Abstract
In latent Epstein-Barr virus infection, the viral EBNA1 protein binds to specific sites in the viral origin of DNA replication, oriP, to activate the initiation of DNA replication, enhance the expression of other viral latency proteins, and partition the viral episomes during cell division. The DNA binding domain of EBNA1 is required for all three function, and a Gly-Arg-rich sequence between amino acids 325 and 376 is required for both the transcriptional activation and partitioning functions. We have used mutational analysis to identify additional EBNA1 sequences that contribute to EBNA1 functions. We show that EBNA1 amino acids 8 to 67 contribute to, but are not absolutely required for, EBNA1 replication, partitioning, and transcriptional activation functions. A Gly-Arg-rich sequence (amino acids 33 to 53) that is similar to that of amino acids 325 to 376 and lies within the 8-to-67 region was not responsible for the functional contributions of residues 8 to 67, since deletion of amino acids 34 to 52 alone did not affect EBNA1 functions. We also found that deletion of amino acids 61 to 83 eliminated the transcriptional activity of EBNA1 without affecting partitioning. This mutant also exhibited an increased replication efficiency that resulted in the maintenance of oriP plasmids at a copy number approximately fourfold higher than for wild-type EBNA1. The results indicate that the three EBNA1 functions have overlapping but different sequence requirements. Transcriptional activation requires residues 61 to 83 and 325 to 376 and is stimulated by residues 8 to 67; partitioning requires residues 325 to 376 and is stimulated by residues 8 to 67; and replication involves redundant contributions of both the 325-to-376 and 8-to-67 regions.
Epstein-Barr virus (EBV) persists in the nuclei of latently infected human B lymphocytes as double-stranded circular DNA episomes (51). These episomes undergo one round of DNA replication per cell cycle and are efficiently partitioned during cell division, enabling the viral genomes to be maintained at a constant copy number (1, 32, 53, 54). The replication and partitioning of the EBV episomes require two viral components: the latent origin of DNA replication, oriP, and the EBNA1 protein (54). EBV DNA replication initiates within the dyad symmetry (DS) element of oriP, which contains four EBNA1 binding sites (17, 37, 40, 50). The partitioning of the episomes involves EBNA1 binding to the family-of-repeats (FR) element of oriP, which contains 20 EBNA1 recognition sites (32, 40). In addition, EBNA1 binding to the FR element enhances the expression of other viral latency proteins (16, 39).
The mechanism by which EBNA1 binds to its recognition sites in oriP is reasonably well understood. EBNA1 binds to each site as a dimer, and the dimers assemble cooperatively on the four sites in the DS element (15, 21, 47). EBNA1 amino acids 459 to 607 are responsible for DNA binding and dimerization, and the structural basis for DNA recognition and dimerization has been revealed through X-ray crystallography and biochemical approaches (2, 5, 6, 9, 10, 47). EBNA1 complexes bound to the FR and DS elements have been observed to interact with each other, resulting in the looping out of the intervening DNA and the linking of multiple oriP DNA molecules together (14, 36, 46). EBNA1 residues 40 to 89 and 327 to 377 are responsible for these looping and linking interactions (3, 13, 18, 30, 33).
The mechanisms by which EBNA1 activates DNA replication and transcription are less well defined. Since EBNA1 lacks enzymatic activity, these mechanisms likely involve the recruitment of cellular proteins to oriP and might also involve remodeling of the EBV chromatin structure. The latter possibility is suggested by the ability of EBNA1 to bind and destabilize nucleosomes formed on the oriP DS element, a property that is intrinsic to the EBNA1 DNA binding and dimerization domain (4). While the nature of the cellular transcription factors that are recruited by EBNA1 is not yet known, recent studies have shown that the cellular origin recognition complex (ORC) and minichromosome maintenance (MCM) complex are present at the DS element and that ORC might be recruited through interaction with EBNA1 (8, 12, 41). As a first step to uncovering the molecular basis for these EBNA1-cellular-protein interactions, it is important to identify the EBNA1 regions that mediate replication and transcriptional activities. To date it is known that both processes involve EBNA1 residues between 1 and 377, and in the case of transcription, a requirement for the Gly-Arg-rich region between amino acids 325 and 376 has been demonstrated (7, 27, 52).
Considerable evidence indicates that EBNA1 governs the partitioning of EBV episomes and FR-containing constructs by mediating their attachment to cellular mitotic chromosomes. First, EBNA1, EBV episomes, and oriP-containing constructs have all been observed to associate with the condensed cellular chromosomes in mitosis (11, 19, 20, 38, 44). Second, the association of oriP plasmids with mitotic chromosomes has been shown to be EBNA1 dependent (25). Third, the FR component of oriP has been shown to be responsible for both efficient segregation and mitotic-chromosome attachment of DNA constructs (25, 28). Fourth, deletion of EBNA1 amino acids 325 to 376 has been found to disrupt both the segregation of FR-containing plasmids and the association of EBNA1 with mitotic chromosomes without affecting the interphase nuclear localization or DNA replication activity of the protein (43, 49). Fifth, the N-terminal half of EBNA1, which mediates chromosome attachment and oriP plasmid maintenance, can be functionally replaced by chromosome binding sequences from HMG-1 and histone H1 (23).
Recent evidence suggests that EBNA1 attaches to cellular mitotic chromosomes by binding to the human EBP2 protein on the chromosomes (43, 49). EBP2 is a component of the nucleolus that, like EBNA1, coats the condensed cellular chromosomes in mitosis. EBNA1 binding to EBP2 requires amino acids 325 to 376, which are essential for the segregation activity and chromosome attachment of EBNA1. The importance of EBP2 in the EBNA1-mediated partitioning of FR-containing plasmids has been demonstrated by its requirement in a reconstituted EBV segregation system in budding yeast (26). A yeast replicating plasmid containing the EBV segregation element was efficiently partitioned in yeast only when both EBNA1 and EBP2 were expressed. Stable maintenance of these plasmids also required the FR segregation element and was not supported by an EBP2-binding mutant of EBNA1 or by an EBNA1-binding mutant of EBP2.
Attempts to identify EBNA1 amino acids that are important for segregation function and chromosome attachment have yielded varied results. Marechal et al. (35) looked for fragments of EBNA1 that would bind to mitotic chromosomes when fused to the enhanced green fluorescent protein (EGFP). They concluded that three EBNA1 polypeptides, spanning amino acids 8 to 67 (CBS-3), 72 to 84 (CBS-1), and 328 to 365 (CBS-2), could independently mediate EBNA1 attachment to mitotic chromosomes, indicating that CBS-1, -2, and -3 make redundant contributions to EBNA1 segregation activity. Similar experiments were conducted by Hung et al. (23), who found that EBNA1 fragments 1 to 89 and 323 to 386 each gave weak mitotic-chromosome association when fused to EGFP and that an EBNA1 fragment spanning both regions gave stronger chromosome binding. These results also support the idea that multiple EBNA1 sequences contribute to segregation activity but suggest that their contributions may be additive. One caveat for the above approach, where polypeptides are examined out of context of the folded protein, is that it can generate false-positive results. Due to changes in structure and/or increased accessibility, polypeptides that are excised from a protein sometimes mediate interactions that they do not mediate in the context of the folded protein.
Our approach to identifying functionally important EBNA1 sequences has been to delete EBNA1 residues within the context of the full-length protein and examine effects on EBNA1 activities. In this way we have shown that the deletion of amino acids 325 to 376 disrupts DNA segregation activity and profoundly reduces the association of EBNA1 with mitotic chromosomes (43, 49). This result strongly supports the EGFP fusion experiments for sequences from this region (CBS-2) but appears to be inconsistent with the hypothesis that N-terminal EBNA1 sequences (CBS-1 and -3) are sufficient for chromosome attachment. However, the possibility that CBS-1 and -3 contribute to the partitioning or other activities of EBNA1 has not been addressed. To address these possibilities, we have generated EBNA1 deletion mutants lacking CBS-1 and all or part of CBS-3 and have assessed the effects of the mutations on DNA replication, plasmid maintenance, plasmid partitioning, transcriptional activation, and cellular localization.
