Abstract
Mammalian reovirus infection results in perturbation of host cell cycle progression. Since reovirus infection is known to activate cellular transcription factors, we investigated alterations in cell cycle-related gene expression following HEK293 cell infection by using the Affymetrix U95A microarray. Serotype 3 reovirus infection results in differential expression of 10 genes classified as encoding proteins that function at the G1-to-S transition, 11 genes classified as encoding proteins that function at G2-to-M transition, and 4 genes classified as encoding proteins that function at the mitotic spindle checkpoint. Serotype 1 reovirus infection results in differential expression of four genes classified as encoding proteins that function at the G1-to-S transition and three genes classified as encoding proteins that function at G2-to-M transition but does not alter any genes classified as encoding proteins that function at the mitotic spindle checkpoint. We have previously shown that serotype 3, but not serotype 1, reovirus infection induces a G2-to-M transition arrest resulting from an inhibition of cdc2 kinase activity. Of the differentially expressed genes encoding proteins regulating the G2-to-M transition, chk1, wee1, and GADD45 are known to inhibit cdc2 kinase activity. A hypothetical model describing serotype 3 reovirus-induced inhibition of cdc2 kinase is presented, and reovirus-induced perturbations of the G1-to-S, G2-to-M, and mitotic spindle checkpoints are discussed.
Perturbation of cell cycle regulation is a characteristic of infection by viruses belonging to a diverse group of viral families. The reasons viruses stimulate proliferation or induce cell cycle arrest are not completely understood. In some cases, virus replication may depend on the availability of host cell precursors, whose abundance varies in a cell cycle-specific manner. In other cases, kinases critical in regulating cell cycle progression may be essential for phosphorylating viral proteins (reviewed in reference 58).
Reoviruses infect a variety of mammalian hosts and serve as an important experimental system for studying the molecular bases of viral pathogenesis (reviewed in reference 80). Reoviruses also provide a valuable model for studying virus-induced perturbations in cell cycle regulation, since reovirus infection has been associated with G1 arrest, G2/M arrest, and disruption of the mitotic spindle apparatus (see below). Inhibition of host cell DNA synthesis is one of the earliest cytopathic effects observed following serotype 3 reovirus infection in cultured cells (reviewed in reference 53). It was originally suggested that serotype 3 reovirus-induced inhibition of cellular proliferation resulted from inhibition of the initiation of DNA replication (14, 27, 68). However, the degree of cell culture synchronization prior to infection was either incomplete or not specified in these studies, impeding accurate identification of the cell cycle phase affected (14, 68).
Serotype 3 prototype strains type 3 Dearing (T3D) and type 3 Abney (T3A) inhibit cellular DNA synthesis to a greater extent than the serotype 1 prototype strain type 1 Lang (T1L) in a variety of cell lines (20, 23, 75, 82). Studies using T1L × T3D and T1L × T3A reassortant viruses indicate that the serotype 3 S1 gene is the primary determinant of differences in the capacity of reovirus strains to inhibit DNA synthesis (75, 82). The reovirus S1 gene segment is bicistronic, encoding the viral attachment protein, σ1, and a non-virion-associated protein, σ1s, from overlapping, alternative open reading frames (53).
Studies using purified recombinant serotype 3 σ1 protein and an anti-idiotype antibody (87.92.6) generated against the T3D σ1-specific monoclonal antibody 9BG5 suggest that inhibition of DNA synthesis in some cells may result from engagement of a cell surface receptor by σ1 (23, 70-72). For example, treatment of R1.1 thymoma cells with purified T3D σ1 results in a reversible G1-to-S transition arrest (70, 71). The mechanism for T3D σ1-induced G1-to-S arrest is not clear but may involve inhibition of p21ras (Ha-ras), since overexpression of Ha-ras prevents T3D σ1-induced G1-to-S transition arrest (71, 72).
We have shown that reovirus infection inhibits cellular proliferation by inducing a G2/M phase cell cycle arrest in a variety of cell types (63). T3A and T3D induce G2/M phase cell cycle arrest to a greater extent than T1L (63). Like strain-specific differences in the capacity of reovirus to inhibit DNA synthesis (82), strain-specific differences in the capacity of reovirus to induce G2/M phase cell cycle arrest are determined by the serotype 3 S1 gene (63). The S1-encoded σ1s protein is both necessary and sufficient to induce G2/M arrest, since a σ1s-deficient reovirus mutant fails to induce G2/M arrest and inducible expression of σ1s results in accumulation of cells in the G2/M phase of the cell cycle (63).
