-mediated membrane fusion. The only examples of nonenveloped viruses that induce syncytium formation are all members of the family Reoviridae, a diverse group of nonenveloped viruses with double-stranded RNA genomes (5). We have shown that two fusogenic orthoreoviruses, avian reovirus (ARV) and Nelson Bay reovirus (NBV), encode homologous 95- to 98-amino-acid 10-kDa fusion proteins, termed p10 (39). Unlike enveloped virus fusion proteins, which are structural components of the virus particle involved in the penetration of host cells, the p10 proteins are nonstructural, accessory viral proteins and are not directly involved in virus entry or exit (8, 39). The p10 FAST proteins are the smallest known viral fusion-inducing proteins, and they assume an unusual type I surface membrane topology in virus-infected and p10-transfected cells with a central signal-anchor sequence that positions ca. 40 amino acids on either side of the membrane. Although the ectodomain of p10 contains a potential fusion peptide motif, the limited size of the ectodomain and the absence of extended heptad repeat structures capable of forming coiled coils appear to be incompatible with a model of membrane fusion that would require extensive conformational changes to drive the fusion reaction (39).
Baboon reovirus (BRV) represents a distinct species of orthoreovirus that shares with the other fusogenic reoviruses the unusual ability to induce syncytium formation (7). Previous sequence analysis indicated extensive sequence diversity between the gene products of BRV and the homologous gene products of other orthoreoviruses (5). With the expectation that this level of evolutionary diversity might offer additional insights into FAST protein structure and function, we undertook the identification of the BRV fusion-inducing protein. Our present analysis indicates that the BRV S4 genome segment is functionally bicistronic, encoding two 140- to 141-amino-acid BRV-specific gene products. The gene product of the 5"-proximal open reading frame (ORF) of S4, p15, was identified as the protein responsible for BRV-induced syncytium formation. Sequence analysis revealed that BRV p15 shares no sequence or predicted structural similarity to the p10 proteins of ARV or NBV and, as such, represents a novel member of the FAST protein family.
MATERIALS AND METHODS
Viruses and cells.
BRV was isolated from the brain tissue of a baboon with meningoencephalomyelitis (7, 23) and was obtained from Julia Hilliard (Southwest Foundation for Biomedical Research, San Antonio, Tex.). The virus was plaque purified, and high titer stocks were grown in African green monkey (Vero) cells as previously described (7). The Vero cells were purchased from the ATCC (CCL-81) and maintained at 37°C in a 5% CO2 atmosphere in medium 199 with Earle's salts containing 5% heat-inactivated fetal bovine serum and 100 U of penicillin and streptomycin per ml.
Cloning.
The procedures for cDNA cloning and sequencing of the BRV S4 genome segment presented in this report (GenBank accession number AF406787) have been described previously (5). The full-length cDNAs corresponding to the S1, S2, S3, and S4 genome segments of BRV were cloned into the pBluescript II SK vector (Stratagene) by using the NotI site. The S4 genome segment and the p15 ORF were separately subcloned into the pcDNA3 vector (Invitrogen) by using the forward primer 5"-CGGGATCCATGGGTCAAAGACATTCAAATAG-3" and the reverse primers 5"-ATAGTTTAGCGGCCGCTCAGTTAACGAGCCCAACCTGCTCAGTC-3" or 5"-ATAGTTTAGCGGCCGCTCAGTTAACGTTGATACTTCCATCTGG-3" (underlined sequences are complementary to the 3" ends of the p16 or p15 ORFs, respectively). The forward primer added a BamHI site to the 5" end of the S4- and p15-pcDNA3 constructs. The reverse primers added 3"-terminal HpaI and NotI sites to the S4- and p15-pcDNA3 constructs, as well as additional C-terminal Val and Phe residues to p16 or an additional C-terminal Asn residue to p15 (the insertion of the HpaI site before the termination codons was intended to facilitate future in-frame insertions; the additional residues do not affect function of p15). The p16 ORF was subcloned from the S4 genome segment in pBluescript II, by PstI and NotI restriction digestion. The p16 ORF in pBluescript II was subsequently subcloned into pcDNA3 by HindIII and NotI restriction digestion.
Radiolabeled cell lysates.
Vero cells were infected with BRV at a multiplicity of infection of 5 PFU per cell in medium containing 1% fetal bovine serum. To prepare radiolabeled cell lysates, uninfected or BRV-infected monolayers were labeled 12 h postinfection for 1 h with 100 μCi of [3H]leucine (Amersham Pharmacia Biotech) per ml of leucine-free medium, prepared from the MEM Select-Amine kit (Life Technologies) according to the manufacturer's instructions. Alternatively, uninfected or BRV-infected monolayers were labeled 12 h postinfection for 4 h with 20 μCi of [3H]myristate (Amersham Pharmacia Biotech) per ml of Eagle MEM (Life Technologies). Radiolabeled monolayers were then rinsed three times with phosphate-buffered saline (PBS; 140 mM NaCl, 2.5 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4) and lysed in cold radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 8; 150 mM NaCl, 1 mM EDTA, 1% IGEPAL CA-630 [Sigma], 0.5% sodium deoxycholate; 0.1% sodium dodecyl sulfate [SDS]) containing protease inhibitors (200 nM aprotinin, 1 μM leupeptin, and 1 μM pepstatin [all from Sigma]). Cell lysates were harvested, and the nuclei were pelleted for 1 min at 13,000 × g in a bench-top microfuge.
Radiolabeled BRV.
To prepare radiolabeled BRV, infected monolayers were labeled at 10 h postinfection for 2 h with 100 μCi of [3H]leucine per ml of leucine-free medium and then chased with 1.2 μg of leucine/ml for an additional 2 h. Monolayers were rinsed three times with cold PBS, scraped from the tissue culture dishes, and passed through a 26.5-gauge needle 10 times. Since BRV is unstable in CsCl, virus was purified by extraction and differential centrifugation. Briefly, cellular debris was removed by centrifugation (20 min at 10,000 × g), and the virus-containing supernatant was pelleted through a 2-ml sucrose cushion (30% sucrose in medium) for 1 h at 27,000 rpm in an SW40 rotor (Beckman). Concentrated BRV pellets were resuspended in medium.
In vitro transcription and translation.
BRV S1 uncapped transcripts were synthesized from BamHI linearized pBluescript II plasmid by using bacteriophage T3 RNA polymerase. BRV S2 and S3 uncapped transcripts were synthesized from SacII-linearized pBluescript II plasmids by using bacteriophage T7 RNA polymerase. BRV S4, p15, and p16 uncapped transcripts were synthesized from XhoI-linearized pcDNA3 plasmids by using bacteriophage T7 RNA polymerase. All transcription reactions were performed with 1 μg of template DNA, 2.5 mM concentrations of nucleoside triphosphates, and 125 U of polymerase and then incubated 2 h at 37°C. All transcription reagents were obtained from Life Technologies. RNA (250 ng per 50-μl reaction) was translated in the presence of [3H]leucine (1 μCi per 50-μl reaction) by using nuclease-treated rabbit reticulocyte lysates (Promega) according to the instructions provided by the manufacturer.