MATERIALS AND METHODS
EBNA1 expression constructs.
EBNA1 proteins were expressed in human cells from the cytomegalovirus (CMV) promoter in the pcDNA3 plasmid (Invitrogen, Carlsbad. Calif.) that also contained the EBV oriP sequence between the BglII and NruI sites of the plasmid. The construction of these plasmids expressing wild-type EBNA1 (lacking most of the Gly-Ala repeat; pc3oriPEBNA1), EBNA1 lacking amino acids 41 to 376 (pc3oriPΔ41-376), or amino acids 325 to 376 (pc3oriPΔ325-376), or no EBNA1 (pc3oriP) has been described previously (43). Plasmids expressing EBNA1 mutants lacking amino acids 8 to 67 (pc3oriPΔ8-67), 34 to 54 (pc3oriPΔ34-54), 61 to 83 (pc3oriPΔ61-83), and 330 to 354 (pc3oriPΔ330-354) were generated from the pc3oriPEBNA1 construct by QuickChange site-directed mutagenesis (Stratagene, La Jolla, Calif.) using oligonucleotides designed to delete the appropriate coding sequences. Plasmids expressing the EBNA1 mutants lacking amino acids 8 to 67 in addition to either 330 to 354 (Δ8-67/Δ330-354) or 325 to 376 (Δ8-67/Δ325-376) were generated by QuickChange mutagenesis of pc3oriPΔ330-354 and pc3oriPΔ325-376 constructs by using oligonucleotides designed to delete the EBNA1 sequences encoding amino acids 8 to 67. The sequences of all of the EBNA1 mutants were verified by DNA sequencing.
EBNA1 proteins were expressed in yeast from the MET25 promoter in plasmid p416MET25, which contains a URA3 marker. The generation of constructs expressing wild-type EBNA1 (lacking most of the Gly-Ala repeat) or the Δ325-376 EBNA1 mutant has been described previously (26). Plasmids expressing EBNA1 mutants lacking amino acids 8 to 67 or 61 to 83 were generated by PCR amplification of the EBNA1 sequences in pc3oriPΔ8-67 or pc3oriPΔ61-83, respectively, and insertion of these EBNA1 sequences into the SmaI site of p416MET25.
Plasmid maintenance assays.
Human C33A cells were plated in 1 well of a 6-well plate at 5 × 105 cells/well and grown for 24 h prior to transfection. The cells were transfected with 1 μg of pc3oriP plasmids expressing either EBNA1, an EBNA1 mutant, or no EBNA1 by using Lipofectamine 2000 (Gibco-BRL) according to the manufacturer's method. After a 5-h incubation with the DNA, cells were washed in phosphate-buffered saline, plated in a 140-mm dish, and grown in a medium containing 400 μg of G418 (Gibco-BRL)/ml to select for cells containing the plasmid. Following 14 days of selection, 5 × 106 cells from each plate were harvested and lysed by the method of Hirt (22). Low-molecular-weight DNA was isolated as described by Ceccarelli and Frappier (7), digested with XhoI and DpnI, separated by agarose gel electrophoresis, Southern blotted, and probed with 32P-labeled pc3oriPEBNA1. Linearized plasmid bands were visualized by autoradiography and quantified by phosphorimager analysis using ImageQuant software (Molecular Dynamics).
Transient replication assays.
C33A cells were plated in 60-mm dishes at 2.5 × 106 cells/plate and, 24 h later, were transfected with 10 μg of pc3oriP plasmids expressing either EBNA1, an EBNA1 mutant, or no EBNA1 by using Lipofectamine 2000 (Gibco-BRL). After a 5-h incubation with the DNA, the cells were washed in phosphate-buffered saline, replated in a 140-mm plate, and grown without selection for 72 h. At 72 h posttransfection, the low-molecular-weight DNA was prepared from 9/10 of the cells on the plate (1/10 of the cells were used for Western blot analysis), digested with XhoI and DpnI, and analyzed by Southern blotting as for plasmid maintenance assays. Linearized, DpnI-resistant plasmid bands were visualized by autoradiography and quantified by phosphorimager analysis using ImageQuant software (Molecular Dynamics).
Transcriptional activation assays.
C33A cells were plated in 1 well of a 6-well plate as for the plasmid maintenance assays, and the cells were transfected with 5 μg of pc3oriPEBNA1 and 2 μg of the pFRTKCAT reporter construct (kindly provided by Bill Sugden) by using Lipofectamine 2000 (52). After a 5-h incubation with the DNA, the cells were washed and then given fresh medium. Twenty-four hours later, lysates were prepared from the cells and assayed for chloramphenicol acetyltransferase (CAT) activity as previously described (7). Briefly, 50 μg of each lysate was incubated with acetyl coenzyme A and [14C]chloramphenicol for various lengths of time, and reaction products were separated by thin-layer chromatography and quantified by PhosphorImager analysis. The amount of acetylated product produced at each time point was used to determine the acetylation rate for each lysate.
Partitioning assays in yeast.
Plasmid partitioning assay were performed with yeast strain KY320.hEBP2 (MATa leu2-PET56::LEU2-hEBP2 ura3-52 trp1-Δ1 lys2-801am ade2-101oc his3-Δ200 GAL+), which constitutively expresses human EBP2 from an integrated copy of this gene, as described by Kapoor et al. (26). Briefly, KY320.hEBP2 cells were transformed to Trp prototrophy with the YRp7FR plasmid, containing the EBV FR and yeast autonomously replicating sequence (ARS) elements, and to Ura prototrophy with a p426MET25 vector expressing either EBNA1, an EBNA1 mutant, or no EBNA1. Positive transformants were first grown in selective medium (synthetic complete medium lacking Trp and Ura [SC−Trp−Ura]) until early- to mid-log phase and then diluted 100-fold into a medium that did not select for YRp7FR (SC−Ura) and grown for 11 generations. Tenfold serial dilutions of the cultures were then spotted onto plates that were selective (SC−Trp−Ura) and nonselective (SC−Ura) with respect to YRp7FR. To quantify the percentage of cells that retained YRp7FR, equal amounts of diluted cultures were also spread onto selective and nonselective plates and the resulting colonies were counted. Plasmid stability was measured as a ratio of the number of colonies on selective versus nonselective plates.
Western blotting.
Expression of EBNA1 proteins in transfected C33A cells was determined from 1/10 of the cells collected for the transient replication assays. Cells were lysed in 500 mM NaCl-20 mM Tris-HCl (pH 8)-0.1% Triton-0.5 mM EDTA-1 mM phenylmethylsulfonyl fluoride-1 mM benzamidine, and lysates were sonicated and clarified by centrifugation. Thirty micrograms of each lysate was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a membrane, and probed with the K67 anti-EBNA1 rabbit serum (kindly supplied by Jaap Middeldorp, Free University Hospital, Amsterdam, The Netherlands) as previously described (7). The blots were developed for enhanced chemiluminescence (NEN Inc.) or for enhanced chemifluorescence (ECF; Amersham, Inc.) by the method of the manufacturer. ECF-reactive bands were quantified by using a Storm 860 scanner and ImageQuant software (Molecular Dynamics). Expression of EBNA1 proteins in yeast was determined by Western blot analysis of the KY320.hEBP2 yeast strain containing a p426MET25 plasmid expressing either EBNA1 or an EBNA1 mutant. Approximately 2 × 107 of these log-phase cells were harvested and boiled in 62 mM Tris-HCl (pH 6.8)-10% glycerol-2% SDS-5% β-mercaptoethanol-0.1% bromophenol blue. After removal of insoluble debris by centrifugation, the proteins in the lysate were separated by SDS-PAGE, Western blotted, and probed with the K67 anti-EBNA1 rabbit polyclonal antibody. The anti-EBNA1 antibody was visualized by enhanced chemiluminescence as described above.
Immunofluorescence.
