Abstract
For all enveloped viruses, the actual mechanism by which nascent virus particles separate or “pinch off” from the cell surface is largely unknown. In the case of retroviruses, the Gag protein drives the budding process, and the virus release step is directed by the late (L) assembly domain within Gag. A PPPPY motif within the L domain of Rous sarcoma virus (RSV) was previously characterized as being critical for the release of virions and shown to interact in vitro with the WW domain of Yes-associated protein (Yap). To determine whether WW domain-L domain interactions can occur in vivo, we attempted to interfere with the host cell machinery normally recruited to the site of budding by inserting this WW domain in different locations within Gag. At a C-terminal location, the WWYap domain had no effect on budding, suggesting that the intervening I domains (which provide the major region of Gag-Gag interaction) prevent its access to the L domain. When positioned on the other side of the I domains closer to the L domain, the WWYap domain resulted in a dramatic interference of particle release, and confocal microscopy revealed a block to budding on the plasma membrane. Budding was restored by attachment of the heterologous L domain of human immunodeficiency virus type 1 Gag, which does not bind WWYap. These findings suggest that cis expression of WW domains can interfere with RSV particle release in vivo via specific, high-affinity interactions at the site of assembly on the plasma membrane, thus preventing host factor accessibility to the L domain and subsequent virus-cell separation. In addition, they suggest that L domain-specific host factors function after Gag proteins begin to interact.
Retroviral Gag proteins direct the budding of enveloped virus-like particles when expressed alone (i.e., in the absence of all the other viral products), and these particles are of the proper size, density, and morphology (21). Detailed mapping of the functional elements of Rous sarcoma virus (RSV) and human immunodeficiency virus (HIV) Gag proteins has allowed the identification of three small modular domains that are essential components of the budding apparatus (7, 21). These are the M (membrane-binding) domain, the I (interaction) domain, and the L (late) domain (Fig. 1). After Gag proteins reach the plasma membrane and begin to interact, buds emerge from the cell surface and are transiently tethered via thin, membranous stalks. For “pinching off” to occur, the opposing membranes in each stalk must fuse to release the particle, and it is the L domain that somehow directs this virus-cell separation step.
FIG. 1.
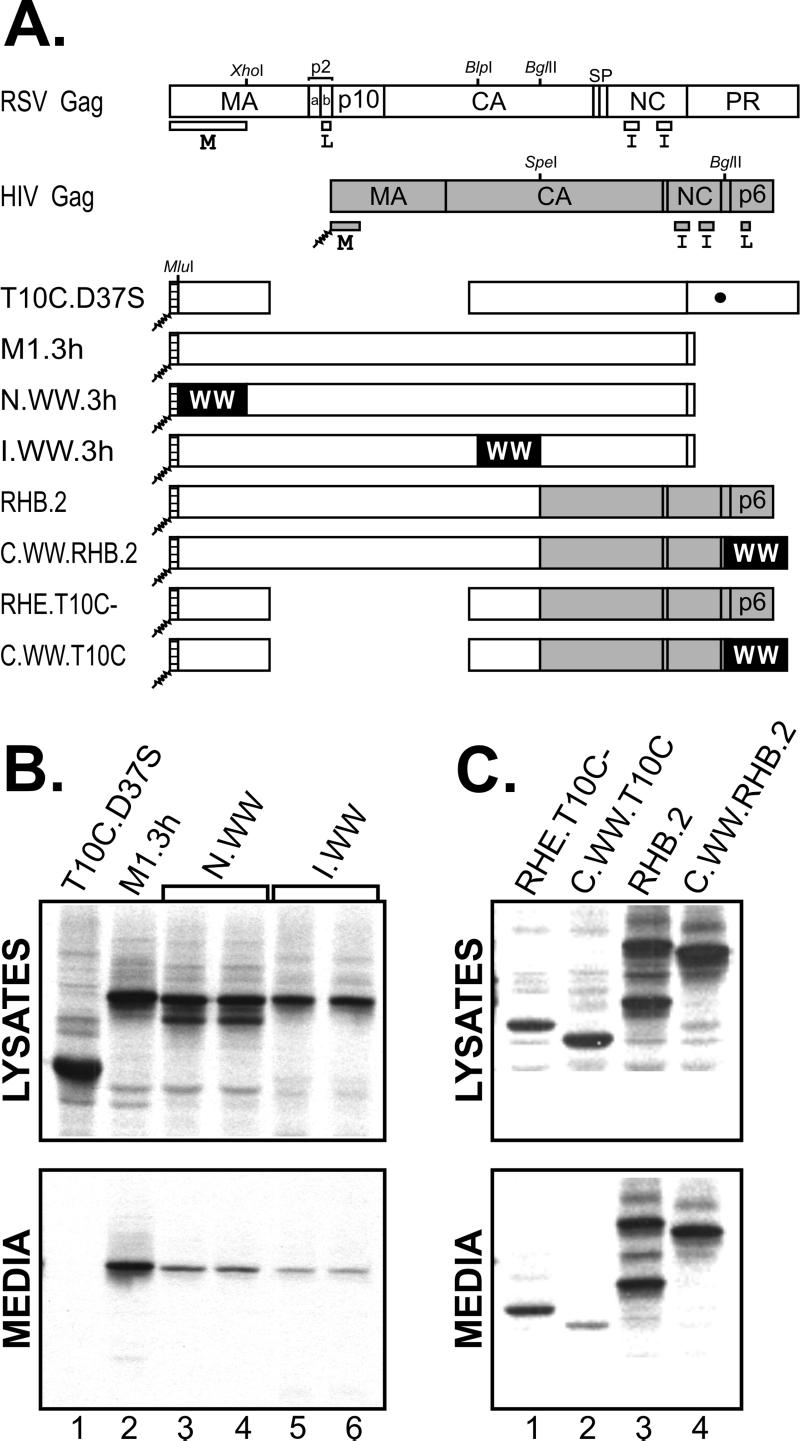
Effects of cis-expressed WW domains on the release of Gag from mammalian cells. (A) The wild-type RSV (open boxes) and HIV-1 (gray boxes) Gag proteins are shown along with sites that are cleaved to release the mature products (vertical lines). The small domains required for budding (M, L, and I) are indicated below each Gag protein. An active site mutation that prevents proteolysis in RSV Gag is represented by a large black dot. The small hatched boxes at the N termini of the chimeras represent the plasma membrane-binding domain from the Src oncoprotein and its associated myristate (squiggly lines). The positions of inserted WW domains are labeled, but the black boxes representing them are not drawn to scale. Relevant restriction endonuclease sites used in the cloning procedures are indicated at their positions relative to DNA. COS-1 cells were transfected with the constructs, and 48 h later they were labeled with [35S]methionine for 2.5 h. Viral proteins in detergent lysates of the cells and growth medium were collected by immunoprecipitation with anti-RSV serum, separated by electrophoresis through a sodium dodecyl sulfate-12% polyacrylamide gel, and visualized by fluorography. (B) Analysis of duplicate N-terminal and internal Gag.WW clones. (C) Analysis of C-terminal Gag.WW chimeras.