G2-to-M transition requires the formation and activation of the p34cdc2/cdk1 (cdc2)-cyclin B heterodimeric complex (reviewed in references 38 and 55). Activation of the cdc2-cyclin B complex is regulated by inhibitory phosphorylation of cdc2 (76). We showed that cdc2 kinase activity is inhibited following serotype 3, not serotype 1, reovirus infection (62). Inhibition of cdc2 kinase activity is, in part, due to serotype 3 σ1s-dependent phosphorylation of cdc2 (62). However, the pathway(s) leading to cdc2 kinase inhibition following serotype 3 reovirus infection remains unclear.
Since G2 phase arrest cannot easily be distinguished from M phase arrest by flow cytometry using DNA intercalating dyes, it is possible that both G2-to-M checkpoint arrest and mitotic spindle checkpoint arrest contribute to reovirus-induced G2/M phase cell cycle arrest. Electron microscopic studies have shown that reovirus particles align along parallel arrays of microtubules and have the capacity to bind microtubules in vitro (3). Serotype 3 reovirus particles associate with microtubules of the mitotic spindle apparatus in infected cells (15) and require microtubule stability for fast axonal transport (81). In addition to association with microtubules, reovirus infection results in progressive disruption and reorganization of the vimentin (intermediate) filament network (74). The binding of reovirus to microtubules and the disruption of vimentin filaments may lead to changes in microtubule tension or binding to the kinetochore. Reovirus-induced disruption of the microtubule-kinetochore association could lead to activation of the mitotic spindle checkpoint and M phase cell cycle arrest (2, 78).
Serotype 3 reovirus infection activates the cellular transcription factors NF-κB (4, 13) and c-Jun (P. Clarke, personal communication). Activation of transcription factors following reovirus infection suggests that gene expression is critical in reovirus-induced pathogenesis. To identify potential pathways leading to serotype 3 reovirus-induced inhibition of proliferation, we conducted experiments to investigate the transcriptional response following serotype 3 and serotype 1 reovirus infections. High-throughput screening of more than 12,000 genes using oligonucleotide microarrays has identified potential reovirus-induced signaling pathways involved in cell cycle control. Of the genes differentially expressed following reovirus infection, several encode regulators of the critical G2-to-M transition kinase cdc2.
MATERIALS AND METHODS
Infection.
Human embryonic kidney (HEK293) cells (ATCC CRL1573) were plated in T75 flasks (Becton Dickinson, Franklin Lakes, N.J.) at a density of 5 × 106 cells per flask in a volume of 12 ml of Dulbecco's modified Eagle's medium supplemented to contain 10% heat-inactivated fetal bovine serum, 2 mM l-glutamine (Gibco-BRL, Gaithersburg, Md.), 1 mM sodium pyruvate (Gibco-BRL), and 100 U of penicillin per ml and 100 μg of streptomycin per ml (Gibco-BRL). After 24 h of incubation, when cells were 60 to 70% confluent, the medium was removed, and cells were infected with viral strain T3A at a multiplicity of infection (MOI) of 100 PFU per cell in a volume of 2 ml at 37°C for 1 h. A high MOI was used to ensure that all susceptible cells were infected, and studies in our laboratory indicate that a high MOI enhances the reproducibility of gene expression studies. Following infection, 10 ml of Dulbecco's modified Eagle's medium was added and flasks were incubated at 37°C. Cells used for control infections were inoculated with a virus-free cell lysate control. In order to identify differentially regulated transcripts important for serotype 3 reovirus-induced cell cycle perturbation, an identical experiment using T1L was performed. UV light-inactivated virus was generated for reverse transcription-PCR (RT-PCR) experiments, by exposing T3A virion stocks to short-wave (254-nm) UV light under conditions sufficient to generate an intensity of 560 μW/cm2 for 15 min (energy level determined with a Blak-Ray J-225 short-wave UV intensity meter). These conditions produce inactivated virus stocks devoid of infectious virus, as determined by plaque assay (<10 PFU/ml). The inoculum of UV virus used was calculated based on the number of infectious particles present in T3A stock prior to UV inactivation.
cRNA target preparation and hybridization.