Indirect immunofluorescence was performed using C33A cells that expressed either EBNA1 or an EBNA1 mutant. These cells were generated by transfecting C33A cells with pc3oriP plasmids expressing either EBNA1 or an EBNA1 mutant and growing the cells under selection for the plasmid for 2 weeks. The cells were first blocked in mitosis by incubation of log-phase cultures with 0.1 μg of Colcemid/ml for 5 h and then swollen in hypotonic buffer, dropped onto slides, fixed in paraformaldehyde, permeabilized with 0.5% Triton X-100, and blocked as previously described (49). The permeabilized cells were incubated with a mouse monoclonal antibody (OT1x) that recognizes EBNA1 amino acids 424 to 448 (kindly supplied by Jaap Middeldorp), followed by a Texas Red-conjugated goat anti-mouse antibody (Molecular Probes) and counterstaining with 4",6"-diamidino-2-phenylindole (DAPI). The slides were mounted in 5 μl of antifade solution and observed at 630-fold magnification using a Leica DMR microscope and Openlab software. For indirect immunofluorescence of interphase cells, log-phase cells growing on coverslips were fixed and permeabilized as described above and then stained with the purified K67 rabbit polyclonal antibody, followed by a fluorescein isothiocyanate-conjugated goat anti-rabbit antibody. After DAPI counterstaining, the slides were observed at 400-fold magnification as described above.
RESULTS
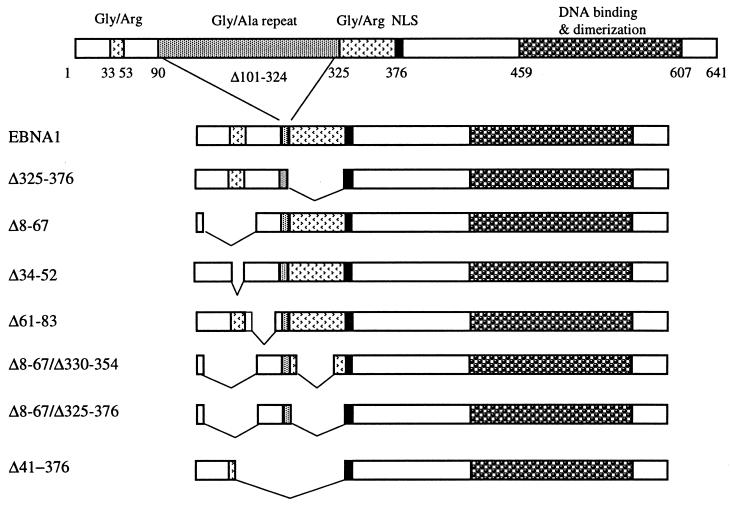
EBNA1 mutants designed to test the functional contribution of the CBS-1 and CBS-3 regions were constructed by deleting amino acids 61 to 83 (Δ61-83) and 8 to 67 (Δ8-67), respectively, in the context of a functional version of EBNA1 that lacks most of the Gly-Ala repeats (Fig. 1). Previous studies have shown that lack of the Gly-Ala repeats does not affect EBNA1 DNA replication, segregation, or transcriptional activation activities (52, 54). The 8-to-67 deletion includes a Gly-Arg-rich sequence (amino acids 33 to 53) similar to the Gly-Arg-rich sequence that lies between amino acids 325 and 376 and that is essential for EBNA1 segregation and transcriptional activation activities (7, 43). This similarity has led to speculation that the 33-to-53 sequence also contributes to segregation and transactivation. To specifically address the functional role of this Gly-Arg-rich sequence, an EBNA1 mutant lacking amino acids 34 to 52 was generated (Δ34-52). All of the EBNA1 mutants were expressed in human C33A cells from a CMV promoter on a plasmid that also contained oriP. Expression of all EBNA1 mutants was confirmed by Western blot analysis of cell extracts prepared 72 h after transfection with the EBNA1-expressing plasmids (Fig. 2). Deletion of amino acids 8 to 67 decreased the EBNA1 expression level approximately 2.5-fold, but mutants containing this deletion were still present in excess over the amount needed to bind the oriP elements in the plasmids (see Discussion). All other EBNA1 mutants were expressed at levels at or above (Δ325-376) that of wild-type EBNA1.
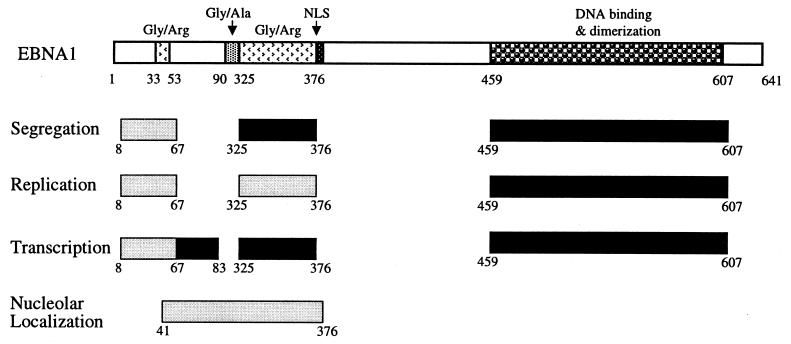
FIG. 1.
EBNA1 mutants. This schematic representation of the EBNA1 mutants shows pertinent regions of the protein including the nuclear localization signal (NLS).
FIG. 2.
Expression of EBNA1 mutants in human cells. C33A cells were transfected with pc3oriP-based plasmids expressing the EBNA1 proteins indicated or no EBNA1 (pc3oriP), and cell lysates were prepared 72 h posttransfection. Western blots from equal amounts of total protein are shown after probing with an anti-EBNA1 antibody.
Plasmid maintenance activity.
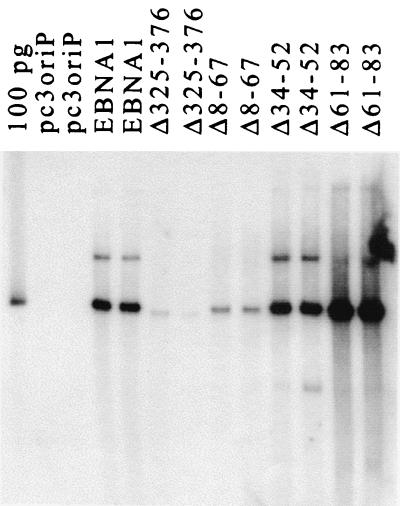
The EBNA1 deletion mutants were first tested for their abilities to maintain the oriP-containing plasmid, on which they were expressed, in long-term culture. To this end, C33A cells were transfected with the expression plasmid and then grown under selection for that plasmid for 14 days. Plasmids were then recovered from the cells, linearized, detected by Southern blotting, and quantified by phosphorimager analysis (Fig. 3 and Table 1). In these experiments, wild-type EBNA1 maintained the oriP plasmids at a level of 9 copies per cell on average, while no plasmids were recovered in the absence of EBNA1. As previously reported, the removal of amino acids 325 to 376 dramatically decreased plasmid maintenance. The Δ8-67 mutant also had a decreased ability to maintain plasmids, but the defect was on average eightfold less severe than that for Δ325-376. These results indicate that amino acids 8 to 67 contribute to plasmid maintenance but are less important for this activity than amino acids 325 to 376. The plasmid maintenance contribution of Δ8-67 was not, however, due to the Gly-Arg-rich sequence within this fragment, as the deletion of the Gly-Arg region alone (Δ34-52) had no effect on plasmid maintenance ability.
FIG. 3.
Plasmid maintenance assay. C33A cells transfected with pc3oriP-based plasmids expressing the EBNA1 proteins indicated or no EBNA1 (pc3oriP) were grown under selection for the plasmids for 14 days. Then the plasmids were recovered, linearized, and quantified by Southern blotting. Results for duplicate samples are shown, as is a 100-pg marker of linearized pc3oriPEBNA1 (100 pg).
TABLE 1.