L domains are composed of short sequence motifs and were named because their alteration causes a block late in the budding pathway (i.e., resulting in the accumulation of virus-like particles attached to the cell surface by means of the stalk). The L domain of RSV is located near the amino terminus of Gag in the p2b region and has the core sequence PPPPY (24, 27). A similar sequence has been found in the p12 sequence of murine leukemia virus (29), the pp16 protein of Mason-Pfizer monkey virus (28), the matrix protein of rhabdoviruses (6, 11, 13), and the VP40 protein of Ebola virus (10, 18). In contrast, the L domains of HIV and equine infectious anemia virus are located near the carboxy termini of their Gag proteins and have the sequences PTAP (9, 12) and YPDL (15, 17), respectively. L domains are functionally exchangeable between unrelated viruses (6, 11, 15, 18) and can also be moved to ends of Gag opposite their normal location, indicating that they are positionally independent, too (15, 27).
It is widely believed that L domains recruit host proteins to the site of budding to facilitate virus-cell separation; however, the composition of this cellular machinery remains to be determined. The PY motifs of the sort found in RSV Gag resemble ligands for WW domains, which are approximately 38-amino-acid modules containing two widely spaced, conserved tryptophans that are found in a wide variety of signaling, regulatory, and cytoskeletal proteins (19, 20). For RSV, vesicular stomatitis virus, and Ebola virus, the L domains have been shown to interact in vitro with WW domains from Yes-associated protein (Yap), a signal-transducing molecule, and NEDD4, an E3 ubiquitin ligase (8, 10, 11). In contrast, the unrelated L domains of HIV and equine infectious anemia virus do not interact with WWYap.
As an initial step towards elucidation of the virus-host cell interactions that mediate pinching off, a cis approach was utilized to determine whether WW domains can specifically interact with RSV L domains in vivo and thereby interfere with the late steps of assembly. The results demonstrate that the interaction does occur, resulting in a block to budding which can be largely overcome by attachment of a heterologous HIV type 1 (HIV-1) L domain to the C terminus of RSV Gag.
MATERIALS AND METHODS
Construction of Gag chimeras.
A previously described simian virus 40 (SV40)-based expression vector (25) was used to express gag alleles in simian (COS-1) cells. Several previously described RSV-HIV gag chimeras were used for some of the DNA manipulations including pSV.RHB (1), pSV.RHB.T10C (15), pSV.RHE.T10C (15), pSV.RHp6 (1), and pSV.RHB.2 (1).
To create N-terminal WW-Gag chimeras, pCEV15, containing the human yap gene (19), was used as a template for PCR, and the following upstream and downstream primers were utilized (the underlined sequence in each oligonucleotide corresponds to the particular restriction site [in parentheses] used for cloning): 5′-CTATACACGCGTCTCAGTCTTCTTTTGAGATACCT-3′ (MluI) and 5′-TACGACCTCGAGGACTGGTGGGGGCTGTGACGTTCA-3′ (XhoI). The PCR products were digested with MluI and XhoI and then inserted in place of the wild-type MluI-XhoI fragment in pSV.M1.3h (22), pSV.RHp6, pSV.RHB, pSV.RHB.2, and pSV.RHE.T10C to generate plasmids encoding N-terminal WW-Gag chimeras (pSV.N.WW.3h, pSV.N.WW.RHp6, pSV.N.WW.RHB, pSV.N.WW.RNB.2, and pSV.N.WW.T10C, respectively).
To create internal WW-Gag chimeras, a similar PCR approach was used, and the following upstream and downstream primers were utilized: 5′-CTATACGCTTAGCTCAGTCTTCTTTTGAGATACCT-3′ (BlpI) and 5′-TACGACAGATCTGGACTGGTGGGGGCTGTGACGTTC-3′ (BglII). The PCR product was digested with BlpI and BglII and inserted into the pSV.M1.3h and pSV.RHp6 constructs to create pSV.I.WW.3h and pSV.I.WW.RHp6, respectively.
The C-terminal WW-Gag chimeras were created using the following upstream and downstream primers: 5′-CTATACAGATCTCTCAGTCTTCTTTTGAGATACCT-3′ (BglII) and 5′-TACGACGCGCGCCTAACTGGTGGGGGCTGTGACGTTCA-3′ (BssHII). The PCR product was digested with BglII and BssHII and inserted into pSV.RHB.2 and pSV.RHE.T10C to create pSV.C.WW.RHB.2 and pSV.C.WW.T10C, respectively.
WW-Gag-GFP chimeras.
To localize RSV Gag proteins within living avian (QT6) cells, the green fluorescence protein (GFP) was fused to its C terminus. Because the SV40-based vector does not express gag in avian cells, we made use of a previously described cytomegalovirus (CMV) promoter-based vector named pCMV.Gag.GFP (3). A derivative of this construct that lacks the L-domain coding sequence, pT10C.GFP, has also been described previously (4). To link GFP to the N-terminal WW chimera, pSV.N.WW.3h and pCMV.Gag.GFP were digested with SstI and BspE1. The small fragment from the former and the large fragment from the latter were gel purified and ligated to generate pCMV.N.WW.GFP.