Cells were harvested at 6, 12, and 24 h postinfection and washed with phosphate-buffered saline, and total RNA was isolated by using the RNeasy Mini Kit (Qiagen, Valencia, Calif.). Ten micrograms of RNA was converted to cDNA by using SuperScript Choice (Gibco-BRL), substituting high-performance liquid chromatography-purified T7-oligo-d(T)24 for random primers. Second-strand synthesis was performed using T4 DNA polymerase, and cDNA was isolated by phenol-chloroform extraction using phase lock gels. Isolated cDNA was in vitro transcribed by using the BioArray High Yield RNA Transcript Labeling Kit (Enzo Biochem, New York, N.Y.) supplied with biotin-labeled UTP and CTP to produce biotin-labeled cRNA. Labeled cRNA was isolated by using an RNeasy Mini Kit column (Qiagen) and quantified for purity and yield. cRNA was fragmented in 100 mM potassium acetate-30 mM magnesium acetate-40 mM Tris-acetate (pH 8.1) for 35 min at 94°C, and hybridization performance was analyzed by using Test 2 arrays (Affymetrix, Santa Clara, Calif.). Target cRNA was hybridized to the U95A microarray (Affymetrix) according to Affymetrix protocols. Briefly, 15 μg of fragmented cRNA was hybridized for 16 h at 45°C with constant rotation (60 rpm). Microarrays were washed and stained with streptavidin-conjugated phycoerythrin (SAPE) by using the Affymetrix GeneChip Fluidic Station 400. Staining intensity was antibody amplified by using a biotinylated anti-streptavidin antibody at a concentration of 3 μg per ml followed by a second SAPE stain and was visualized at 570 nm. All hybridization steps were performed at the University of Colorado Health Sciences Center (UCHSC) Cancer Center Microarray Facility.
Data analysis.
Each gene on the U95A array is represented by 20 different 25-base cDNA oligonucleotides complementary to a cRNA target transcript (perfect match). As a hybridization specificity control, an oligonucleotide containing a single base substitution corresponding to each perfect-match cDNA oligonucleotide (mismatch) is represented on the array. The combination of perfect-match and mismatch cDNA oligonucleotides for each gene is termed a probe set. By using Affymetrix-defined absolute mathematical algorithms describing perfect-match and mismatch intensities, each gene was defined as absent or present and assigned a value. Binding intensity values were scaled to evaluate differential expression following reovirus infection. By using Affymetrix-defined comparison mathematical algorithms, a reovirus-induced transcript was classified as not changed, marginally increased, marginally decreased, increased, or decreased, and a fold change in expression was calculated. Nucleotide accession numbers for transcripts cited in the text that were not changed appear in Table 2. Since each condition was performed in duplicate, we used two additional criteria, as follows, to classify a gene as significantly up- or downregulated following reovirus infection. (i) Reovirus-induced expression of a gene at each time point must be classified as increased or marginally increased (or decreased or marginally decreased) in each virus-infected condition compared to each mock-infected condition. (ii) The mean fold change in reovirus-induced gene expression must be greater than 2. Mean transcriptional expression of a given gene at a given time point was calculated as the sum of the fold change in gene expression for each virus-infected condition compared to each mock-infected condition divided by 4. Standard errors in mean transcriptional expression of a gene were also calculated.
TABLE 2.
Nucleotide accession numbers for genes mentioned in the text that were not changed following T3A or TIL infection
| Gene | Accession numbera |
|---|---|
| ATM | U26455 |
| B-myb | X13293 |
| BUB1 | AF053305 |
| cdc25C | M34065 |
| cdc6 | U77949 |
| cdk4 | U37022 |
| Cyclin A | X51688, U66838b |
| Cyclin B | M25753 |
| Cyclin D1 | M64349 |
| Cyclin D2 | X68452 |
| Cyclin D3 | M92287 |
| Dihydrofolate reductase | J00146, J00140, J00139b |
| E2F1 | U47677, M96577b |
| E2F2 | L22846 |
| E2F4 | S75174 |
| E2F5 | U15642 |
| E2F6 | AF041381 |
| Ha-ras | J00277 |
| MAD2 | AJ000186, U65410b |
| MAD3 | AF053306 |
| pRb | M15400, L49219, L49229, L41913, L41870b |
| p107 | L14812 |
| p130 | X74594, X76061b |
| p15INK4b | AF004819, L36844b |
| p16INK4a | U26727 |
| p19INK4d | U40343 |
| p21WAF1/CIP1 | U03106 |
| p27KIP1 | U10906 |
| p57KIP2 | D64137, U22398b |
| Thymidine kinase | K02581, M15205, U80628b |
Corresponds to the nucleotide accession number assigned to an Affymetrix probe set on the U95A microarray.
Represented multiple times on the U95A microarray.
RT-PCR.