Antibodies.
To generate p16- and p15-specific antisera, p16 and the C-terminal domain of p15 (residues 90 to 140) were cloned in frame with maltose-binding protein (MBP) by using the XmnI and BamHI sites of the pMAL-c2 vector (New England Biolabs) and transformed into E. coli DH5α cells. The MBP-p16 and MBP-p15-C chimeric proteins were separately expressed and purified from Escherichia coli on amylose affinity columns, according to the protocol suggested by the suppliers of the pMal-c2 vector, and then used to immunize rabbits. Each rabbit was injected with 0.1 mg of p15-C or 0.5 mg of p16 in Freund complete adjuvant, administered at three sites (two intramuscular and one subcutaneous), and then boosted three more times with protein in Freund incomplete adjuvant.
Membrane association analysis.
To investigate the nature of p15-membrane interactions, p15 transcripts were translated in vitro in the presence of canine pancreatic microsomes (Promega; 4 μl per 50-μl reaction). The translation reaction was diluted 1:10 in 20 mM Tris-HCl (pH 7.4) and separated into the soluble and membrane-containing insoluble fractions by centrifugation (30 min at 100,000 × g by using a Beckman TLS 55 rotor) through a 200-μl sucrose cushion (250 mM sucrose in 20 mM Tris-HCl [pH 7.4]). The insoluble fraction was subsequently extracted with 0.1 M sodium carbonate (pH 11.5) and recentrifuged as described above.
The association of p15 with membranes was also examined in BRV-infected cells. [3H]leucine-labeled, BRV-infected or mock-infected cell lysates were separated into soluble and membrane fractions by centrifugation in a Beckman TLS-55 rotor at 100,000 × g for 30 min through a 200-ml sucrose cushion as previously described (39). Proteins associated with the membrane pellet were solubilized with RIPA buffer, and the contaminating virus particles were removed by further centrifugation as described above.
Immune precipitation.
Aliquots of [3H]leucine-labeled, in vitro-translated protein (1/10 translation reaction) or [3H]leucine-labeled cell lysate (200,000 cpm) were immune precipitated in RIPA buffer for 1 h by using a 1:100 dilution of anti-p15, anti-p16, or normal rabbit sera. Immune complexes were recovered by using fixed Staphylococcus aureus cells (14), washed extensively with RIPA buffer, and released by boiling in SDS protein sample buffer (22) prior to SDS-15% polyacrylamide gel electrophoresis (PAGE) with polyacrylamide gels and fluorography (2).
Cell staining.
Vero cell monolayers grown in six-well plates were transfected with pcDNA3-encoded BRV genes (S4, S4 ORF1, and S4 ORF2) by using Lipofectamine (Life Technologies) and stained 18 h later with Wright-Giemsa stain (Diff-Quik) according to the manufacturer's instructions (VWR Scientific) to visualize cell nuclei and polykaryon formation. Images of stained cells were visualized on a Nikon Diaphot inverted microscope at ×200 magnification and captured by using computer software Image-Pro Plus (v.4.0).
RESULTS
BRV σ-class protein profile.
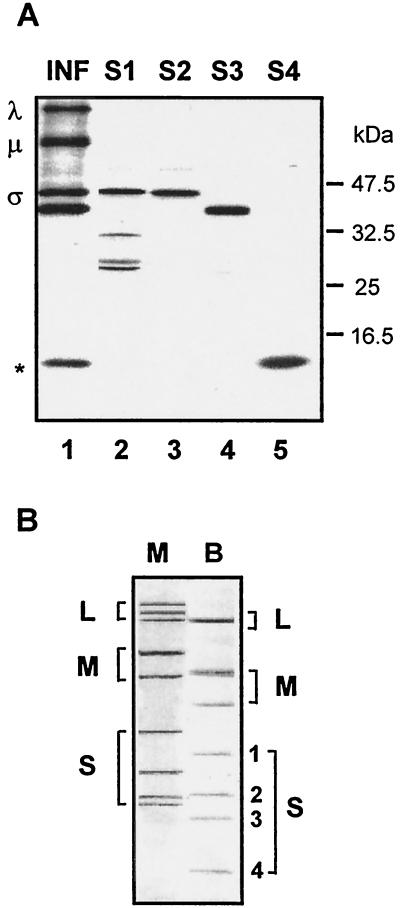
We sought to determine whether BRV encoded a p10 homologue within its S-class genome segments. Sequence analysis previously revealed that the BRV S1, S2, and S3 genome segments encoded the major σ-class core, outer capsid, and nonstructural proteins of the virus (5). We therefore undertook a molecular analysis of the remaining S-class genome segment of BRV, S4. As shown in Fig. 1A, the gene products of the S1, S2, and S3 genome segments, derived by in vitro transcription and translation, correspond to the dominant σ-class proteins present in [3H]leucine-labeled BRV-infected cell lysates. The products of the S1 ORF (major core protein; 413 codons) and the S2 ORF (major outer capsid protein; 396 codons) comigrate on SDS-15% polyacrylamide gels (Fig. 1A, lanes 2 and 3), slightly behind the product of the S3 ORF (major nonstructural protein; 353 codons; Fig. 1A, lane 4). Interestingly, a low-molecular-mass polypeptide of ca. 15 kDa was detected in the BRV-infected cell lysates that comigrated with a product of the S4 gene (Fig. 1A, lane 5). The migration pattern of the S4-specific polypeptide was surprising since the relative gel mobility of the BRV S4 genome segment (Fig. 1B) suggested a molecular size of ca. 800 to 900 bp, sufficient to encode a protein at least twice the size of the 15-kDa polypeptide generated by in vitro translation of S4.
FIG. 1.
(A) Identification of the σ-class proteins encoded by the BRV S-class genome segments. The relative gel mobilities of the in vitro translation products of the BRV S1, S2, S3, and S4 genome segments, resolved by SDS-15% PAGE, are compared to the dominant viral bands present in [3H]leucine-labeled BRV-infected cell lysate (INF). The positions of the λ-, μ-, and σ-class reovirus proteins are shown at the left. The asterisk denotes the location of a ca. 15-kDa polypeptide present in the infected cell lysate that comigrates with the translation product(s) of the S4 genome segment. The relative mobilities of molecular mass markers are indicated at the right. (B) Double-stranded RNA genome segment profiles of mammalian reovirus type 3, Dearing (M), and baboon reovirus (B) resolved by SDS-10% PAGE and detected by silver staining. The positions of the BRV S1 (1,311 nt), S2 (1,253 nt), S3 (1,150 nt), and S4 genome segments are indicated on the right.
BRV S4 is functionally bicistronic.