To generate pCMV.I.WW.GFP, pSV.I.WW.3h was used as a template to PCR amplify a fragment containing the WW domain. The following upstream and downstream primers were utilized: 5′-GATCTCGAGCTCTACTGCAGGGAGCCC-3′ (SstI) and 5′-TACGACGGGCCCGGGCCACGGCCCCGAAGA-3′ (ApaI). The PCR product was digested with SstI and ApaI and inserted into pCMV.Gag.GFP to generate pCMV.I.WW.GFP.
To link GFP to a membrane-binding mutant of N.WW.3h, plasmids pCMV.N.WW.GFP and pSV.Myr1(−) (2) were digested with SstI and MluI. The small fragment from pSV.Myr1(−) and the large fragment from pCMV.N.WW.GFP were gel purified and ligated to generate pCMV.M(−).N.WW.GFP.
Transfections.
COS-1 cells were grown in 35-mm-diameter dishes in Dulbecco's modified Eagle medium supplemented with 3% fetal bovine serum (FBS) and 7% calf serum. The DEAE-dextran-chloroquine method was used to transfect these cells with XbaI-digested and ligated plasmids as previously described (25). Typically, 0.75 μg of DNA was applied to each monolayer, and Gag expression was analyzed 48 h after transfection.
The QT6 cells were grown in F10 medium supplemented with 8.5% tryptose phosphate broth, 5.1% FBS, 1.0% chicken serum, and 0.1% penicillin-streptomycin. They were transfected by the calcium phosphate precipitation method. Approximately 1 h before transfection, the primary growth medium was replaced with Dulbecco's modified Eagle medium containing 10% FBS. Typically, 10 μg of DNA was applied to each monolayer, and expression was analyzed 18 h after transfection.
Budding assay.
Transfected cells were metabolically labeled with [35S]methionine for 2.5 h as previously described (25). The cells and growth medium from each labeled culture were separated and mixed with lysis buffer containing protease inhibitors, and the Gag proteins were collected by immunoprecipitation at 4°C. In all cases, a rabbit antiserum against whole RSV was used (22). The immunoprecipitated proteins were separated by electrophoresis in sodium dodecyl sulfate-12% polyacrylamide gels and detected by fluorography. Phosphorimager analysis was used to compute the budding efficiency, which was calculated as the amount of protein in the medium divided by the total in the lysates and medium. The effect of the WW domain on budding was then determined by computing the ratios of budding efficiency in the absence and presence of the WW domain unless otherwise indicated.
Confocal microscopy.
Duplicate plates were transfected for each construct. One plate from each pair was used to visualize the subcellular location of chimeric proteins by confocal microscopy. The second plate was metabolically labeled with [35S]methionine and subsequently processed as described above to determine the expression and budding efficiencies of the chimeric proteins.
RESULTS
As an initial step towards understanding the proposed role of L domain function in vivo, we attempted to interfere with RSV budding by cis overexpression of the WW domain from the Yap protein. We chose this WW domain because it has been shown to bind RSV Gag in vitro (8); however, there is no evidence to suggest that Yap is the normal binding partner for Gag in vivo, and the biologically relevant molecule(s) is uncertain. The WWYap domain was inserted at three locations: the N terminus of Gag within MA, in the middle of Gag within CA, or at the C terminus of Gag (Fig. 1A). All of these constructs have the membrane-binding domain of the Src oncoprotein replacing the first 10 amino acids of the M domain. This manipulation renders the M domain dispensable since the Src sequence provides the necessary plasma membrane targeting and binding information (2, 26). The Gag derivatives were expressed in COS-1 (mammalian) cells by using a previously described SV40-based vector (25) and in QT6 (quail) cells by using a previously described CMV promoter-based vector (4). Similar results were obtained in both systems.
To assay for budding interference, cells were transfected with the N-terminal, internal, and C-terminal WWYap expression vectors and radiolabeled with [35S]Met for 2.5 h approximately 48 h posttransfection. The cells and growth media were separated and mixed with lysis buffer, and the Gag proteins were collected by immunoprecipitation at 4°C by using rabbit antiserum against whole RSV. As a positive control, we used M1.3h (22), which lacks the RSV protease but is released into the medium with normal efficiency (Fig. 1B, lane 2). As a negative control, we used T10C.D37S (23, 24), which is defective for budding because it lacks the L domain (Fig. 1B, lane 1). When the WW domain was inserted into Gag at an N-terminal or internal position, the chimeras were defective for particle release (Fig. 1B, lanes 3 to 6). Phosphorimager analysis showed that the N.WW.3h and I.WW.3h chimeric proteins were released at 35.8 ± 7.9% (n = 4) and 14.2 ± 2.7% (n = 7) of the level of M1.3h, respectively.
To insert the Yap WW domain at the C terminus of Gag, we made use of an RSV-HIV chimera, RHB.2, because the gene for this chimera contains a convenient restriction endonuclease (BglII) site near the 3′ end (Fig. 1A). As previously reported (1), this chimera buds with high efficiency (Fig. 1C, lanes 3). To our surprise, when the WW domain was inserted into the C terminus of RHB.2 (replacing HIV-1 p6Gag which contains the HIV L domain), the resulting C.WW.RHB.2 chimera was released at essentially normal efficiency (Fig. 1C, lanes 4), suggesting that the WW domain is unable to access the RSV L domain when placed at the C-terminal location (see Discussion). Phophorimager analysis revealed that this C-terminal WW.Gag chimera was released at 85.4 ± 16.9% (n = 4) of the level of RHB.2. When the RSV L domain was removed to create C.WW.T10C, budding was lost as expected (Fig. 1C, lanes 2), but it was restored by replacing the WW domain with p6 and its associated L domain to create RHE.T10C− (Fig. 1C, lanes 1). Phosphorimager analysis revealed that C.WW.T10C was released at only 3.9 ± 2.8% (n = 4) relative to RHE.T10C−.