HEK293 cells were infected as described above. Additional cells were infected with UV-inactivated T3A virus (replication incompetent) as a control to assess whether active replication was required for the effects observed with T3A-infected cells (see “Infection” above for description of UV-inactivated T3A virus). Cells were harvested at 12 and 24 h postinfection and washed with phosphate-buffered saline, and RNA was isolated. Five micrograms of RNA was converted to cDNA by using the SuperScript Preamplification System (Gibco-BRL) supplied with oligo-d(T)12-18 primer. Reverse transcription was performed at 42°C for 1 h. PCR was performed using primers generated against chk1 (AF016582) (forward primer, 5′-CTG AAG AAG CAG TCG CAG TG-3′; reverse primer, 5′-TTC CAC AGG ACC AAA CAT CA-3′), wee1 (U10564) (forward primer, 5′-AAC CTC AAT CCC AAA TGC TG-3′; reverse primer, 5′-TTG CCA TCT GTG CTT TCT TG-3′), the growth arrest and DNA damage-inducible protein 45 (GADD45) (M60974) (forward primer, 5′-TGC GAG AAC GAC ATC AAC AT-3′; reverse primer, 5′-TCC CGG CAA AAA CAA ATA AG-3′), and β-actin (BC004251) (forward primer, 5′-GAA ACT ACC TTC AAC TCC ATC-3′; reverse primer, 5′-CGA GGC CAG GAT GGA GCC GCC-3′), yielding product sizes of 495, 506, 200, and 219 bp, respectively. PCR cycle conditions were 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min for 25 cycles. Dilutions of cDNA were performed to determine the linear range for each primer pair. PCR products were resolved on a 2% agarose gel containing ethidium bromide and were visualized with UV light. Densitometric analysis was performed using a FluorS MultiImager System and Quantity One software (Bio-Rad, Hercules, Calif.).
RESULTS
Reovirus-induced alterations in gene expression.
Previous work has established that serotype 3, not serotype 1, reovirus infection induces perturbations in cell cycle regulation in multiple cell lines, including HEK293 cells. To identify potential pathways exploited by serotype 3 reovirus to deregulate the host cell cycle, we evaluated the profile of HEK293 cellular gene expression following T3A infection at 6, 12, and 24 h by using the Affymetrix U95A microarray. We found that serotype 3 reovirus infection induced significant changes in the expression of 25 genes encoding proteins regulating cell cycle progression. Ten genes (40%) were classified as encoding proteins that function at the G1-to-S transition, 11 genes (44%) were classified as encoding proteins that function at the G2-to-M transition, and 4 genes (16%) were classified as encoding proteins that function at the mitotic spindle checkpoint (Table 1). Twenty cell cycle-related transcripts were increased following serotype 3 reovirus infection, and five transcripts were decreased. Four of the five downregulated genes were classified as transcripts encoding proteins regulating G2-to-M transition. No cell cycle-related genes were differentially expressed at 6 h postinfection. At 12 h, five cell cycle-related genes were found to be differentially expressed following serotype 3 reovirus infection, two were upregulated, and three were downregulated. Four of the five genes altered at 12 h postinfection were classified as transcripts encoding proteins regulating the G2-to-M transition. At 24 h postinfection, 23 cell cycle-related genes were found to be differentially expressed, 19 were upregulated, and 3 were downregulated. One transcript found to be upregulated and one found to be downregulated at 12 h post-T3A infection were increased and decreased at 24 h postinfection, respectively. Histone H1, histone H2A, and wee1 were represented twice on the array, and histone H2B was represented six times. Fold changes in gene expression for replicates and for multiple representations of a gene were highly reproducible.
TABLE 1.