Summary of the functional activities of EBNA1 mutants
| EBNA1 protein | % of wild-type activity ± SD (n)a
|
|||
|---|---|---|---|---|
| Plasmid maintenance | Transient replication | Transcription activation | Plasmid partitioning (yeast)b | |
| Wild type | 100 | 100 | 100 | 100 |
| Δ325-376 | 3 ± 1 (4) | 100 ± 38 (9) | 0 (1)c | 1.6 |
| Δ8-67 | 25 ± 20 (6) | 87 ± 31 (12) | 17 ± 3 (5) | 32 |
| Δ34-52 | 97 ± 39 (6) | 129 ± 19 (4) | 104 ± 34 (4) | NDd |
| Δ61-83 | 362 ± 117 (6) | 276 ± 63 (6) | 1.4 ± 1.7 (4) | 90 |
| Δ8-67/Δ330-351 | ND | 66 ± 29 (8) | 2 ± 2 (5) | ND |
| Δ8-67/Δ325-376 | ND | 26 ± 16 (7) | 1 ± 1 (4) | ND |
Activities are shown as a percentage of the activity observed for wild-type EBNA1 after subtraction of any background activity seen in the absence of EBNA1. Activities and standard deviations are followed by and the number of experiments (n) performed to generate the values (in parentheses).
For the plasmid partitioning activities in the yeast system, assays were performed twice with very similar results, and the results of the experiment from Fig. 5 are shown. In this experiment the 100% partitioning activity of the wild-type EBNA1 represents retention of 70% of the segregation plasmids after the 11-generation assay period.
This value is from the experiment for which results are shown in Fig. 7. Results from multiple experiments for Δ325-376 have been reported previously (7).
ND, not determined.
The contribution of amino acids 61 to 83 to oriP plasmid maintenance was also assessed. The Δ61-83 EBNA1 mutant was consistently found to maintain oriP plasmids at levels three- to fourfold higher than wild-type EBNA1. Thus, amino acids 61 to 83 are not required for plasmid maintenance but may play a regulatory role.
DNA replication activity.
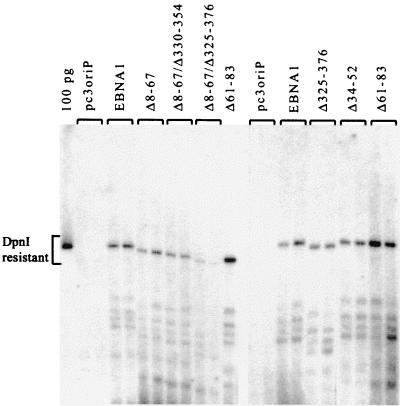
The plasmid maintenance activity measured above is a combination of the abilities of EBNA1 to activate DNA replication from oriP and to partition the oriP plasmids during cell division. To determine whether the plasmid maintenance effects of EBNA1 mutations were due to effects on DNA replication, we conducted transient replication assays using the oriP plasmids expressing EBNA1 mutants. These plasmids were used to transfect C33A cells, harvested from the cells after 72 h, linearized, and then digested with DpnI. Under the conditions used, DpnI digests the input plasmid from Escherichia coli, which is methylated, but does not digest the unmethylated products of replication in human cells. Full-length and digested plasmids were detected by Southern blotting, and DpnI-resistant plasmid bands were quantified as a measure of DNA replication activity. EBNA1-dependent replication was consistently detected by this assay, and deletion of amino acids 325 to 376, 8 to 67, or 34 to 52 had no significant effect on replication activity (Fig. 4 and Table 1). Therefore, the reduced plasmid maintenance abilities of Δ325-376 and Δ8-67 were not due to replication defects. Deletion of amino acids 61 to 83 consistently increased replication activity two- to threefold, suggesting that the increase in plasmid maintenance ability associated with Δ61-83 is due, at least in part, to increased replication efficiency.
FIG. 4.
Transient replication assay. C33A cells transfected with the same plasmids as those in Fig. 3 were harvested 72 h posttransfection. Plasmids recovered from the cells were linearized, digested with DpnI to remove any unreplicated plasmid, and analyzed by Southern blotting. Results of two experiments with duplicate samples are shown.
While neither the Δ8-67 nor the Δ325-376 mutation individually affected DNA replication, we also made double mutants to test the possibility that the two EBNA1 regions make redundant contributions to DNA replication. Deletion of amino acids 8 to 67 in combination with a 330-to-351 or a 325-to-376 deletion (Fig. 1) resulted on average in 34 and 74% decreases in replication activity, respectively (Fig. 4 and Table 1). These results suggest that either the 8-to-67 or the 325-to-376 region can contribute to replication efficiency.
Plasmid partitioning.
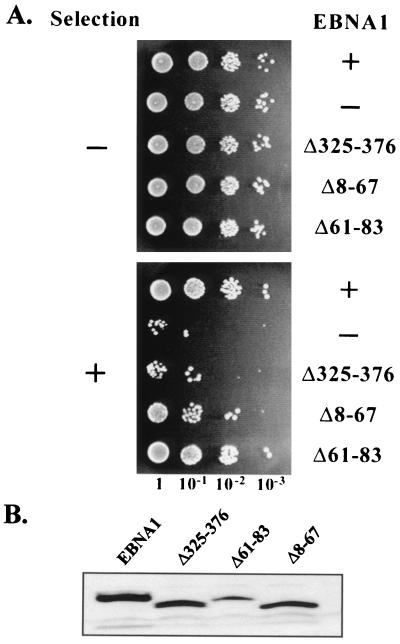
We have recently developed a system in which the segregation activity of EBNA1 can be directly measured and quantified (26). The segregation activity is measured in yeast using an FR/ARS plasmid (YRp7FR) that replicates from a yeast origin of replication, segregates using the EBV FR element, and contains a selectable marker. This plasmid is stably maintained in yeast when both EBNA1 and the human EBP2 proteins are expressed. The Δ325-376 EBNA1 mutation, which disrupts segregation in human cells, also disrupts FR-plasmid partitioning in the yeast system. To directly test the ability of the EBNA1 Δ8-67 and Δ61-83 mutants to partition plasmids, we expressed these proteins from plasmids in a yeast strain that constitutively expresses human EBP2 and contains YRp7FR. We then removed selection for YRp7FR for 11 generations and plated serial dilutions of the cultures on plates that do and do not select for this plasmid. Efficient partitioning, such as occurs in the presence but not in the absence of wild-type EBNA1, was indicated by the growth of a similar number of colonies on selective and nonselective plates (Fig. 5A) and was quantified by counting the colonies and determining the percentage of yeast cells that retained YRp7FR (Table 1).
FIG. 5.
Plasmid partitioning assay. (A) Yeast cells expressing hEBP2 were transformed with the YRp7FR plasmid containing the EBV segregation element and with a plasmid expressing either EBNA1 (+), no EBNA1 (−), or the EBNA1 mutants indicated. After 11 cell doublings in the absence of selection for YRp7FR, serial dilutions of the cultures were spotted on plates that do (+) and do not (−) select for cells containing YRp7FR. (B) Western blot from equal numbers of yeast cells showing expression of the EBNA1 proteins from the plasmids used in panel A.
Analysis of the Δ8-67 mutant in this system indicated a threefold reduction in its ability to partition plasmids compared to wild-type EBNA1. This mutant is, however, 20-times more active for partitioning than Δ325-376. These results are in agreement with the relative abilities of these mutants to maintain plasmids in human cells and indicate that the plasmid maintenance defects are due to decreases in segregation ability. Analysis of Δ61-83 indicated only a slight (10%) decrease in partitioning ability, which might have been due to the lower expression level of this protein in yeast compared to the other EBNA1 proteins analyzed (Fig. 5B).
Mitotic localization of Δ8-67.