There are numerous ways that insertion of the WW domain at the N-terminal and internal positions of Gag could possibly interfere with budding, some of which would not be interesting. For example, if intermolecular interactions occurred between the WW domain in Gag and PY motifs on cellular protein(s) or PYs on other Gag proteins before the chimeric molecules reached the plasma membrane, then tangled aggregates might form in the cytoplasm. In this case, the block to budding would occur early in the pathway and in a manner that is irrelevant to L domain activity. Similarly, intramolecular interactions between the inserted WW domain and the L domain within each Gag molecule might result in a cytoplasmic accumulation of assembly-incompetent, misfolded molecules which fail to enter the budding pathway. However, it was also possible that the chimeric Gag proteins would be properly folded and capable of reaching the plasma membrane, with the observed block to budding occurring late due to masking of the PPPPY motif by the inserted WW domain (either inter- or intramolecularly). This would be a more-interesting event that would result in the chimeras accumulating at the plasma membrane during steady state.
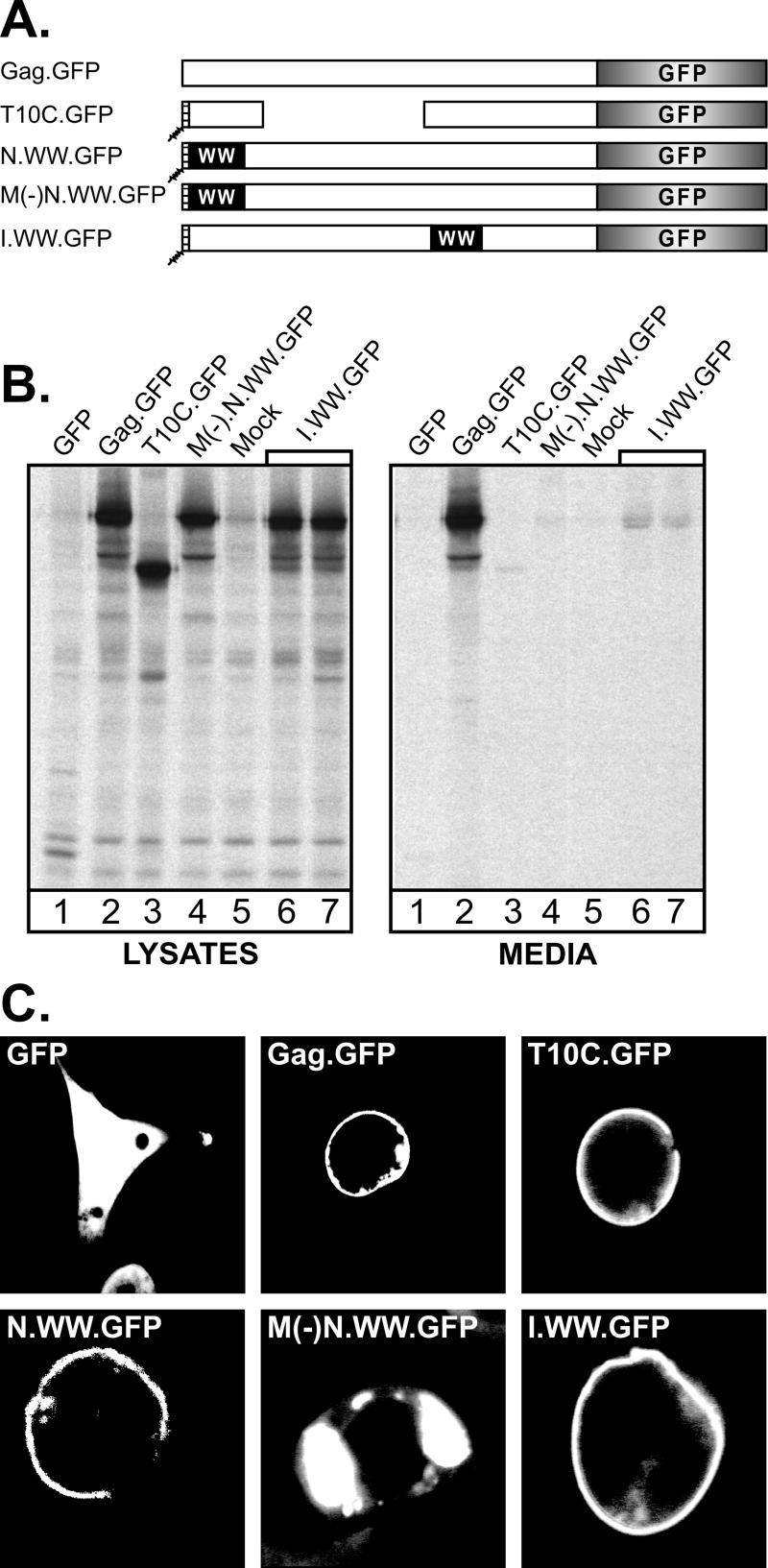
To ascertain the subcellular location of the WW chimeras, the GFP sequence was linked to the C termini of N.WW.3h and I.WW.3h (Fig. 2A). As a positive control, we used Gag.GFP (3), which is released into the medium with normal efficiency (Fig. 2B, lanes 2). As a negative control, we used T10C.GFP (4) which lacks the L domain and is defective for budding (Fig. 2B, lanes 3). Both of the Gag.WW constructs expressed proteins of the appropriate size, and as expected, N.WW.GFP (not shown) and I.WW.GFP (Fig. 2B, lanes 6 and 7) demonstrated severe budding defects, like the parental constructs lacking GFP. Confocal microscopy (Fig. 2C) revealed that both N.WW.GFP and I.WW.GFP localized to the plasma membrane, resembling the L domain mutant (T10C.GFP), which also accumulates at the cell surface. Inactivation of the M domain of N.WW.GFP [to create M(−)N.WW.Gag.GFP; Fig. 2A], resulted in a loss of membrane binding and the accumulation of large cytoplasmic aggregates. These results show that the budding defects in the N-terminal and internal WW.Gag chimeras are late in the assembly-release pathway.