Consistent with the in vitro translation results, the cDNA sequence of the BRV S4 genome segment indicated the presence of two sequential partially overlapping ORFs rather than a single ORF spanning most of the genome segment (Fig. 2). Two independent clones yielded the same sequence, confirming the bicistronic gene arrangement of BRV S4. The first ORF contains 140 codons and encodes a predicted 15-kDa protein (molecular mass of 15,223 Da), termed p15. The second ORF encodes a predicted 16-kDa protein (molecular mass of 16,723 Da), p16, from one of two adjacent in-frame potential initiator methionine codons. Both methionine codons are in a preferred context for an initiator methionine with purines in the −3 and +4 positions (20, 21), and it is unclear which functions as the initiation site for the 140- to 141-amino-acid p16 protein.
FIG. 2.
The BRV S4 genome segment contains two partially overlapping ORFs. The cDNA sequence of the BRV S4 genome segment is shown along with the predicted amino acid sequences encoded by two sequential ORFs. ORF1 (nt 25 to 447) encodes p15, a 140-amino acid protein with a predicted molecular mass of 15 kDa. ORF2 (nt 413 to 835) encodes p16, a 140- to 141-amino-acid protein with a predicted molecular mass of 16 kDa. The locations of the p15 start codon, the two adjacent potential p16 start codons, and the other two methionine codons that precede the p16 ORF, are underlined. The nucleotides comprising the 5"- and 3"-terminal noncoding regions are in lowercase. Nucleotide and amino acid numbering is indicated at the right.
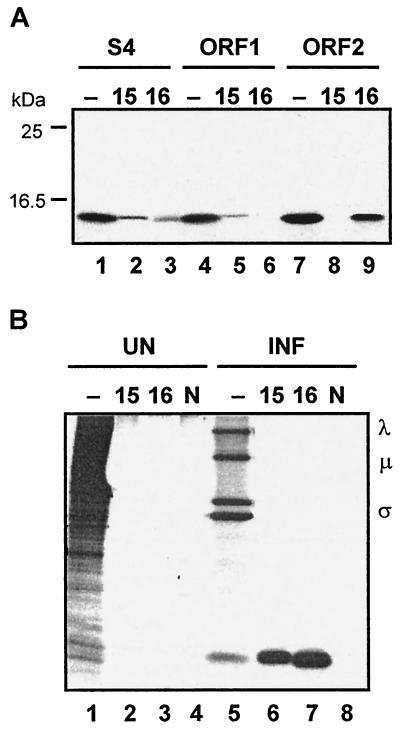
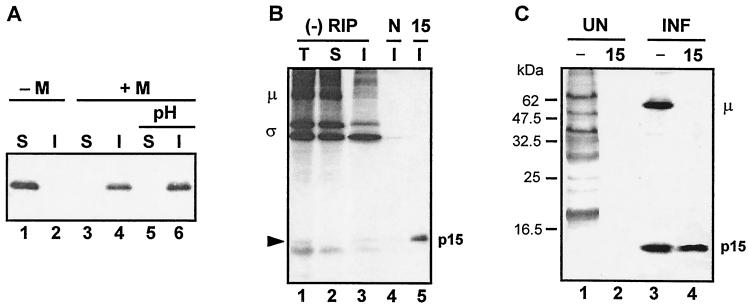
To determine which of the two S4 ORFs produced the ca. 15-kDa band evident in the σ-class protein profile (Fig. 1, lane 5), the individual S4 ORFs were subcloned and separately translated in vitro. The presence of specific translation products was detected by immune precipitation with polyclonal rabbit antiserum raised against the products of either ORF1 or ORF2. While both antisera recognized an ca. 15-kDa polypeptide produced in the S4 translation mixture (Fig. 3A, lanes 1 to 3), only the anti-p15 serum reacted with the product of ORF1 (Fig. 3A, lanes 4 to 6), while only the anti-p16 serum reacted with the product of ORF2 (Fig. 3A, lanes 7 to 9). The specific antisera were also used to confirm that both ORFs were functional in virus-infected cells (Fig. 3B). Concurrent with the strong inhibition of host cell translation and abundant synthesis of the major λ-, μ-, and σ-class proteins evident in BRV-infected cell lysates, synthesis of both p15 and p16 was detected in cells productively infected with BRV (Fig. 3B, lanes 5 to 8). The specificity of the immune precipitations was evident by the absence of the 15-kDa polypeptides in uninfected cells (Fig. 3B, lanes 1 to 4) and by the inability of normal rabbit serum to precipitate these proteins (Fig. 3B, lanes 4 and 8). These results clearly indicated that the S4 genome segment of BRV is functionally bicistronic, encoding two distinct BRV-specific proteins that comigrate on SDS-15% polyacrylamide gels.
FIG. 3.
The BRV S4 genome segment is functionally bicistronic. (A) In vitro translation products of the entire S4 genome segment (lanes 1 to 3), S4 ORF1 (lanes 4 to 6), or S4 ORF2 (lanes 7 to 9) were resolved by SDS-15% PAGE and detected by fluorography. Translation reactions were fractionated without prior immunoprecipitation (−) or after immunoprecipitation with polyclonal antiserum raised against the product of ORF1, p15 (15), or the product of ORF2, p16 (16). The relative mobilities of molecular mass markers are indicated on the left. (B) [3H]leucine-labeled uninfected (UN) and BRV-infected (INF) cell lysates were fractionated by SDS-15% PAGE either without prior immune precipitation (−) or after immunoprecipitation with anti-p15 serum (15), anti-p16 serum (16), or normal rabbit serum (N). The positions of the λ-, μ-, and σ-class reovirus proteins are indicated on the right.
BRV S4 ORF1 encodes a nonstructural membrane fusion-inducing protein.
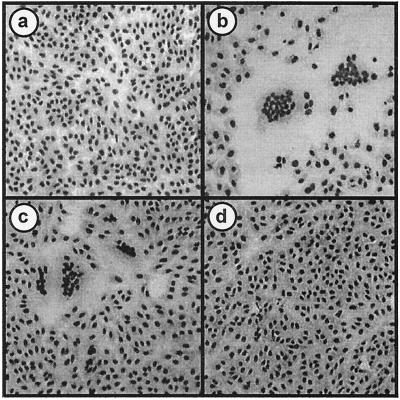
Transfection of Vero cells with the S4 genome segment by itself induced syncytium formation (Fig. 4b), while the other S-class genome segments failed to promote cell-cell fusion (data not shown). Clearly, one or both of the S4 gene products is responsible for virus-induced syncytium formation. Expression of ORF1 resulted in syncytium formation equivalent to that induced by the entire S4 genome segment (Fig. 4c), while expression of ORF2 induced no cell fusion (Fig. 4d). Furthermore, transfection of increasing concentrations of the ORF1 expression plasmid generated more-extensive syncytium formation (data not shown). These results indicated that the gene product of the first ORF of the S4 genome segment is the only BRV protein required to induce cell-cell fusion. Sequence comparisons of p15 with the p10 FAST proteins of ARV and NBV revealed no significant sequence similarity (data not shown). Similarly, BLAST searches failed to identify any obvious sequence similarity to other proteins in the database. The BRV p15 protein, therefore, appears to represent a new member of the FAST protein family.