Reovirus-induced alterations in expression of genes known to regulate the cell cycle
| Genea | Accession no.b | Fold change in expressionc at the indicated time after infection with:
|
||||
|---|---|---|---|---|---|---|
| T3A
|
TIL (24 h) | |||||
| 12 h | 24 h | |||||
| G1-S checkpoint | ||||||
| N-Ras | X02751 | 2.1 ± 0.2 | ||||
| Cyclin E | M73812 | −2.8 ± 0.1 | ||||
| Cyclin E2 | AF091433 | 2.1 ± 0.2 | ||||
| PP2C-α | AF070670 | 5.3 ± 0.4 | ||||
| C-1d | U41816 | 2.2 ± 0.1 | ||||
| ANAe | D64110 | 2.9 ± 0.1 | 2.5 ± 0.1 | |||
| E2F6 | AF041381 | 2.1 ± 0.1 | ||||
| Histone H1 | X03473f | 2.7 ± 0.1 | 2.3 ± 0.2 | |||
| 3.1 ± 0.2 | 2.1 ± 0.2 | |||||
| Histone H2A | LI9779g | 2.0 ± 0.1 | ||||
| AI885852 | 2.2 ± 0.1 | |||||
| Histone H2B | X00088g | 2.0 ± 0.1 | ||||
| Z80782 | 2.8 ± 0.1 | |||||
| Z80780 | 3.1 ± 0.3 | |||||
| Z80779 | 3.2 ± 0.2 | |||||
| AA873858 | 5.8 ± 0.6 | |||||
| Histone H4 | X00038 | 4.7 ± 0.5 | ||||
| G2-M checkpoint | ||||||
| wee-1 | U10564g | 2.4 ± 0.1 | 1.9 ± 0.1h | |||
| X62048 | 2.5 ± 0.1 | 2.0 ± 0.1 | ||||
| chk-1 | AF016582 | 3.2 ± 0.1 | ||||
| GADD45 | M60974 | 3.3 ± 0.2 | 4.9 ± 0.1 | 4.4 ± 0.1 | ||
| B56-β subunit of PP2A | L42374 | −3.1 ± 0.3 | ||||
| SG2NA | U17989 | 2.4 ± 0.2 | 2.5 ± 0.2 | |||
| FRP1 | U49844 | −2.1 ± 0.1 | ||||
| PCTAIRE-2 | X66360 | 2.6 ± 0.1 | ||||
| Trap | AB025254i | 2.0 ± 0.3 | ||||
| C-2K | X80230 | −2.1 ± 0.1 | ||||
| SAK | Y13115 | 3.8 ± 0.5 | ||||
| Ki-67 antigenj | X65550 | −1.9 ± 0.0 | −2.3 ± 0.2 | |||
| Mitotic spindle checkpoint | ||||||
| BUB-3 | AF047472 | 2.2 ± 0.1 | ||||
| MPP11 | X98260 | 2.0 ± 0.1 | ||||
| HZWINT1 | AF067656 | 2.6 ± 0.0 | ||||
| Nuc-2k | S78234 | 2.1 ± 0.2 | ||||
Genes that are not addressed in the text are described in footnotes.
Corresponds to the nucleotide accession number assigned to the Affymetrix probe set on the U95A microarray.
Values are means ± standard errors of the means from two independent experiments. A negative number indicates that the transcript was downregulated. Where no value is given, there was no change.
A possible transcription factor with activity at the G1-to-S transition (37).
Overexpression leads to retardation of cell cycle progression through the G1 phase (85).
Represented twice on the U95A microarray.
Represented multiple times on the U95A microarray.
This gene was not considered upregulated as defined in Materials and Methods, because it was found to be increased in three of four analyses.
Affymetrix-assigned GenBank accession number refers to partial coding sequence for Trap. The complete coding sequence is at AB030644.
To identify differentially regulated transcripts important for serotype 3 reovirus-induced cell cycle perturbation, we compared the profile of HEK293 cellular gene expression following T1L infection at 24 h postinfection using the Affymetrix U95A microarray. We found that serotype 1 reovirus infection induced significant changes in the expression of six genes encoding proteins regulating cell cycle progression. Three genes (50%) were classified as encoding proteins that function at the G1-to-S transition, and three genes (50%) were classified as encoding proteins that function at the G2-to-M transition (Table 1). T1L infection did not alter any genes classified as encoding proteins that function at the mitotic spindle checkpoint at 24 h postinfection (Table 1). Each cell cycle-related transcript that was differentially regulated following serotype 1 reovirus infection was increased. T1L infection resulted in differential regulation of 23% of transcripts differentially regulated by T3A at 24 h postinfection. The transcript encoding E2F6 was the only cell cycle-related transcript differentially regulated by T1L infection that was not differentially regulated by T3A infection. Of the nine T3A-regulated transcripts encoding proteins that function at the G1-to-S transition at 24 h postinfection, two (22%) were differentially regulated by T1L. Of the nine T3A-regulated transcripts encoding proteins that function at the G2-to-M transition at 24 h postinfection, three (33%) were differentially regulated by T1L. These results indicate that serotype 3 reovirus infection perturbs the differential expression of genes encoding proteins that regulate the G1-to-S, G2-to-M, and mitotic spindle checkpoints and that T1L infection results in differential regulation of a subset of these transcripts. In addition, the earliest detectable changes in gene expression following T3A infection were for genes encoding G2-to-M regulatory proteins, suggesting that deregulation of the G2-to-M transition is an early event following serotype 3 reovirus infection.
Reovirus-induced alterations in genes encoding proteins that regulate G1-to-S progression.
Treatment of specific cell types, including R1.1 thymoma cells, with purified T3D σ1 protein leads to a reversible G1-to-S transition arrest that depends on the inhibition of Ha-ras (70-72). Ten genes encoding proteins that function at the G1-to-S transition were altered following serotype 3 reovirus infection. The level of Ha-ras gene expression was not changed following reovirus infection. Expression of the gene encoding N-ras was also not inhibited and was, in fact, increased 2.1-fold ± 0.2-fold in serotype 3 reovirus-infected cells at 24 h postinfection (Table 1).