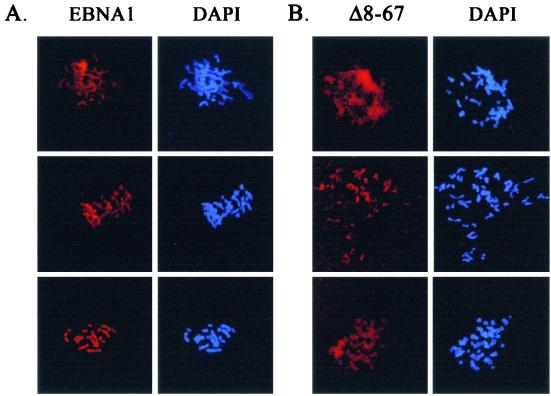
It has been well established that EBNA1-mediated plasmid partitioning in human cells involves the attachment of EBNA1 to the cellular chromosomes in mitosis, and we have previously shown that the 325-to-376 deletion disrupts both partitioning and mitotic-chromosome binding. Since the 8-to-67 mutation decreases the efficiency with which EBNA1 partitions a plasmid but does not completely disrupt this process, we were interested in determining whether Δ8-67 attached to the cellular chromosomes in mitosis. Mitotic-chromosome spreads of C33A cells expressing EBNA1 or the Δ8-67 mutant are shown in Fig. 6A and B, respectively. Like EBNA1, Δ8-67 is closely associated with the condensed cellular chromosomes, but we consistently found the chromosomal staining of Δ8-67 to be more diffuse than that of wild-type EBNA1 (compare Fig. 6A and 6B). The more diffuse staining is not due to increased expression of Δ8-67, since this protein was expressed at somewhat lower levels than the wild-type protein (Fig. 2). The staining pattern suggests that Δ8-67 binds to mitotic chromosomes but may be less tightly bound than wild-type EBNA1. The Δ34-52 and Δ61-83 mutants, which did not have impaired segregation function, gave mitotic-chromosome staining patterns that were indistinguishable from that of EBNA1 (data not shown).
FIG. 6.
. Mitotic-chromosome localization of EBNA1 and Δ8-67. C33A cells expressing EBNA1 (A) or Δ8-67 (B) were blocked in mitosis, and the chromosomes were spread for microscopy. EBNA1 proteins were detected with a monoclonal antibody, and the chromosomes were visualized by DAPI staining. Images were captured by immunofluorescence microscopy using a 630-fold magnification.
Transcriptional activation activity.
We also tested the contribution of EBNA1 N-terminal sequences to the transcriptional activation activity of EBNA1. We have shown previously that the 325-to-376 deletion severely affects transactivation activity, indicating a major transcriptional role for this Gly-Arg-rich region (7). To determine if N-terminal sequences also contribute to this process, C33A cells were cotransfected with the constructs described above that express Δ8-67, Δ34-53, or Δ61-83 mutants and with a reporter construct that contains the CAT gene under the control of the FR enhancer element. Cell lysates were prepared 24 h posttransfection, and equal amounts of protein were used to determine the rates of chloramphenicol acetylation.
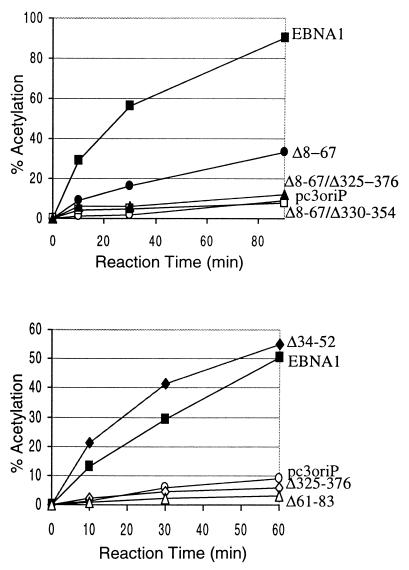
As shown in Fig. 7 and Table 1, like Δ325-376, the Δ61-83 mutant completely lacked transcriptional activation activity, implicating amino acids 61 to 83 in this process. The 8-to-67 deletion reduced transcriptional activity approximately sixfold but was more active than Δ325-376 or Δ61-83. The low level of transactivation by Δ8-67 was virtually eliminated by a further deletion of all or part of the 325-to-376 region (Δ8-67/Δ330-354 and Δ8-67/Δ325-376). The similarity of the Gly-Arg-rich sequences between amino acids 325 to 376 and amino acids 34 to 52 suggested that residues 34 to 52 might be responsible for the transcriptional activity of the 8-to-67 region. However, when the transactivating activity of the Δ34-52 mutant was tested, it was found to be equivalent to that of wild-type EBNA1, indicating that sequences elsewhere in the 8-to-67 region are responsible for the decreased transcriptional activation seen in the Δ8-67 mutant.
FIG. 7.
Transcriptional activation assays. Lysates were prepared from C33A cells cotransfected with a CAT reporter plasmid in which the CAT gene is under the control of the EBV FR element and with pc3oriP plasmids expressing EBNA1 (solid squares), Δ8-67 (solid circles), Δ8-67/Δ325-376 (solid triangles), Δ8-67/Δ330-354 (open squares), Δ34-52 (solid diamonds), Δ61-83 (open triangles), Δ325-376 (open diamonds), or no EBNA1 (pc3oriP; open circles). Equal amounts of lysates were tested for chloramphenicol acetylation rates as a measure of CAT expression levels. Percentages of chloramphenicol acetylated after various reaction times are shown.
Nuclear localization.
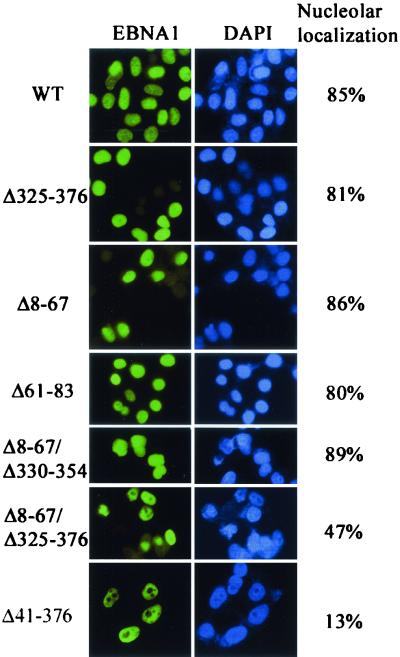
In interphase, EBNA1 is nuclear and, unlike the other EBV nuclear antigens, is observed throughout the nucleus, including the nucleolus (38). The interphase localization of the EBNA1 mutants was examined by immunofluorescent staining of C33A cells expressing the mutants, using an EBNA1 monoclonal antibody (Fig. 8). While all of the mutants localized to the nucleus, we found that some of the mutations decreased the ability of EBNA1 to enter the nucleolus. This effect was quantified by comparing the number of cells expressing each EBNA1 mutant in which nucleolar exclusion or nucleolar staining was observed (Fig. 8). For wild-type EBNA1, nucleolar staining was observed in 85% of the cells scored. Deletion of amino acids 325 to 376, 8 to 67, or 61 to 83 did not significantly affect nucleolar localization, nor did deletion of part of the 325-to-376 region in combination with the 8-to-67 deletion (Δ8-67/Δ330-351). A decreased frequency of nucleolar localization was observed, however, when the complete 325-to-76 sequence was deleted in the context of the Δ8-67 mutant (Δ8-67/Δ325-376). The most dramatic decrease in nucleolar localization occurred when amino acids 41 to 376 were deleted (see Fig. 1), resulting in nucleolar staining in 13% of the cells. These results suggest that multiple EBNA1 sequences between amino acids 41 and 376 contribute to nucleolar localization.
FIG. 8.
Effects of EBNA1 mutations on nucleolar staining. Log-phase C33A cells expressing EBNA1 (WT) or the EBNA1 mutants indicated were stained for EBNA1 using anti-EBNA1 polyclonal antibodies and counterstained with DAPI to visualize the nucleus. Images were captured by immunofluorescence microscopy using a 400-fold magnification. One hundred cells for each sample were scored for nucleolar staining or exclusion, and the percentages of cells in which the nucleolus was stained are indicated.
DISCUSSION
EBNA1 activates viral latent DNA replication and transcription and mediates the partitioning of the viral genomes during cell division by binding to specific recognition sites in oriP. The region of EBNA1 comprising amino acids 325 to 376 is essential for the transcriptional activation and partitioning functions of EBNA1 (7, 43, 52), but the possible involvement of other EBNA1 sequences in these processes is unclear, as are the EBNA1 sequence requirements for DNA replication. Definition of the EBNA1 residues important for replication and partitioning has been complicated by the fact that, in many published assays, the two activities have not been cleanly distinguished (oriP plasmid “replication” assays performed at 4 days posttransfection or later are actually measuring a combination of replication and segregation functions). Here we have investigated the contribution of EBNA1 N-terminal sequences to EBNA1 functions.