FIG. 4.
ORF1 of the S4 genome segment encodes the membrane fusion-inducing protein of BRV. Vero cells were transfected with pcDNA3 (a), pcDNA3-S4 (b), pcDNA3-S4 ORF1 (c), or pcDNA3-S4 ORF2 (d). Transfected monolayers were Wright-Giemsa stained 18 h posttransfection to detect the formation of multinucleated syncytia.
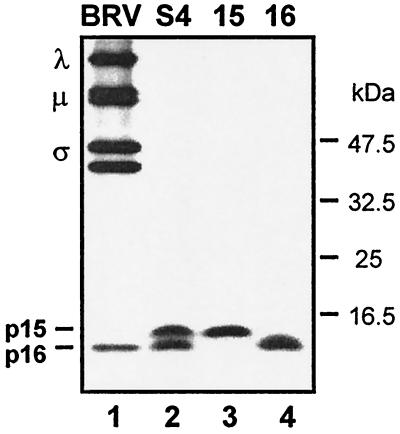
Since the p10 fusion proteins of ARV and NBV are nonstructural proteins (39), we sought to determine whether the BRV p15 fusion protein is also a nonstructural viral protein. Concentrated, radiolabeled virus particles prepared from BRV-infected cell lysates revealed the characteristic reovirus protein profile with the major λ-, μ-, and σ-class proteins in addition to an ca. 15-kDa polypeptide (Fig. 5, lane 1). Since the S4 gene products comigrate by SDS-15% PAGE, we used Tricine-SDS-16.5% PAGE to better resolve p15 and p16. Under such conditions, the electrophoretic mobility of in vitro translated p16 is slightly greater than that of p15, and p16 comigrated with the smallest σ-class protein observed in BRV particles (Fig. 5, lane 2). Immunoprecipitation of detergent-solubilized BRV virions with the p15- and p16-specific antisera confirmed the identification of the smallest σ-class particle-associated protein as being p16 and not p15 (S. Dawe and R. Duncan, unpublished data). These results indicated that BRV p15, as with the p10 proteins of ARV and NBV, is a nonstructural membrane fusion-inducing protein.
FIG. 5.
BRV p15 is a nonstructural viral protein. The relative gel mobilities of the radiolabeled in vitro translation products of BRV S4, S4 ORF1 (15), and S4 ORF2 (16) were resolved by Tricine-SDS-16.5% PAGE and compared to the viral bands present in concentrated [3H]leucine-labeled BRV particles. The positions of the λ-, μ-, and σ-class reovirus proteins and of the p15 and p16 proteins encoded by the BRV S4 genome segment are shown on the left. The migration of molecular mass markers is indicated on the right.
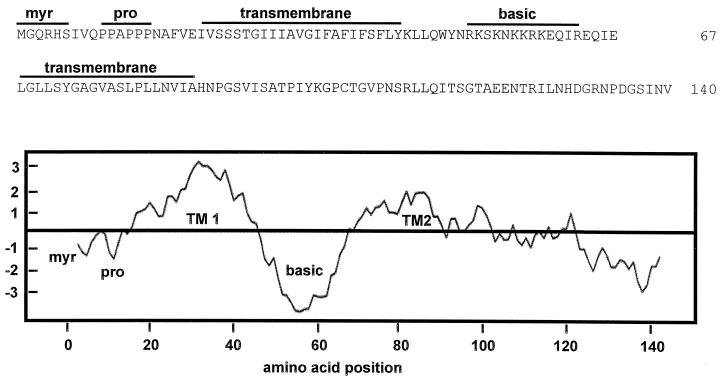
Sequence-predicted structural motifs in p15.
Several structural motifs predicted from the p15 amino acid sequence are indicated in Fig. 6. Two stretches of predominantly apolar amino acids were identified, between residues 21 and 43 and between residues 68 and 87. Both of these apolar regions were identified as presumptive transmembrane domains (TM domains) by using the TMpred (30), TMAP (31), SOSUI (15), and HMMTOP (46) algorithms. The intervening region (residues 44 to 63) between the two hydrophobic domains is highly basic. This region, containing 9 of the 14 positively charged residues present in p15, contributes substantially to the overall basic character of p15 (pI = 9.8). Additional motifs identified in p15 include an N-terminal myristylation consensus sequence (MGXXXS/T) and a proline-rich motif (PPAPPP; residues 10 to 15). The absence of extended heptad repeat sequences implies that p15 does not contain coiled-coil regions, suggesting that p15 cannot promote membrane fusion according to the proposed model for class I viral fusion proteins. Furthermore, aside from the predicted TM domains, the hydropathy profile of p15 reveals no additional hydrophobic clusters of sufficient size or hydrophobicity (i.e., >15 residues, hydrophobic index of >0.5) (51) that might function as a fusion peptide, a motif common to the majority of enveloped virus fusion proteins.
FIG. 6.
Sequence-predicted structural motifs and hydropathy profile of p15. The amino acid sequence of p15 is presented in the top panel, along with the locations of several predicted structural motifs, including a myristylation consensus sequence (myr), a polyproline helix (pro), a basic region, and two predicted TM domains. The bottom panel shows the hydropathy profile of p15 as determined by using the hydrophobicity scale of Kyte and Doolittle, averaged over a window of 11 residues. Regions above the horizontal line are hydrophobic, while those below the line are hydrophilic.
p15 is a myristylated integral membrane protein.
Of the motifs predicted from the primary structure of p15, the predicted TM domains and the myristylation consensus sequence potentially offer the greatest functional information since these motifs are known to play a role in protein subcellular localization and membrane interactions (26, 33). The TM domains imply that p15 is an integral membrane protein. We confirmed this prediction by in vitro translation of p15 in the presence of canine pancreatic microsomes. While p15 translated in the absence of membranes partitioned exclusively to the soluble fraction (Fig. 7A, lanes 1 and 2), all of the p15 translated in the presence of microsomal membranes was associated with the pelleted membrane fraction (Fig. 7A, lanes 3 and 4). Extraction of the membrane pellet with a high-pH buffer failed to extract p15 (Fig. 7A, lanes 5 and 6) consistent with p15 assuming an integral transmembrane topology and suggesting that p15 contains at least one TM domain.
FIG. 7.