The G1-to-S transition requires activation of cyclin-dependent kinases (cdk) (57). cdk2, -4, and -6 kinase activity results in phosphorylation of the retinoblastoma protein (pRb) and the pRb-related proteins p107 and p130. Phosphorylation of pRb leads to activation of the transcription factor E2F (31). The level of cdk4 gene expression was not changed following reovirus infection (Table 2). cdk2 and -6 were not represented on the U95A microarray. Levels of pRb, p107, p130, and E2F1, -2, -4, -5, and -6 gene expression were not changed following reovirus infection (Table 2), except for the transcript encoding E2F6, which was upregulated 2.1-fold ± 0.1-fold following T1L infection (Table 1). Activation of cdk2, -4, or -6 requires binding to cyclin E or cyclin D (31). Cyclin E expression was reduced 2.8-fold ± 0.1-fold in serotype 3 reovirus-infected cells. However, the level of cyclin E2 (a cyclin E isoform) gene expression was increased 2.1-fold ± 0.2-fold in serotype 3 reovirus-infected cells (Table 1). Levels of cyclin D1, -2, and -3 expression were not changed following reovirus infection (Table 2).
Activation of cdk2 depends on its phosphorylation state (57). The level of protein phosphatase 2C (PP2C) gene expression at 24 h after infection with T3A was 5.3-fold ± 0.4-fold greater than that for a mock-infected control. cdk-cyclin complex activity is known to be inhibited by cdk inhibitors (CDKI) (29, 31). Levels of CDKI expression, including p21WAF1/CIP1, p27KIP1, p57KIP2, p16INK4a, p15INK4b, and p19INK4d were not changed following reovirus infection (Table 2). Since active cdk2, -4, or -6 kinase results in E2F-dependent transcription, the transcriptional level of E2F-responsive genes necessary for the G1-to-S transition, including cyclin E, cyclin A, cdc2, cdc25C, p21WAF1/CIP1, c-myc, B-myb, p107, E2F1, E2F2, dihydrofolate reductase, thymidine kinase, DNA polymerase α, histone H2A, and cdc6, serve as a measure of cdk2, -4, or -6 kinase activity (26). Levels of these E2F-responsive transcripts were not changed following reovirus infection except for cyclin E, c-myc, and DNA polymerase α, which were downregulated 2.8-fold ± 0.1-fold, 2.0-fold ± 0.0-fold, and 2.5-fold ± 0.2-fold following serotype 3 reovirus infection, respectively, and histone H2A and cdc2, which were upregulated 2.0-fold ± 0.1-fold or 2.2-fold ± 0.1-fold and 1.8-fold in serotype 3 reovirus compared to mock-infected cells. The failure to detect a coherent pattern of gene expression encoding key G1-to-S checkpoint control proteins is consistent with our observation that reovirus infection does not induce G1 arrest in HEK293 cells. However, we did find changes in the expression of genes encoding histones, cyclin E and E2, and some E2F-responsive transcripts following serotype 3 reovirus infection.
Reovirus-induced alterations in genes encoding proteins that regulate the G2-to-M progression.