FIG. 2.
Subcellular localization of WW-Gag chimeras. (A) GFP was inserted in place of the RSV PR sequence and the last 6 residues of NC to create the illustrated Gag.GFP derivatives. The open boxes represent the wild-type Gag protein. Hatched boxes with squiggly lines at the N termini of some of the constructs represent the myristylated Src membrane-binding domain. The large deletion in T10C.GFP removes the RSV L domain, and a mutation in M(−)N.WW.Gag inactivates the membrane-binding domain by preventing myristylation. (B) Duplicate plates of QT6 cells were transfected with the GFP chimeras. Approximately 24 h posttransfection, one set of plates was metabolically labeled as described in the legend to Fig. 1, and the viral proteins from cell lysates and growth medium were analyzed as described in the legend to Fig. 1. (C) Cells from the second set of plates were examined by confocal microscopy.
To examine the presumptive WW-L domain interaction, we attempted to weaken it with amino acid substitutions designed by using the previously described structure of the WWYap domain in complex with a proline-rich peptide (5, 14). Of the mutants we examined, two with single (H192F or P202A) and one with triple (H192F/W199F/P202A) substitutions in the WW domain resulted in an increase in budding for the I.WW chimera (Table 1). The magnitude of the increase was modest, but this is not surprising since the 1,500 Gag molecules that come together to make a particle are tightly associated by means of their interaction domains, resulting in high local concentrations of WW and L domains. Thus, a weakened WW domain might be expected to still have substantial inhibitory activity in this context. Another substitution (W199F) had no effect on budding by itself and negated the positive effect seen with H192F (Table 1), suggesting that it has an adverse effect on the structure of the WW domain. The failure of this mutant to behave as expected may be a limitation of the available structural information, which was obtained by using a proline-rich motif (GTPPPPYTVG) that is similar to, but distinct from, the sequence found in RSV Gag (SAPPPPYVGS). Nevertheless, these results collectively suggest that a specific interaction does occur between the WW and L domains in Gag.
TABLE 1.
cis and trans rescue of I.WW
| Experiment | Construct(s) | Fold enhancement of buddingb |
|---|---|---|
| cis WW substitution(s)a | H192F | 2.0 |
| W199F | 0.9 | |
| P202A | 1.5 | |
| H192F/W199F | 0.9 | |
| H192F/W199F/P202A | 1.5 | |
| trans complementationc | I.WW + Gag | 2.0 |
COS-1 cells were transfected with I.WW.3h mutants having the indicated amino acid substitutions in the human Yap WW domain.
Viral proteins from cell lysates and growth medium were analyzed as described in the legend to Fig. 1 and the release of the I.WW chimeras relative to that of the controls was calculated.
QT6 cells were cotransfected with 5 μg of pCMV.I.WW.GFP plus 5 μg of either empty vector (negative control) or pCMV.Gag (minus GFP).
To directly address the possibility that I.WW.3h fails to bud because it is misfolded, we attempted to rescue this chimera by trans complementation. This approach is based on the finding that functional L domains are needed on only a few molecules in a population of Gag proteins (2, 24). If I.WW.3h fails to bud because of structural defects resulting from the presence of the WW domain (rather than the specific interaction of WW with the late domain), then coexpression of a budding-competent Gag molecule would not rescue it into particles. On the other hand, if I.WW.3h fails to bud because its L domain is bound up with the WW domain, then rescue should occur when additional L domains are provided in trans. We found that I.WW.3h can indeed be rescued into particles (Table 1), further supporting the hypothesis that a specific interaction occurs between the WW domain and the L domain in Gag.
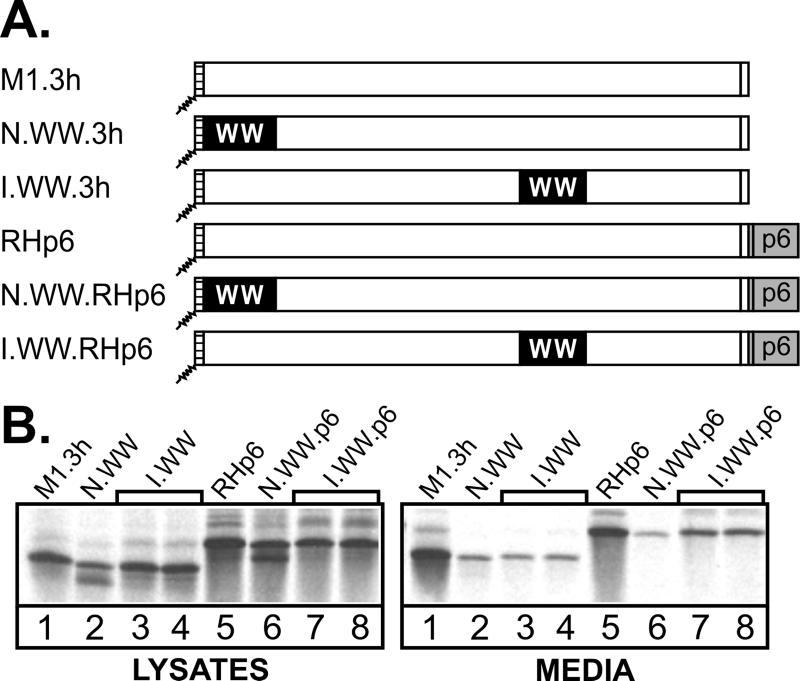
Further evidence that insertion of the WW domain does not result in grossly misfolded Gag proteins was obtained by using a heterologous L domain. If a specific interaction between the RSV L domain and the WWYap domain occurs at the site of assembly on the plasma membrane to block the recruitment of host machinery, then this defect should be eliminated by attachment of a foreign L domain having a completely different sequence. To test this, we utilized the L domain of HIV, which is located near the C terminus of Gag, in the p6 region (9, 12), and does not interact with WWYap in vitro (8). The p6Gag sequence was attached to the C-terminal ends of N.WW.3h and I.WW.3h to create N.WW.RHp6 and I.WW.RHp6 (Fig. 3A). As predicted, I.WW.RHp6 showed an increase (∼4-fold) in particle release relative to the corresponding chimeras without HIV-1 p6 (Fig. 3B, compare lanes 3 and 4 with lanes 7 and 8). In contrast, the N.WW.RHp6 chimera was still severely defective for particle release (Fig. 3B, compare lanes 2 and 6) and was released even more poorly than N.WW.3h (∼2-fold).