BRV p15 is a myristylated integral membrane protein. (A) BRV p15 was translated in vitro without or with microsomal membranes (−M or +M) in the presence of [3H]leucine. The translation products were separated into the soluble fraction (S) and the insoluble membrane fraction (I) by centrifugation. Membrane fractions were subsequently extracted with a high-pH buffer (pH) to removal peripheral membrane-associated proteins and refractionated into the soluble and insoluble fractions by centrifugation. (B) BRV-infected cells were radiolabeled with [3H]leucine, and cell lysates were fractionated into the total (T), soluble (S), and insoluble membrane (I) fractions. Samples were resolved by SDS-PAGE either without prior immunoprecipitation (−RIP) or after immunoprecipitation with normal preimmune rabbit serum (N) or anti-p15 rabbit immune serum (15). The locations of the major μ and σ proteins of BRV are indicated on the left, and the location of p15 detected by immunoprecipitation is indicated on the right. The arrow indicates a faint polypeptide present in the total and insoluble fractions that comigrates with p15. (C) Uninfected (UN) and BRV-infected (INF) [3H]myristate-labeled cell lysates were resolved by SDS-15% PAGE either without prior immunoprecipitatation (−) or after immunoprecipitatation with anti-p15 serum (15). The positions of the myristylated μ-class reovirus protein and p15 are shown on the right. The locations of molecular mass markers are indicated on the left.
The membrane association property of p15 was confirmed in vivo. Examination of total cell lysates from radiolabeled BRV-infected cells by SDS-PAGE, as well as the isolated soluble and insoluble membrane fractions, revealed a faint band present in the total and insoluble membrane fractions (Fig. 7B, lanes 1 and 3) that migrated with the expected molecular mass of p15. The presence of p15 in the membrane fraction was confirmed by immunoprecipitation with the p15-specific antiserum (Fig. 7B, lane 5). The presence of the soluble m-class viral protein in the soluble fraction and its absence from the insoluble fraction (Fig. 7B, lanes 2 and 3) was an indication that the presence of p15 in the membrane fraction was unlikely to be simply the result of contamination of the membrane fraction by soluble p15. These results are consistent with the in vitro translation results and with the sequence analysis, and imply that p15 is an integral membrane protein.
To determine whether the N-terminal myristylation consensus sequence of p15 is functional, uninfected and infected cells were labeled with [3H]myristate and immunoprecipitated with p15-specific antiserum. The p15 antiserum specifically precipitated a 15-kDa radiolabeled polypeptide from infected cell lysates that was absent from uninfected cell lysates (Fig. 7C). Compared to the [3H]leucine-labeled protein profile of uninfected and infected cells (Fig. 3B, lanes 1 and 4), only a subset of cellular and viral proteins were [3H]myristate labeled (Fig. 7C, lanes 1 and 3), indicating that the radiolabeled myristate moiety was not metabolized into amino acids that would nonspecifically label viral proteins. Furthermore, substitution of the penultimate N-terminal glycine residue with alanine eliminated p15 labeling with [3H]myristate (data not shown). These results indicate that p15 is a myristylated protein. BRV-infected cell lysates also contained a myristylated polypeptide in the μ-class size range which likely corresponds to the structural protein μ1 of MRV and its homologue in ARV, μ2, which are known to be myristylated (27, 47).
DISCUSSION
The unique nature of the BRV S4 genome segment.
Previous sequence analysis determined that the BRV S1, S2, and S3 genome segments encode the major σ-class core, outer capsid, and nonstructural proteins of the virus, homologous to the products of the S2, S3, and S4 genome segments of other reoviruses (5). We speculated that the remaining BRV S-class genome segment, S4, might represent a truncated version of the S1 genome segment of other orthoreoviruses. The MRV S1 genome segment is bicistronic, encoding the σ1 cell attachment protein and the small nonstructural protein, σ1NS, implicated in cell cycle arrest (10, 18, 32, 35). The S1 genome segments of the fusogenic ARV and NBV are tricistronic, containing a 5"-proximal ORF encoding the fusion protein in addition to the downstream genes encoding the σ1 cell attachment protein homologue, σC, and the small nonstructural protein, p17, of unknown function (40). BRV S4 shares with the S1 segment of the other reoviruses its polycistronic nature and, like the other fusogenic reoviruses, contains a 5"-proximal cistron encoding a small hydrophobic fusion protein. However, there is no significant sequence similarity between p15 and p10 and these two proteins possess a unique repertoire of structural motifs (see below), suggesting that p15 is a new member of the FAST protein family. Similarly, analysis of the p16 protein encoded by the downstream ORF in the BRV S4 genome segment revealed p16 shares no sequence or structural similarity to either the orthoreovirus σ1/C cell attachment proteins nor the small nonstructural proteins σ1NS or p17, the other gene products encoded by the S1 genome segments of the other orthoreoviruses (Dawe and Duncan, unpublished). Therefore, the BRV S4 genome segment encodes two novel BRV-specific proteins with no identified homology to any other viral or cellular proteins.
Bicistronic gene arrangement of BRV S4.
The p15 and p16 ORFs are organized on the bicistronic BRV S4 genome segment in an unusual sequential, partially overlapping fashion with start codons that are separated by 388 nucleotides (nt) and whose reading frames overlap by only 38 nt. A similar situation exists for the functionally tricistronic S1 genome segments of ARV and NBV that contain three sequential ORFs with minimal overlap (40). This situation is unlike the majority of polycistronic viral mRNAs, which contain two or more translation start codons located relatively close together resulting in significant overlap between the ORFs (37). Leaky scanning (20, 21) accounts for the ability of ribosomes to initiate translation at two alternate start sites in the majority of these bicistronic viral mRNAs.
Despite the considerable distance separating the two translation start sites on the BRV S4 genome segment, leaky scanning may account for the translation of p16. Although rare, ribosomes are known to scan as much as 900 nt (more than twice the distance separating p15 and p16 on S4) in order to initiate translation (24). The initiator methionine codon for p15 (UAC AUGG) lies within a suboptimal context, with a pyrimidine at the −3 position, which may enable preinitiation complexes to scan through the p15 ORF in order to initiate translation of p16 at one of two potential tandem initiator methionine codons, both of which exist in a preferred context for translation initiation (purines at −3 and +4). Scanning preinitiation complexes would also need to bypass two additional suboptimal AUG codons in the +1 reading frame relative to the p15 ORF (Fig. 2). These AUG codons precede two minicistrons encoding four or six amino acids. Extended scanning past multiple upstream AUGs has been previously reported. For example, leaky scanning through 350 nt, including five upstream AUGs, accounts for the translation of the G protein from the borna disease virus unspliced M/G transcript (41).
Although leaky scanning may be solely responsible for translation of the bicistronic BRV S4 genome segment, alternative translation initiation mechanisms are operative with other bicistronic mRNAs whose ORFs are arranged in a sequential manner with minimal overlap. For example, translation of the hepatitis B virus P protein from the bicistronic C/P mRNA is influenced by translation of an upstream seven-amino-acid minicistron and by ribosome backscanning following translation termination of the C protein (11, 17). A similar backscanning mechanism contributes to expression of the ORF-2 protein of the human respiratory syncytial virus bicistronic M2 gene (1). Functional translation analysis of the BRV S4 genome segment is required to determine whether leaky scanning alone accounts for p16 expression or whether alternate forms of translational control may be operative.
p15 is a nonstructural membrane fusion-inducing protein.