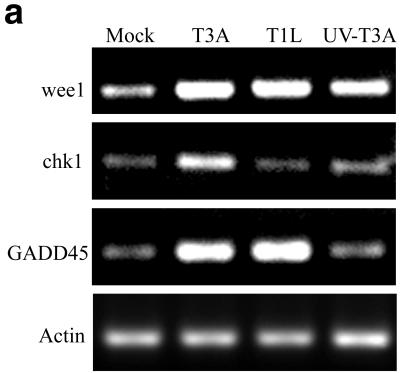
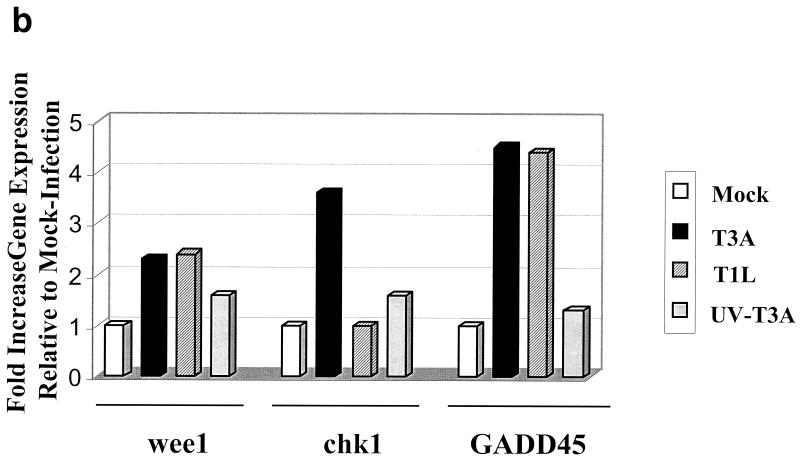
We have previously shown that serotype 3 reovirus-induced G2/M phase cell cycle arrest is due to inhibitory phosphorylation of cdc2 (62). Eleven genes encoding proteins that function at the G2-to-M transition were altered following serotype 3 reovirus infection. These changes were among the earliest changes in gene expression identified in serotype 3 reovirus-infected cells. The level of cdc2 gene expression at 24 h post-T3A infection was 1.8-fold greater than that in a mock-infected control. We have not reproducibly detected changes in levels of cdc2 protein in reovirus-infected cells; however we have consistently found an increase in the proportion of hyperphosphorylated cdc2 following serotype 3 reovirus infection (62). An increase in cdc2 phosphorylation could result from an increase in wee1 kinase activity, which directly phosphorylates and inactivates cdc2 (60). wee1 was determined to be upregulated at 24 h postinfection by using the U95A microarray (Table 1). Levels of wee1 gene expression at 24 h post-T3A infection were 2.4-fold ± 0.1-fold (for accession number U10564) and 2.5-fold ± 0.1-fold (for accession number X62048) greater than those for a mock-infected control. Levels of wee1 gene expression at 24 h post-T1L infection were 1.9-fold ± 0.1-fold (for accession number U10564) and 2.0-fold ± 0.1-fold (for accession number X62048) greater than those for a mock-infected control. The increases in wee1 gene expression following T3A and T1L infection were confirmed by RT-PCR and were 2.3- and 2.4-fold greater, respectively, than those for a mock-infected control at 24 h postinfection (Fig. 1). RT-PCR analysis of UV-inactivated T3A-infected cells, demonstrated a small but measurable increase in wee1 transcripts compared that for to mock-infected cells (a 1.6-fold increase), indicating that active viral replication is not necessary but may augment the expression of wee1 in reovirus-infected cells (Fig. 1). Increased levels of wee1 kinase may result in inhibitory phosphorylation of cdc2 following serotype 3 reovirus infection, but increases in wee1 are not sufficient for serotype 3 reovirus-induced phosphorylation of cdc2, since T1L infection results in similar increases in wee1 gene expression.
FIG. 1.
wee1, chk1, and GADD45 transcripts are increased following reovirus infection. (a) HEK293 cells were either mock infected or infected with T3A, T1L, or UV-inactivated T3A at an MOI of 100 PFU per cell. mRNA was collected at 24 h postinfection and analyzed for chk1, wee1, and GADD45 transcripts by RT-PCR to confirm microarray analyses and also to assess the importance of replication competence for the observed changes in gene expression. β-Actin was used as a control in each RT-PCR to ensure matched total mRNA and equivalent amplication conditions for each experimental sample. (b) Quantitative densitometric analysis of specific mRNA abundance in each sample.
Increased inhibitory phosphorylation of cdc2 following reovirus infection could also result from inactivation of the cdc2-specific phosphatase cdc25C (reviewed in reference 38). Following reovirus infection, the level of cdc25C gene expression was not changed (Table 2). The kinase chk1 can enhance wee1 kinase activity (56) and inhibit cdc25C phosphatase activity (22). The level of chk1 gene expression at 24 h post-T3A infection was 3.2-fold ± 0.1-fold greater than that for a mock-infected control and was unchanged in T1L-infected cells (Table 1). The increase in chk1 gene expression following T3A infection was confirmed by RT-PCR and was 3.6-fold greater than that for a mock-infected control at 24 h postinfection (Fig. 1). In contrast, chk1 transcripts were not altered following T1L infection, as assessed by RT-PCR. chk1 transcripts were slightly increased in UV-inactivated T3A-infected cells compared to mock-infected cells (1.6-fold increase), again suggesting that active viral replication is not required but may augment differential expression of chk1 following T3A infection (Fig. 1). These results suggest that increased levels of chk1 kinase may result in inhibitory phosphorylation of cdc25C on Ser-216 and activation of wee1 following serotype 3 reovirus infection.