FIG. 3.
Suppression of WW-inhibitory effects by HIV p6. (A) The p6 sequence and its heterologous L domain were attached to the C termini of the N-terminal and internal Gag.WW chimeras to create the illustrated chimeras. (B) COS-1 cells were transfected with the indicated DNAs, and 48 h later the cells were labeled with [35S]methionine for 2.5 h. Viral proteins in the cell lysates and growth medium were analyzed as described in the legend to Fig. 1.
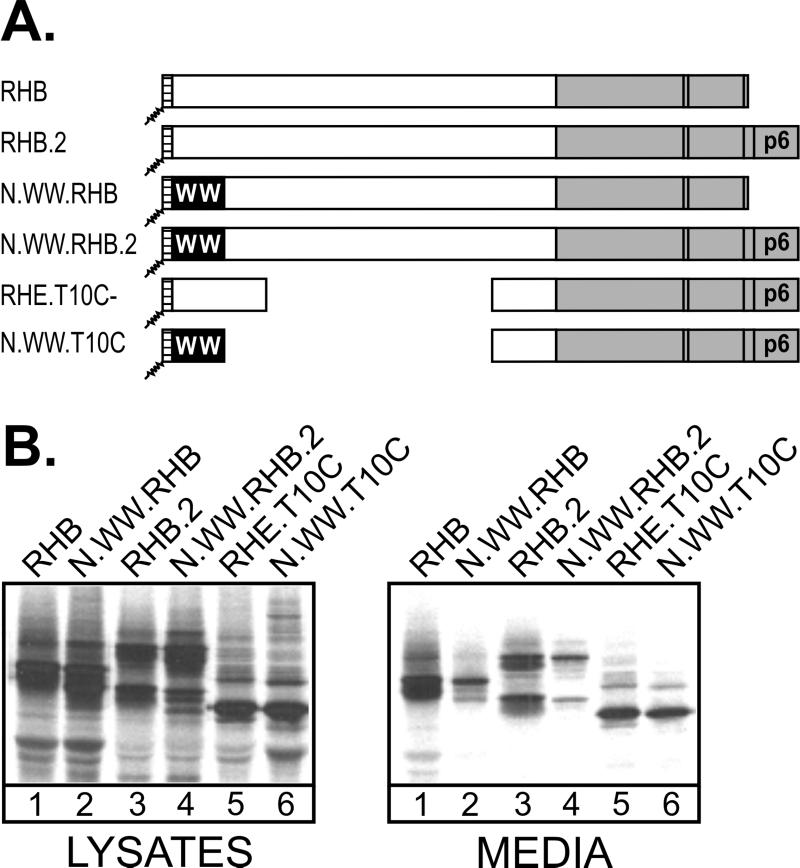
We considered two possible explanations for the lack of rescue seen with N.WW.RHp6. If the close proximity of the WW domain to the plasma membrane at the N-terminal location sterically interferes with budding, then removal of the RSV L domain from the p6 chimera (to create N.WW.T10C Fig. 4A) would not increase budding because the WW domain would still be present. On the other hand, if an interaction between the WW domain and the RSV L domain results in a conformational bulge that sterically interferes with budding, then removal of the L domain would prevent that interaction and restore budding even though the WW domain remains. As a positive control in this experiment, we used RHB (Fig. 4A), an RSV-HIV chimera which lacks p6Gag and has been shown (1) to be released into the medium with an efficiency similar to that of the p6Gag-containing parental construct, RHB.2 (Fig. 4B, compare lanes 1 and 3). As a negative control, we used N.WW.RHB, in which the WW domain is inserted at the N terminus of RHB. As expected, this recombinant was severely defective for particle release (Fig. 4B, lanes 2), and attachment of p6Gag (to make N.WW.RHB.2) did not restore budding (Fig. 4B, lanes 4), as discussed earlier. Budding was restored when the RSV L domain was deleted (to create N.WW.T10C), even though the WW domain was present (Fig. 4B, lanes 6). Quantitation revealed that N.WW.T10C was released at the same efficiency as previously described for RHE.T10C (15), a matched construct that lacks the WW domain (Fig. 4B, lanes 5). These results support the idea that specific WW-L domain interactions at a position close to the membrane produce a conformational bulge that interferes with budding.
FIG. 4.
Restoration of N.WW.RHp6 budding by deletion of the RSV L domain. (A) The top two chimeras have no inserted WW domains. The middle two chimeras have N-terminal WW domains but differ in the presence of the p6 sequence. The bottom two chimeras have the p6 sequence and its associated L domain, but both lack the RSV L domain as a result of large deletions. (B) COS-1 cells were transfected with the indicated DNAs, and viral proteins from cell lysates and growth medium were analyzed as described in the legend to Fig. 1.