Transfection analysis identified p15 as only the second example of a membrane fusion-inducing protein encoded by a nonenveloped virus. The absence of detectable p15 in concentrated BRV particles (Fig. 5) suggests that p15 is a nonstructural protein of the virus, a situation similar to that previously reported for the FAST proteins of ARV and NBV (39). The reovirus FAST proteins are, therefore, distinguished from the fusion proteins of enveloped viruses in both their nonstructural nature and their derivation from nonenveloped viruses. In the case of ARV, the primary purpose of the FAST proteins appears to be the triggering of cell-cell fusion and syncytium formation as a means of promoting a rapid lytic response and an enhanced rate of virus release, properties that correlate with the relative pathogenicity of the virus (8, 6). In view of the potential pathogenicity of BRV in nonhuman primates (7, 23), the syncytium-inducing property of p15 may similarly contribute to the pathogenic nature of BRV.
p15 is a rare example of a myristylated fusion-inducing protein.
The N-terminal myristylation consensus sequence of p15 is functional, as demonstrated by the ability of p15 to be specifically labeled by [3H]myristate in infected cell lysates (Fig. 7B). Recent analysis of a myristylation-minus mutant (G2A) also indicates that myristylation is required for the fusogenic activity of p15 (Dawe and Duncan, unpublished). While fatty acylation of fusion proteins is common, the vast majority of viral fusion proteins that are acylated contain palmitate attached to membrane-proximal cysteine residues rather than myristate attached to an N-terminal glycine residue (3, 28, 38, 48, 52). The relatively short 14-C myristate moiety is known to reversibly associate with lipid bilayers, a property that facilitates the reversible membrane association of numerous cytosolic myristylated proteins (33). Consequently, the myristylated N terminus of p15 may reversibly associate with cell membranes.
The only other example of a viral protein involved in membrane fusion that contains an N-terminal myristate moiety is the hepadnavirus large surface antigen (L protein) which contains a myristylated pre-S domain (29). The mechanism of hepadnavirus-mediated membrane fusion remains unclear but may involve proteolytic cleavage of L to remove the myristylated pre-S domain thereby exposing the potential fusogenic S protein (36). Immunoprecipitation analysis of p15-transfected or BRV-infected cells by using polyclonal antisera specific for the C-terminal domain of p15 (Fig. 3) provided no evidence to suggest that a similar precursor protein relationship exists for BRV p15. The fusogenic form of p15, therefore, is likely represented by the myristylated species of the protein. It is not clear whether the myristate moiety contributes directly to the fusion reaction, or whether it indirectly influences cell-cell fusion, for example, by affecting p15 subcellular localization to lipid microdomains (33).
p15 is a basic integral membrane protein.
Sequence analysis of p15 identified two potential TM domains, suggesting that p15 is an integral membrane protein. In vitro and in vivo analysis supported the transmembrane nature of the protein (Fig. 7). The fact that association of p15 with the membrane fraction resisted extraction with high pH indicated that p15 membrane association was unlikely to be peripheral and mediated by either ionic interactions with the polybasic region or by reversible membrane insertion of the myristate moiety. These results suggest that either or both of the predicted TM domains in p15 are functional.
Sequence analysis also revealed that the p15 protein is a basic protein with the majority of positively charged residues residing in a discrete centrally located polybasic region that separates the two predicted TM domains (Fig. 6). The presence of a polybasic region in p15 is shared by the ARV and NBV p10 FAST proteins that contain a similar region on the carboxy-proximal side of an internal signal-anchor sequence (39). Interestingly, TM domains with an adjacent polybasic region are the hallmark features of a diverse group of small hydrophobic proteins, termed viroporins, that are encoded by numerous enveloped and nonenveloped viruses (4). Although the viroporins are not known to induce membrane fusion, they are involved in interactions that destabilize membranes, leading to cytopathic effects and enhanced virus release. The combination of the TM domains and adjacent basic domains in the reovirus FAST proteins may contribute to destabilization of the donor membrane as part of the FAST-mediated membrane fusion reaction.
Topological and functional implications of the p15 structural motifs.
As expected for a membrane fusion protein and as demonstrated (Fig. 7), p15 appears to be an integral membrane protein. The presence of two potential membrane-spanning domains, identified by numerous different search algorithms, implies that p15 adopts a polytopic membrane topology. Such a topology is rarely displayed by enveloped virus fusion proteins. The hepadnavirus surface antigen is currently the only known example of a polytopic viral protein involved in membrane fusion (13, 45). Since myristylated proteins are localized almost exclusively to the cytoplasmic leaflet (33), the N-terminal myristylation of p15 suggests an N-terminus-in topology. However, the myristylated N terminus of a fraction of the hepadnavirus L proteins is posttranslationally translocated across the membrane (12). In addition, since the polybasic region of p15 resides on the C-proximal side of the first predicted TM domain (Fig. 6), the basic inside rule (25, 49) supports an external topology for the myristylated N terminus of p15. Consequently, the orientation of p15 in the membrane is unclear. It is also unclear whether p15 is a bitopic or polytopic membrane protein. If both of the hydrophobic regions of p15 function as TM domains, then p15 lacks an essential feature of almost all viral membrane fusion proteins, namely, a fusion peptide motif. Aside from the two predicted TM domains, no other significant stretch of hydrophobic residues in p15 shares properties characteristic of a fusion peptide (51). If one of the two predicted TM domains actually functions as a fusion peptide, then the relative hydrophobicities, calculated by using the Eisenberg normalized consensus scale (9), and the glycine-alanine content suggest the second TM domain may serve as a fusion peptide (hydrophobicity of 0.59 with 30% glycine-alanine content). Direct topological analysis of p15 is clearly required if we are to understand the mechanisms by which this unusual fusion-inducing protein promotes membrane fusion. Such studies are currently under way.
BRV p15 represents a new class of reovirus fusion protein within the FAST protein family.
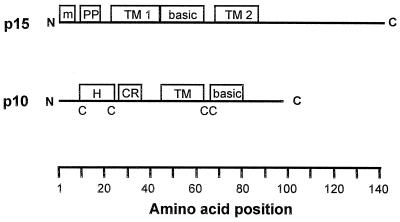
The only known examples of nonenveloped viruses that induce syncytium formation are all members of the family Reoviridae (5, 7). Furthermore, the other S-class genome segment-encoded proteins of BRV share clear sequence similarity to the homologous proteins encoded by other orthoreoviruses (5). It was, therefore, extremely surprising to find that BRV p15 not only shares no significant sequence similarity with the p10 fusion proteins of ARV and NBV (data not shown) but that the predicted domain organizations of these different reovirus FAST proteins share little similarity. A schematic comparison of the sequence-predicted domains of p15 and p10 is shown in Fig. 8.
FIG. 8.