EBNA1 ensures the partitioning of EBV episomes and oriP plasmids by mediating their attachment to cellular mitotic chromosomes. Thus, one method that has been used to identify EBNA1 sequences that may mediate EBV partitioning involved determining which EBNA1 fragments can bind to mitotic chromosomes when fused to EGFP. This approach identified amino acids 8 to 67 and 72 to 84, in addition to the 325-to-376 region, as potentially important for segregation activity (23, 35). We have directly tested the contributions of these sequences to EBNA1 segregation function and found that amino acids 72 to 84 (deleted in the Δ61-83 mutant) do not contribute to the segregation activity of EBNA1 in the context of the functional protein. This result indicates either that the chromosome binding ability of this sequence is redundant in the presence of the other EBNA1 chromosome attachment regions or that the 72-to-84 sequence does not contact the mitotic chromosomes in the context of the folded EBNA1 protein.
A role for amino acids 8 to 67 in EBNA1-mediated partitioning, however, was supported by mutational analysis. While not essential for EBNA1-mediated segregation, residues 8 to 67 were found to increase the efficiency of this process, as indicated by effects on oriP plasmid maintenance in human cells and on the partitioning of FR/ARS plasmids in a reconstituted yeast system. The degrees to which partitioning was affected by the Δ8-67 mutation were very similar in the human and yeast systems. This deletion increased the rate of plasmid loss, on average, by 5 and 6% per generation in the human and yeast systems, respectively. Since plasmid partitioning in the yeast system is completely dependent on the human EBP2 protein, the results continue to support the role of EBP2 in EBNA1-mediated segregation in human cells. Given the ability of the 8-to-67 polypeptide to bind mitotic chromosomes (32) and the slightly more diffuse staining of Δ8-67 than of wild-type EBNA1 on mitotic-chromosome spreads, we believe that the 8-to-67 region contributes to partitioning by increasing the strength of the EBNA1-mitotic-chromosome interaction. This interpretation is consistent with the results of Hung et al. (23), who found that the mitotic-chromosome association of an EBNA1 fragment containing amino acids 1 to 89 and 323 to 382 was reduced by deletion of residues 1 to 89. The 8-to-67 region may contribute to mitotic-chromosome interactions by increasing the binding affinity for EBP2 on these chromosomes. In keeping with this hypothesis, we have found that, while Δ8-67 binds EBP2 in a yeast two-hybrid assay, the interaction is weaker than that observed with wild-type EBNA1 (as measured by the activation of both lacZ and HIS3 reporter genes [P. Kapoor and L. Frappier, unpublished data]).
In addition to its effects on segregation, the Δ8-67 mutation decreased transcriptional activation and contributed to a reduction in replication efficiency when combined with deletions of the 325-to-376 region. These results suggest that the 8-to-67 region might also stabilize interactions needed for replication and transcription, either directly or indirectly. It has recently been demonstrated that the cellular ORC is recruited to the DS element, presumably through interaction with EBNA1 (8, 12, 41). Therefore, the redundant contribution of the 8-to-67 and 325-to-376 sequences to replication might be in recruitment of ORC. The expression of Δ8-67 in human cells was somewhat lower than that of wild-type EBNA1, but the reduced level of Δ8-67 is unlikely to account for the functional reductions associated with this mutant for the following reasons. First, a previously studied EBNA1 mutant, Δ367-376, is expressed at levels similar to those of Δ8-67 yet has wild-type replication, partitioning, and transcriptional activities (7), indicating that wild-type EBNA1 levels exceed those necessary to fill the oriP functional elements. Second, Δ8-67 has wild-type replication activity, indicating that the expression level of this mutant is sufficient to bind the oriP DS element. Since EBNA1 has a lower affinity for the DS element than for the FR element (15, 24), the FR must also be bound by Δ8-67 in the functional assays. Third, the decreased partitioning activity of Δ8-67 was also seen in the yeast system, where Δ8-67 was expressed at the same levels as wild-type EBNA1.
The EBNA1 325-to-376 sequence, which is essential for the EBNA1 partitioning and transcriptional activation functions, is extremely rich in Gly and Arg residues (44 out of 52 amino acids). The presence of a similar Gly-Arg-rich sequence between amino acids 34 and 53 has suggested that this region might also contribute to EBNA1 partitioning and transactivation functions. Deletion of the 34-to-52 sequence, however, had no detectable effect on any EBNA1 function, indicating that this sequence is not functionally equivalent to the 325-to-376 region. Similar results were obtained by Van Scoy et al. (48), using a Δ33-53 EBNA1 mutant that possessed plasmid maintenance and transcriptional activation activities close to wild-type levels. Differences in the functionalities of the two Gly-Arg-rich regions might be due to differences in the structures adopted and/or differences in the amino acid sequence. Unlike the 34-to-52 sequence, amino acids 325 to 376 contain an octameric repeat (GGRGRGGS), which includes a Ser residue that is predicted to be phosphorylated (30). It is possible that the octameric sequence or the presence of phosphorylated serines is important for the functionality of the 325-to-376 sequence.
We have shown that deleting either amino acids 61 to 83 or amino acids 325 to 376 eliminates the transcriptional activation function of EBNA1. These regions lack extensive sequence similarity but, like the 325-to-376 sequence, residues 61 to 83 contain several arginines (five) and serines that are predicted to be phosphorylated (two). The 61-to-83 and 325-to-376 regions are separated by the Gly-Ala repeat region of EBNA1, which varies in length in different EBNA1 isolates. We have used a version of EBNA1 with a very short Gly-Ala repeat, and it will be interesting to determine whether the same results are obtained with an EBNA1 protein containing full-length Gly-Ala repeats. There are several models that might account for the apparent contributions of the 61-to-83 and 325-to-376 regions to transcriptional activation. First, the two sequences might mediate interactions with different cellular proteins, both of which are required for transactivation. Second, the two regions might contact the same cellular protein, where both contacts are required for a stable interaction. Third, the 61-to-83 region alone might mediate the interaction(s) needed for transcriptional activation, and the 325-to-376 deletion might affect the folding of the 61-to-83 region. Fourth, the 325-to-376 region alone might mediate the interaction(s) needed for transcriptional activation, and the 61-to-83 deletion might affect the folding of the 325-to-376 region. The last possibility is unlikely because the 61-to-83 deletion has no effect on the partitioning function of EBNA1, which is also mediated by residues 325 to 376.
Studies by Langle-Rouault et al. (31) suggest that the transcriptional enhancement activity of EBNA1 involves two components: (i) the ability of EBNA1 to increase the uptake of FR-containing plasmids from the cytoplasm to the nucleus and (ii) the ability of EBNA1 to mediate interactions with as yet undefined factors in the cell nucleus. For the following reasons, we believe that the effects reported here on EBNA1 transcriptional activation are due to effects on nuclear protein interactions and not to nuclear uptake. First, the ability of EBNA1 to promote plasmid uptake into the nucleus is thought to be due to the nuclear localization signal of EBNA1 (amino acids 379 to 386 [2]), which was not altered in any of our EBNA1 mutants. Second, all of our EBNA1 mutants were observed by immunofluorescence staining to be in the nucleus and not in the cytoplasm (Fig. 8), indicating that they were not impaired in nuclear entry. Third, all three of the EBNA1 functions measured here (replication, segregation, and transcription) require the test plasmids to be in the nucleus, and therefore, a mutation that inhibits plasmid uptake would be expected to inhibit all three EBNA1 functions. In contrast, the EBNA1 mutations shown here to decrease transcriptional activity (Δ8-67, Δ61-83, and Δ325-376) did not decrease DNA replication.