DISCUSSION
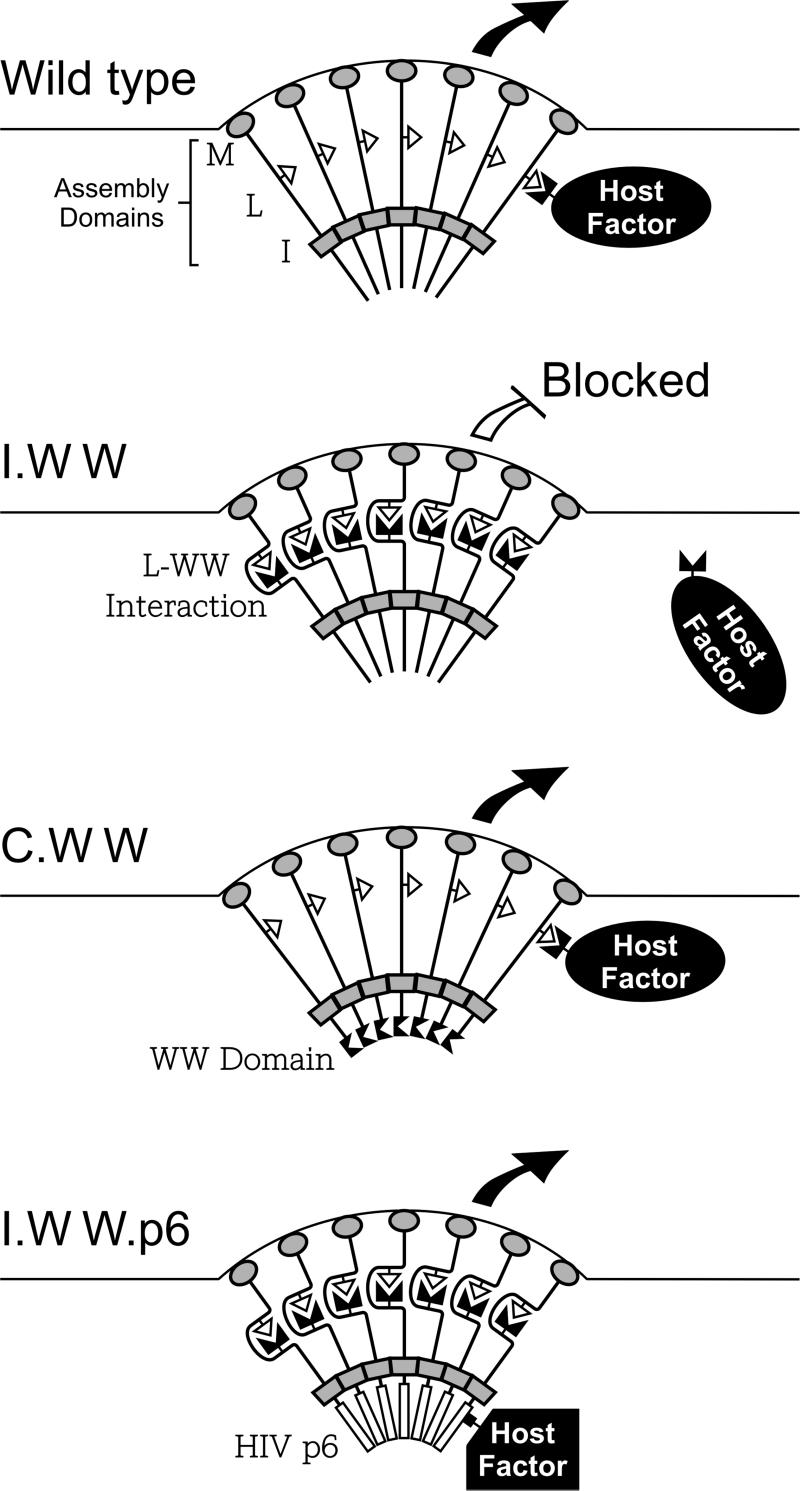
In this study, we attempted to interfere with retrovirus budding by inserting a WWYap domain at three different locations within Gag. This foreign sequence did not prevent any of the chimeras from reaching the plasma membrane. At the internal position of Gag, the WW domain dramatically interfered with particle release, but at the C terminus it did not. This positional dependence can be explained by examining the location of the inserted WW domains relative to the assembly domains of the wild-type Gag protein (M, L, and I). The major sites of Gag-Gag interaction are provided by interaction (I) domains, which form a physical barrier between the N and C termini of Gag, and the L domain is positioned on the side near the membrane where it interacts with host factors (Fig. 5, top panel). In the case of the internal chimera (Fig. 5, second panel), the L and WW domains are located on the same side of this barrier, which enables them to interact, thereby preventing the recruitment of host factors needed for budding. In the case of the C-terminal chimera (Fig. 5, third panel), the L and WW domains are on opposite sides of the barrier, which precludes their interaction and the inhibitory effect. The particle release defect seen with the internal chimera can be largely rescued by HIV-1 p6Gag because the late domain it contains does not interact with WWYap (Fig. 5, bottom panel). If these interpretations are correct, then it must follow that the host proteins recruited by L domains function after Gag proteins begin to interact and the I domain barrier is established. However, at this point we cannot rule out other possible explanations for the inability of the WW domain to block budding at the C-terminal location (e.g., improper folding).
FIG. 5.
Models for in vivo WW-Gag interactions. Assembly domains M (gray ovals), L (open triangles), and I (gray rectangles) are indicated. The horizontal lines in each panel denote cell membranes with emerging buds. WW domains are represented by paired black triangles. Host factors specifically recruited by late domains are indicated. The ability of the chimeric Gag proteins to be released from the cell surface is represented by a large black arrow (no budding interference) or an open headless arrow (inhibition of release). See the text for further discussion.
In the case of the N-terminal chimera (not illustrated in Fig. 5), the WW and L domains are positioned on the same side of Gag, but the block to budding must be more complex because attachment of the p6Gag sequence did not restore particle release. We believe that the WW and L domains do in fact interact in a specific manner in this construct because when the L domain is deleted, the p6Gag sequence is then capable of restoring budding activity even though the WW domain remains. It seems likely that the WW-L interaction would create a distortion in the Gag molecule, and we hypothesize that the close proximity of this bulge to the plasma membrane has an adverse effect on particle release. By moving the interacting domains away from the plasma membrane (as in the internal chimera), this nonspecific effect was eliminated.