Sequence-predicted structural motifs in BRV p15 and the p10 proteins of ARV and NBV. The linear arrangement, drawn to approximate scale, of sequence-predicted structural motifs of the BRV p15 protein and the p10 proteins of ARV and NBV are shown. The amino acid positions are indicated at the bottom of the figure. TM, TM domain; basic, positively charged region; m, myristylation site; PP, polyproline helix; H, hydrophobic peptide; CR, conserved motif present in ARV and NBV p10; C, conserved cysteine residues present in ARV and NBV p10 (the dicysteine motif adjacent to the TM domain in p10 is the site of palmitylation of the protein).
With a single centrally located TM domain, p10 assumes a type I (i.e., N-terminus-out) membrane topology (39). As discussed above, p15 most likely assumes either a polytopic or a type II bitopic topology. While p15 is N-terminally myristylated, p10 is internally palmitylated on two conserved cytoplasmic cysteine residues adjacent to the TM domain (39). The p15 protein contains a single cysteine residue located seventeen residues away from the C-terminal end of the second predicted TM domain (Fig. 6). Since palmitylated cysteine residues generally lie within, or immediately adjacent to, a TM domain, and since a C104A substitution has no effect on p15 fusion activity (data not shown), p15 is unlikely to be palmitylated. The N-proximal region of p15 contains a proline-rich motif (residues 10 to 15) that is absent from p10. Conversely, the N-proximal domain of p10 contains a hydrophobic patch and two additional cysteine residues, all of which are conserved in the p10 proteins of ARV and NBV and influence the fusion activity of the protein (39). There is also an N-proximal stretch of 10 amino acids that is completely conserved between the ARV and NBV p10 proteins; this region is absent in p15. The novel structural motifs of BRV p15 clearly distinguish this protein from the p10 fusion protein of ARV and NBV.
Based on the unique properties of p15, we propose that the BRV p15 protein represents a new member of the reovirus FAST protein family. The features that currently define the reovirus FAST protein family include: (i) their nonstructural nature; (ii) their small size; (iii) the presence of at least one TM domain and a second region of significant hydrophobicity; (iv) a membrane-proximal basic cluster; (v) modification by acylation; (vi) the lack of both an obvious fusion peptide and the coiled-coil motif typical of many enveloped virus fusion proteins; and (vii) the ability to trigger membrane fusion in the absence of any other viral proteins. It remains to be determined whether these shared characteristics contribute to a common mechanism of FAST protein-mediated membrane fusion or whether the unique features of these proteins determine divergent p10- and p15-induced fusion pathways. It seems clear, however, that both p10 and p15 function to promote membrane fusion independent of extensive conformational changes in large, complex, multimeric proteins, the proposed mechanism utilized by the enveloped virus fusion proteins. Further functional analysis of the reovirus FAST proteins is likely to lead to alternative models of protein-mediated membrane fusion.
Acknowledgments
We thank Jingyun Shou for outstanding technical assistance.
This research was funded by grants from the Canadian Institutes of Health Research (CIHR) and from the Natural Sciences and Engineering Research Council of Canada (NSERC). S.D. is the recipient of an NSERC Graduate Studentship and an Izaak Walton Killam (IWK) Graduate Scholarship. R.D. is the recipient of a CIHR Regional Partnership Program Investigator Award.
REFERENCES
- 1.Ahmadian, G., J. S. Randhawa, and A. J. Easton. 2000. Expression of the ORF-2 protein of the human respiratory syncytial virus M2 gene is initiated by a ribosomal termination-dependent reinitiation mechanism. EMBO J. 19:2681-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonner, W. M. 1984. Fluorography for the detection of radioactivity in gels. Methods Enzymol. 104:460-465. [DOI] [PubMed] [Google Scholar]
- 3.Caballero, M., J. Carabana, J. Ortego, R. Fernandez-Munoz, and M. L. Celma. 1998. Measles virus fusion protein is palmitoylated on transmembrane-intracytoplasmic cysteine residues which participate in cell fusion. J. Virol. 72:8198-8204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrasco, L. 1995. Modification of membrane permeability by animal viruses. Adv. Virus Res. 45:61-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duncan, R. 1999. Extensive sequence divergence and phylogenetic relationships between the fusogenic and nonfusogenic orthoreoviruses: a species proposal. Virology 260:316-328. [DOI] [PubMed] [Google Scholar]
- 6.Duncan, R., and K. Sullivan. 1998. Characterization of two avian reoviruses that exhibit strain-specific quantitative differences in their syncytium-inducing and pathogenic capabilities. Virology 250:263-272. [DOI] [PubMed] [Google Scholar]
- 7.Duncan, R., F. A. Murphy, and R. Mirkovic. 1995. Characterization of a novel syncytium-inducing baboon reovirus. Virology 212:752-756. [DOI] [PubMed] [Google Scholar]
- 8.Duncan, R., Z. Chen, S. Walsh, and S. Wu. 1996. Avian reovirus-induced syncytium formation is independent of infectious progeny virus production and enhances the rate, but is not essential, for virus-induced cytopathology and virus egress. Virology 224:453-464. [DOI] [PubMed] [Google Scholar]
- 9.Eisenberg, D. 1984. Three-dimensional structure of membrane and surface proteins. Annu. Rev. Biochem. 53:595-623. [DOI] [PubMed] [Google Scholar]
- 10.Ernst, H., and A. J. Shatkin. 1985. Reovirus hemagglutinin mRNA codes for two polypeptides in overlapping reading frames. Proc. Natl. Acad. Sci. USA 82:48-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fouillot, N., S. Tlouzeau, J. M. Rossignol, and O. Jean-Jean. 1993. Translation of the hepatitis B virus P gene by ribosomal scanning as an alternative to internal initiation. J. Virol. 67:4886-4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallina, A., and G. Milanesi. 1993. Trans-membrane translocation of a myristylated protein amino terminus. Biochem. Biophys. Res. Commun. 195:637-642. [DOI] [PubMed] [Google Scholar]
- 13.Grgacic, E. V. L., and H. Schaller. 2000. A metastable form of the large envelope protein of duck hepatitis B virus: low-pH release results in a transition to a hydrophobic, potentially fusogenic conformation. J. Virol. 74:5116-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Hirokawa, T., S. Boon-Cheng, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378-379. [DOI] [PubMed] [Google Scholar]
- 16.Hughson, F. M. 1997. Enveloped viruses: a common mode of membrane fusion? Curr. Biol. 7:R565-R569. [DOI] [PubMed] [Google Scholar]