The properties of the Δ61-83 EBNA1 mutant are particularly interesting because this deletion increases the efficiency of DNA replication, eliminates transcriptional activation, and does not affect partitioning. The increased replication efficiency of this EBNA1 mutant results in the maintenance of oriP-containing plasmids at higher levels, in at least some human cells, than those with wild-type EBNA1. This property suggests that oriP plasmids can be maintained for longer periods by Δ61-83 than by wild-type EBNA1, which typically does not maintain oriP plasmids for more than a couple of months. This feature makes Δ61-83 a useful alternative to wild-type EBNA1 for gene delivery constructs. An additional attractive feature of the Δ61-83 mutant is its lack of transcriptional activity. Wild-type EBNA1 enhances the expression of EBV latency proteins by binding to the FR element (16, 39) and has also been found to activate the expression of some cellular genes (29, 45). The failure of Δ61-83 to activate gene expression from the FR element suggests that this EBNA1 mutant would be unable to activate cellular gene expression, making it safer for use in gene therapy constructs than wild-type EBNA1.
The effects of deletions on the nuclear localization of EBNA1 in interphase were also examined. Unlike most nuclear proteins, EBNA1 stains the nucleolus as well as other parts of the nucleus, but the functional significance of EBNA1 in the nucleolus is not known. The percentage of cells in which EBNA1 stained the nucleolus was reduced when both the 8-to-67 and 325-to-376 sequences were deleted and was further reduced by the 41-to-376 deletion. These results suggest that sequences 325 to 376 and 41 to 67 make redundant contributions to nucleolar entry and that amino acids 68 to 100 also contribute to this process (Fig. 9). Gly-Arg-rich sequence motifs, referred to as GAR domains (RGGXRGG or RG GXGGR, where X can be F, S, Y, or A), have been identified in several nucleolar proteins and are present in three copies between EBNA1 residues 330 and 353 (42). While these sequences may contribute to the nucleolar association of EBNA1, they are not required for this property, and the other EBNA1 sequence motifs that permit nucleolar entry are not yet clear. The cellular EBP2 protein, which appears to mediate the attachment of EBNA1 to the condensed chromosomes in mitosis, is exclusively nucleolar in interphase, raising the possibility that the nucleolar fraction of EBNA1 might bind to EBP2 in interphase. If this interaction occurs, however, it does not appear to account for the localization of EBNA1 to the nucleolus, as the Δ325-376 mutant, which is severely impaired in its ability to bind EBP2, exhibits nucleolar staining.
FIG. 9.
Summary of the amino acid requirements for the replication, segregation, and transcriptional activation functions and nucleolar staining of EBNA1. Deletions that completely disrupt (dark shading), or that affect but do not abrogate (light shading), the EBNA1 function are indicated. “Nucleolar localization” refers to the staining of the nucleolus by EBNA1 in addition to other parts of the nucleus. The region that contributes to nucleolar staining is shown with light shading to reflect the fact that multiple redundant elements in this region contribute to nucleolar localization.
We have shown previously that the replication and transcriptional activation functions of EBNA1 can be resolved, as can the replication and partitioning functions. However, the sequence requirements for the partitioning and transcriptional activities appeared to be coincident, since the two activities responded in the same way to EBNA1 mutations. Using the Δ61-83 mutant, we have now shown that these two functions can be clearly distinguished. Differences in the responses of the partitioning and transcriptional activities of EBNA1 have also been reported when amino acids 40 to 89 were replaced by residues 328 to 377 (34). While the EBNA1 partitioning and transcriptional activities can be disrupted by deletion of a single EBNA1 region, no single EBNA1 sequence outside of the DNA binding domain has been shown to be essential for replication; rather, the EBNA1 N terminus and the 325-to-376 region play redundant roles in activating replication. The sequence requirements for the replication, segregation, and transcriptional activities of EBNA1 are summarized in Fig. 9. This figure reflects the requirement for DNA binding in addition to the sequences discussed in this paper. While it remains to be determined whether additional EBNA1 sequences also contribute to EBNA1 replication, segregation, and transcription functions, we find that deletion of amino acids 395 to 450 (H. Wu, M. Holowaty, and Lori Frappier, unpublished data) or 607 to 641 (7) does not disrupt these three activities.
Acknowledgments
We thank Derek Ceccarelli for pc3oriP plasmids expressing EBNA1, Δ325-376, and Δ41-376 EBNA1 mutants and for helpful advice on functional assays in human cells. We also acknowledge Kathy Shire for generating the EBNAΔ330-354 mutant, Jaap Middeldorp for anti-EBNA1 antibodies, and Aled Edwards for critical reading of the manuscript.
This work was supported by a grant from the National Cancer Institute of Canada and a Premier's Research Excellence Award to L.F. L.F. is a Medical Research Council of Canada Scientist.
REFERENCES
- 1.Adams, A. 1987. Replication of latent Epstein-Barr virus genomes. J. Virol. 61:1743-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambinder, R. F., M. Mullen, Y. Chang, G. S. Hayward, and S. D. Hayward. 1991. Functional domains of Epstein-Barr nuclear antigen EBNA-1. J. Virol. 65:1466-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avolio-Hunter, T. M., and L. Frappier. 1998. Mechanistic studies on the DNA linking activity of the Epstein-Barr nuclear antigen 1. Nucleic Acids Res. 26:4462-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avolio-Hunter, T. M., P. N. Lewis, and L. Frappier. 2001. Epstein-Barr nuclear antigen 1 binds and destabilizes nucleosomes at the viral origin of latent DNA replication. Nucleic Acids Res. 29:3520-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bochkarev, A., J. Barwell, R. Pfuetzner, E. Bochkareva, L. Frappier, and A. M. Edwards. 1996. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein, EBNA1, bound to DNA. Cell 84:791-800. [DOI] [PubMed] [Google Scholar]
- 6.Bochkarev, A., J. Barwell, R. Pfuetzner, W. Furey, A. Edwards, and L. Frappier. 1995. Crystal structure of the DNA binding domain of the Epstein-Barr virus origin-binding protein EBNA1. Cell 83:39-46. [DOI] [PubMed] [Google Scholar]
- 7.Ceccarelli, D. F. J., and L. Frappier. 2000. Functional analyses of the EBNA1 origin DNA binding protein of Epstein-Barr virus. J. Virol. 74:4939-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaudhuri, B., H. Xu, I. Todorov, A. Dutta, and J. L. Yates. 2001. Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 98:10085-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, M.-R., J. M. Middeldorp, and S. D. Hayward. 1993. Separation of the complex DNA binding domain of EBNA-1 into DNA recognition and dimerization subdomains of novel structure. J. Virol. 67:4875-4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cruickshank, J., A. Davidson, A. M. Edwards, and L. Frappier. 2000. Two domains of the Epstein-Barr virus origin DNA binding protein, EBNA1, orchestrate sequence-specific DNA binding. J. Biol. Chem. 275:22273-22277. [DOI] [PubMed] [Google Scholar]
- 11.Delecluse, H.-J., S. Bartnizke, W. Hammerschmidt, J. Bullerdiek, and G. W. Bornkamm. 1993. Episomal and integrated copies of Epstein-Barr virus coexist in Burkitt's lymphoma cell lines. J. Virol. 67:1292-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhar, S. K., K. Yoshida, Y. Machida, P. Khaira, B. Chaudhuri, J. A. Wohlschlegel, M. Leffak, J. Yates, and A. Dutta. 2001. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell 106:287-296. [DOI] [PubMed] [Google Scholar]
- 13.Frappier, L., K. Goldsmith, and L. Bendell. 1994. Stabilization of the EBNA1 protein on the Epstein-Barr virus latent origin of DNA replication by a DNA looping mechanism. J. Biol. Chem. 269:1057-1062. [PubMed] [Google Scholar]
- 14.Frappier, L., and M. O'Donnell. 1991. Epstein-Barr nuclear antigen 1 mediates a DNA loop within the latent replication origin of Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 88:10875-10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frappier, L., and M. O'Donnell. 1991. Overproduction, purification and characterization of EBNA1, the origin binding protein of Epstein-Barr virus. J. Biol. Chem. 266:7819-7826. [PubMed] [Google Scholar]