Although the results described in this paper provide proof of the importance of the WW-L domain interaction in vivo, they do not shed light on the actual host proteins that are recruited to the sites of budding by the RSV late domain. There is no evidence that the Yap WW domain normally interacts with Gag in vivo, but when taken out of context, it is clearly an efficient inhibitor of budding. This is not surprising since WW domains exhibit broad binding specificities (16), and we see no reason why trans overexpression of an irrelevant WW domain could not have a similar effect. Our results emphasize the difficulty of demonstrating biological relevance for candidate host proteins that are identified on the basis of their ability to bind to a given late domain. Overexpression of such candidates (or fragments containing WW domains) may well interfere with budding even if that protein is not the actual binding partner. Moreover, there is no reason to believe that the actual binding partners of RSV Gag will have WW domains that bind better to its late domain than irrelevant proteins. Therefore, rigorous standards must be applied in attempting to identify and characterize the host proteins involved in the virus-cell separation step.
Acknowledgments
Special thanks go to Rebecca Craven for comments regarding the manuscript.
This work was supported by a grant from the National Institutes of Health (NIH) awarded to J.W.W. (CA47482).
REFERENCES
- 1.Bennett, R. P., T. D. Nelle, and J. W. Wills. 1993. Functional chimeras of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 67:6487-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett, R. P., and J. W. Wills. 1999. Conditions for copackaging Rous sarcoma virus and murine leukemia virus Gag proteins during retroviral budding. J. Virol. 73:2045-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowzard, J. B., R. J. Visalli, C. B. Wilson, E. M. Callahan, R. J. Courtney, and J. W. Wills. 2000. Membrane targeting properties of a herpesvirus tegument protein-retrovirus Gag chimera. J. Virol. 74:8692-8699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Callahan, E. M., and J. W. Wills. 2000. Repositioning basic residues in the M domain of the Rous sarcoma virus Gag protein. J. Virol. 74:11222-11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, H. I., A. Einbond, S.-J. Kwak, H. Linn, E. Koepf, S. Peterson, J. W. Kelly, and M. Sudol. 1997. Characterization of the WW domain of human yes-associated protein and its polyproline-containing ligands. J. Biol. Chem. 272:17070-17077. [DOI] [PubMed] [Google Scholar]
- 6.Craven, R. C., R. N. Harty, J. Paragas, P. Palese, and J. W. Wills. 1999. Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras. J. Virol. 73:3359-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craven, R. C., and L. J. Parent. 1996. Dynamic interactions of the Gag polyprotein, p. 65-94. In H.-G. Krausslich (ed.), Morphogenesis and maturation of retroviruses. Springer-Verlag, Berlin, Germany. [DOI] [PubMed]
- 8.Garnier, L., J. W. Wills, M. F. Verderame, and M. Sudol. 1996. WW domains and retrovirus budding. Nature (London) 381:744-745. [DOI] [PubMed] [Google Scholar]
- 9.Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harty, R. N., J. Paragas, M. Sudol, and P. Palese. 1999. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J. Virol. 73:2921-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jayakar, H. R., K. G. Murti, and M. A. Whitt. 2000. Mutations in the PPPY motif of vesicular stomatitis virus matrix protein reduce virus budding by inhibiting a late step in virion release. J. Virol. 74:9818-9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macias, M. J., M. Hyvonen, E. Baraldi, J. Schultz, M. Sudol, M. Saraste, and H. Oschkinat. 1996. Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature 382:646-649. [DOI] [PubMed] [Google Scholar]
- 15.Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 69:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pirozzi, G., S. J. McConnell, A. J. Uveges, J. M. Carter, A. B. Sparks, B. K. Kay, and D. M. Fowlkes. 1997. Identification of novel human WW domain-containing proteins by cloning of ligand targets. J. Biol. Chem. 272:14611-14616. [DOI] [PubMed] [Google Scholar]
- 17.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strack, B., A. Calistri, M. A. Accola, G. Palú, and H. G. Göttlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97:13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sudol, M., P. Bork, A. Einbond, K. Kastury, T. Druck, M. Negrini, K. Huebner, and D. Lehman. 1995. Characterization of the mammalian YAP (Yes-associated protein) gene and its role in defining a novel protein module, the WW domain. J. Biol. Chem. 270:14733-14741. [DOI] [PubMed] [Google Scholar]
- 20.Sudol, M., H. I. Chen, C. Bougeret, A. Einbond, and P. Bork. 1995. Characterization of a novel protein-binding module-the WW domain. FEBS Lett. 369:67-71. [DOI] [PubMed] [Google Scholar]
- 21.Swanstrom, R., and J. W. Wills. 1997. Retroviral gene expression. II. Synthesis, processing, and assembly of viral proteins, p. 263-334. In R. Weiss, N. Teich, H. Varmus, and J. M. Coffin (ed.), Retroviruses. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y. [PubMed]
- 22.Weldon, R. A., Jr., C. R. Erdie, M. G. Oliver, and J. W. Wills. 1990. Incorporation of chimeric Gag protein into retroviral particles. J. Virol. 64:4169-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weldon, R. A., Jr., and J. W. Wills. 1993. Characterization of a small (25-kilodalton) derivative of the Rous sarcoma virus Gag protein competent for particle release. J. Virol. 67:5550-5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wills, J. W., R. C. Craven, and J. A. Achacoso. 1989. Creation and expression of myristylated forms of Rous sarcoma virus Gag protein in mammalian cells. J. Virol. 63:4331-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wills, J. W., R. C. Craven, R. A. Weldon, Jr., T. D. Nelle, and C. R. Erdie. 1991. Suppression of retroviral MA deletions by the amino-terminal membrane-binding domain of p60src. J. Virol. 65:3804-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiang, Y., C. E. Cameron, J. W. Wills, and J. Leis. 1996. Fine mapping and characterization of Rous sarcoma virus pr76gag late assembly domain. J. Virol. 70:5695-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72:4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan, B., X. Li, and S. Goff. 1999. Mutations altering the Moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18:4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]