- 17.Hwang, W. L., and T. S. Su. 1998. Translational regulation of hepatitis B virus polymerase gene by termination-reinitiation of an upstream minicistron in a length-dependent manner. J. Gen. Virol. 79:2181-2189. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs, B. L., and C. E. Samuel. 1985. The reovirus S1 mRNA encodes two primary translation products. Virology 143:63-74. [DOI] [PubMed] [Google Scholar]
- 19.Kielian, M., and S. Jungerwirth. 1990. Mechanisms of enveloped virus entry into cells. Mol. Biol. Med. 7:17-31. [PubMed] [Google Scholar]
- 20.Kozak, M. 1989. The scanning model for translation: an update. J. Cell Biol. 108:229-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozak, M. 1991. An analysis of vertebrate mRNA sequences: intimations on translational control. J. Cell Biol. 115:887-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.Leland, M. M., G. B. Hubbard, H. T. Sentmore, K. F. Soike, and J. K. Hilliard. 2000. Outbreak of orthoreovirus-induced meningoencephalomyelitis in baboons. Comp. Med. 50:199-205. [PubMed] [Google Scholar]
- 24.Liu, Q., and G. Hobom. 2000. Evidence for translation of VP3 of avian polyomavirus BFDV by leaky ribosomal scanning. Arch. Virol. 145:407-416. [DOI] [PubMed] [Google Scholar]
- 25.Matlack, K. E. S., W. Mothes, and T. A. Rapoport. 1998. Protein translocation: tunnel vision. Cell 92:381-390. [DOI] [PubMed] [Google Scholar]
- 26.McCabe, J. B., and L. G. Berthiaume. 1999. Functional roles for fatty acylated amino-terminal domains in subcellular localization. Mol. Biol. Cell 10:3771-3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nibert, M. L., L. A. Schiff, and B. N. Fields. 1991. Mammalian reoviruses contain a myristylated structural protein. J. Virol. 65:1960-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olsen, K. E. P., and K. B. Anderson. 1999. Palmitoylation of the intracytoplasmic R peptide of the transmembrane envelope protein in Moloney murine leukemia virus. J. Virol. 73:8975-8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Persing, D. H., H. E. Varmus, and D. Ganem. 1987. The preS1 protein of hepatitis B virus is acylated at its amino terminus with myristic acid. J. Virol. 61:1672-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Persson, B., and P. Argos. 1994. Prediction of transmembrane segments in proteins utilising multiple sequence alignments. J. Mol. Biol. 237:182-192. [DOI] [PubMed] [Google Scholar]
- 31.Persson, B., and P. Argos. 1997. Prediction of membrane protein topology utilizing multiple sequence alignments. J. Protein Chem. 16:453-457. [DOI] [PubMed] [Google Scholar]
- 32.Poggioli, G. J., C. Keefer, J. L. Connolly, T. S. Dermody, and K. L. Tyler. 2000. Reovirus-induced G2/M cell cycle arrest requires σ1s and occurs in the absence of apoptosis. J. Virol. 74:9562-9570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Resh, M. D. 1999. Fatty acylation of proteins: new insights into membrane targeting of myristylated and palmitoylated proteins. Biophys. Bichim. Acta 1451:1-16. [DOI] [PubMed] [Google Scholar]
- 34.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 35.Rodgers, S. E., J. L. Connolly, J. D. Chappell, and T. S. Dermody. 1998. Reovirus growth in cell culture does not require the full complement of viral proteins: identification of a σ1s-null mutant. J. Virol. 72:8597-8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez-Crespo, I., E. Nuñez, B. Yélamos, J. Gómez-Gutiérrez, J. P. Albar, D. L. Peterson, and F. Gavianes. 1999. Fusogenic activity of hepadnavirus peptides corresponding to sequences downstream of the putative cleavage site. Virology 261:133-142. [DOI] [PubMed] [Google Scholar]
- 37.Samuel, C. E. 1989. Polycistronic animal virus mRNAs. Prog. Nucleic Acid Res. Mol. Biol. 37:127-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt, M., M. F. G. Schmidt, and R. Rott. 1988. Chemical identification of cysteine as palmitoylation site in a transmembrane protein (Semliki forest virus E1). J. Biol. Chem. 263:18635-18639. [PubMed] [Google Scholar]
- 39.Shmulevitz, M., and R. Duncan. 2000. A new class of fusion-associated small transmembrane (FAST) proteins encoded by the nonenveloped fusogenic reoviruses. EMBO J. 19:902-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shmulevitz, M., Z. Yameen, S. J. Dawe, J. Shou, D. O'Hara, I. Holmes, and R. Duncan. 2002. Sequential partially overlapping gene arrangement in the tricistronic S1 genome segments of avian reovirus and Nelson Bay reovirus: implications for translation initiation. J. Virol. 76:609-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider, P. A., M. Schwemmle, and W. I. Lipkin. 1997. Implication of a cis-acting element in the cytoplasmic accumulation of unspliced Borna disease virus RNAs. J. Virol. 71:8940-8945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh, M., B. Berger, and P. S. Kim. 1999. LearnCoil-VMF: computational evidence for coiled-coil-like motifs in many viral membrane-fusion proteins. J. Mol. Biol. 290:1031-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skehel, J. J., and D. C. Wiley. 1998. Coiled coils in both intracellular vesicle and viral membrane fusion. Cell 95:871-874. [DOI] [PubMed] [Google Scholar]
- 44.Stiasny, K., S. L. Allison, C. W. Mandi, and F. X. Heinz. 2001. Role of metastability and acidic pH in membrane fusion by tick-borne encephalitis virus. J. Virol. 75:7392-7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stirk, H. J., J. M. Thornton, and C. R. Howard. 1992. A topological model for hepatitis B surface antigen. Intervirology 33:148-158. [DOI] [PubMed] [Google Scholar]
- 46.Tusnády, G. E., and I. Simon. 1998. Principles governing amino acid composition of integral membrane proteins: applications to topological prediction. J. Mol. Biol. 283:489-506. [DOI] [PubMed] [Google Scholar]
- 47.Varela, R., J. Martinez-Costas, M. Mallo, and J. Benavente. 1996. Intracellular posttranslational modifications of S1133 avian reovirus proteins. J. Virol. 70:2971-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Veit, M., H. Reverey, and M. F. Schmidt. 1996. Cytoplasmic tail length influences fatty acid selection for acylation of viral glycoproteins. Biochem. J. 318:163-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.von Heijne, G., and Y. Gavel. 1988. Topogenic signals in integral membrane proteins. Eur. J. Biochem. 174:671-678. [DOI] [PubMed] [Google Scholar]
- 50.Weissenhorn, W., A. Dessen, L. J. Calder, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1999. Structural basis for membrane fusion by enveloped viruses. Mol. Membr. Biol. 16:3-9. [DOI] [PubMed] [Google Scholar]
- 51.White, J. M. 1990. Viral and cellular membrane fusion proteins. Annu. Rev. Physiol. 52:675-679. [DOI] [PubMed] [Google Scholar]
- 52.Yang, C., C. P. Spies, and R. W. Compans. 1995. The human and simian immunodeficiency virus envelope glycoprotein transmembrane subunits are palmitoylated. Proc. Natl. Acad. Sci. USA 92:9871-9875. [DOI] [PMC free article] [PubMed] [Google Scholar]