- 16.Gahn, T., and B. Sugden. 1995. An EBNA1-dependent enhancer acts from a distance of 10 kilobase pairs to increase expression of the Epstein-Barr virus LMP gene. J. Virol. 69:2633-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gahn, T. A., and C. L. Schildkraut. 1989. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell 58:527-535. [DOI] [PubMed] [Google Scholar]
- 18.Goldsmith, K., L. Bendell, and L. Frappier. 1993. Identification of EBNA1 amino acid sequences required for the interaction of the functional elements of the Epstein-Barr virus latent origin of DNA replication. J. Virol. 67:3418-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grogan, E. A., W. P. Summers, S. Dowling, D. Shedd, L. Gradoville, and G. Miller. 1983. Two Epstein-Barr viral nuclear neoantigens distinguished by gene transfer, serology and chromosome binding. Proc. Natl. Acad. Sci. USA 80:7650-7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris, A., B. D. Young, and B. E. Griffin. 1985. Random association of Epstein-Barr virus genomes with host cell metaphase chromosomes in Burkitt's lymphoma-derived cell lines. J. Virol. 56:328-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hearing, J., Y. Mulhaupt, and S. Harper. 1992. Interaction of Epstein-Barr virus nuclear antigen 1 with the viral latent origin of replication. J. Virol. 66:694-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell culture. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 23.Hung, S. C., M.-S. Kang, and E. Kieff. 2001. Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc. Natl. Acad. Sci. USA 98:1865-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones, C. H., S. D. Hayward, and D. R. Rawlins. 1989. Interaction of the lymphocyte-derived Epstein-Barr virus nuclear antigen EBNA-1 with its DNA-binding sites. J. Virol. 63:101-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanda, T., M. Otter, and G. M. Wahl. 2001. Coupling of mitotic chromosome tethering and replication competence in Epstein-Barr virus-based plasmids. Mol. Cell. Biol. 21:3576-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kapoor, P., K. Shire, and L. Frappier. 2001. Reconstitution of Epstein-Barr virus-based plasmid partitioning in budding yeast. EMBO J. 20:222-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirchmaier, A. L., and B. Sugden. 1997. Dominant-negative inhibitors of EBNA1 of Epstein-Barr virus. J. Virol. 71:1766-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krysan, P. J., S. B. Haase, and M. P. Calos. 1989. Isolation of human sequences that replicate autonomously in human cells. Mol. Cell. Biol. 9:1026-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kube, D., M. Vockerodt, O. Weber, K. Hell, J. Wolf, B. Haier, F. A. Grasser, N. Muller-Lantzsch, E. Kieff, V. Diehl, and H. Tesch. 1999. Expression of Epstein-Barr virus nuclear antigen 1 is associated with enhanced expression of CD25 in the Hodgkin cell line L428. J. Virol. 73:1630-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laine, A., and L. Frappier. 1995. Identification of Epstein-Barr nuclear antigen 1 protein domains that direct interactions at a distance between DNA-bound proteins. J. Biol. Chem. 270:30914-30918. [DOI] [PubMed] [Google Scholar]
- 31.Langle-Rouault, F., V. Patzel, A. Benavente, M. Taillez, N. Silvestre, A. Bompard, G. Sczakiel, E. Jacobs, and K. Rittner. 1998. Up to 100-fold increase of apparent gene expression in the presence of Epstein-Barr virus oriP sequences and EBNA1: implications of the nuclear import of plasmids. J. Virol. 72:6181-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lupton, S., and A. J. Levine. 1985. Mapping of genetic elements of Epstein-Barr virus that facilitate extrachromosomal persistence of Epstein-Barr virus-derived plasmids in human cells. Mol. Cell. Biol. 5:2533-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mackey, D., T. Middleton, and B. Sugden. 1995. Multiple regions within EBNA1 can link DNAs. J. Virol. 69:6199-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mackey, D., and B. Sugden. 1999. The linking regions of EBNA1 are essential for its support of replication and transcription. Mol. Cell. Biol. 19:3349-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marechal, V., A. Dehee, R. Chikhi-Brachet, T. Piolot, M. Coppey-Moisan, and J. Nicolas. 1999. Mapping EBNA1 domains involved in binding to metaphase chromosomes. J. Virol. 73:4385-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Middleton, T., and B. Sugden. 1992. EBNA1 can link the enhancer element to the initiator element of the Epstein-Barr virus plasmid origin of DNA replication. J. Virol. 66:489-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niller, H. H., G. Glaser, R. Knuchel, and H. Wolf. 1995. Nucleoprotein complexes and DNA 5"-ends at oriP of Epstein-Barr virus. J. Biol. Chem. 270:12864-12868. [DOI] [PubMed] [Google Scholar]
- 38.Petti, L., C. Sample, and E. Kieff. 1990. Subnuclear localization and phosphorylation of Epstein-Barr virus latent infection nuclear proteins. Virology 176:563-574. [DOI] [PubMed] [Google Scholar]
- 39.Reisman, D., and B. Sugden. 1986. trans-Activation of an Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclear antigen 1. Mol. Cell. Biol. 6:3838-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reisman, D., J. Yates, and B. Sugden. 1985. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol. Cell. Biol. 5:1822-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schepers, A., M. Ritzi, K. Bousset, E. Kremmer, J. L. Yates, J. Harwood, J. F. X. Diffley, and W. Hammerschmidt. 2001. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J. 20:4588-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaw, P. J., and E. G. Jordan. 1995. The nucleolus. Annu. Rev. Dev. Biol. 11:93-121. [DOI] [PubMed] [Google Scholar]
- 43.Shire, K., D. F. J. Ceccarelli, T. M. Avolio-Hunter, and L. Frappier. 1999. EBP2, a human protein that interacts with sequences of the Epstein-Barr nuclear antigen 1 important for plasmid maintenance. J. Virol. 73:2587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simpson, K., A. McGuigan, and C. Huxley. 1996. Stable episomal maintenance of yeast artificial chromosomes in human cells. Mol. Cell. Biol. 16:5117-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srinivas, S. K., and J. W. Sixbey. 1995. Epstein-Barr virus induction of recombinase-activating genes RAG1 and RAG2. J. Virol. 69:8155-8158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Su, W., T. Middleton, B. Sugden, and H. Echols. 1991. DNA looping between the origin of replication of Epstein-Barr virus and its enhancer site: stabilization of an origin complex with Epstein-Barr nuclear antigen 1. Proc. Natl. Acad. Sci. USA 88:10870-10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Summers, H., J. A. Barwell, R. A. Pfuetzner, A. M. Edwards, and L. Frappier. 1996. Cooperative assembly of EBNA1 on the Epstein-Barr virus latent origin of replication. J. Virol. 70:1228-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Scoy, S., I. Watakabe, A. R. Krainer, and J. Hearing. 2000. Human p32: a coactivator for Epstein-Barr virus nuclear antigen-1-mediated transcriptional activation and possible role in viral latent cycle DNA replication. Virology 275:145-157. [DOI] [PubMed] [Google Scholar]
- 49.Wu, H., D. F. J. Ceccarelli, and L. Frappier. 2000. The DNA segregation mechanism of the Epstein-Barr virus EBNA1 protein. EMBO Rep. 1:140-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wysokenski, D. A., and J. L. Yates. 1989. Multiple EBNA1-binding sites are required to form an EBNA1-dependent enhancer and to activate a minimal replicative origin within oriP of Epstein-Barr virus. J. Virol. 63:2657-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yates, J. L. 1996. Epstein-Barr virus DNA replication, p. 751-773. In M. L. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 52.Yates, J. L., and S. M. Camiolo. 1988. Dissection of DNA replication and enhancer activation functions of Epstein-Barr virus nuclear antigen 1. Cancer Cells 6:197-205. [Google Scholar]
- 53.Yates, J. L., and N. Guan. 1991. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J. Virol. 65:483-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yates, J. L., N. Warren, and B. Sugden. 1985. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 313:812-815. [DOI] [PubMed] [Google Scholar]