In addition to the inhibition of cdc2 by phosphorylation, serotype 3 reovirus-induced G2/M phase cell cycle arrest could result from dissociation of cdc2 from cyclin B. The level of cyclin B gene expression was not changed following reovirus infection (Table 2), consistent with our studies suggesting that changes in the level of cyclin B protein are not responsible for reovirus-induced G2-to-M transition arrest (62).
GADD45 is known to inhibit cdc2 kinase activity by physically dissociating cdc2 kinase from cyclin B (87). To determine whether GADD45 could be responsible for reovirus-induced inactivation of cdc2, we analyzed the transcriptional expression of GADD45 following serotype 3 reovirus infection. Levels of GADD45 gene expression at 12 and 24 h after infection with T3A were 3.3-fold ± 0.2-fold and 4.9-fold ± 0.1-fold greater than those for a mock-infected control, respectively (Table 1). The increase in GADD45 gene expression following T3A infection was confirmed by RT-PCR and was 4.5-fold greater than that for a mock-infected control at 24 h postinfection (Fig. 1). The level of GADD45 gene expression at 24 h after infection with T1L was 4.4-fold ± 0.1-fold greater than that for a mock-infected control, and this increase was confirmed by RT-PCR (5.2-fold increase [Fig. 1]). A small but measurable increase in the level of GADD 45 transcripts was noted by RT-PCR analysis in cells infected with UV-inactivated T3A compared to mock-infected cells (1.3-fold increase [Fig. 1]). This suggests that increased levels of GADD45 may result in inhibition of cdc2 by physically dissociating cdc2 from cyclin B following serotype 3 reovirus infection, but increases in GADD45 are not sufficient for serotype 3 reovirus-induced phosphorylation of cdc2, since T1L infection results in a similar increase in GADD45 gene expression. Thus, we found that reovirus infection induces changes in expression levels of genes encoding key proteins regulating cdc2 kinase activity, including wee1, chk1, and GADD45, which is consistent with the findings of our previous studies suggesting a central role for cdc2 inhibition in serotype 3 reovirus-induced G2/M phase cell cycle arrest (63).
Reovirus-induced alterations in genes encoding proteins that regulate mitotic spindle checkpoint signaling.
Reovirus particles are capable of binding microtubules, and infection results in progressive disruption of vimentin filaments. Changes in microtubule tension or binding to the kinetochore could lead to reovirus-induced mitotic arrest. Four genes encoding proteins that function at the mitotic spindle checkpoint were altered following reovirus infection. The transcriptional levels of mitotic spindle checkpoint components that monitor spindle abnormalities, including MAD2, MAD3, and BUB1, were not changed following reovirus-infection (Table 2). The level of BUB3 gene expression at 24 h after infection with T3A was 2.2-fold ± 0.1-fold greater than that for a mock-infected control (Table 1). Activation of the mitotic spindle checkpoint leads to inhibition of the anaphase-promoting complex (APC) and M phase cell cycle arrest (2, 78). Expression of the gene encoding Nuc2, an APC component, was 2.1-fold ± 0.2-fold greater than that for a mock-infected control (Table 1). These results suggest that serotype 3 reovirus-induced alterations in gene expression may induce M phase cell cycle arrest.
DISCUSSION
The use of DNA microarray technology following reovirus infection has identified several genes encoding proteins that may play a role in serotype 3 reovirus-induced disruption of the cell cycle. The results presented here provide a comprehensive overview of alterations in expression of genes encoding proteins that regulate cell cycle progression following reovirus infection. Although these alterations are likely to reflect changes in gene expression, it should be noted that DNA microarray technology assesses changes in steady-state levels of specific mRNAs rather than transcriptional activity.
G1-to-S checkpoint: reovirus-induced alterations in G1-to-S restriction checkpoint control.
Treatment with purified T3D σ1 protein leads to a reversible G1-to-S phase transition arrest in R1.1 thymoma cells that is dependent on Ha-ras inhibition (70-72). However, G1-to-S arrest is not a universal feature of reovirus infection and does not occur in reovirus-infected HEK293 cells (63). We were unable to detect changes in expression of p21ras-related genes involved in G1-to-S regulation, and we found that gene expression of the p21ras isoform, N-ras, was selectively increased following serotype 3 reovirus infection.
G1-to-S transition arrest following serotype 3 reovirus infection could result from inactivation of the G1-to-S cyclin dependent kinase cdk2, -4, or -6 by the protein phosphatase PP2C-α. PP2C-α is known to dephosphorylate activating phosphates on cdk2 (9), which may result in inhibition of cdk2 kinase activity. Decreased expression of the cdk2 regulatory cyclin, cyclin E, was counterbalanced by an increase in the expression of the cyclin E isoform, cyclin E2, suggesting that G1 cyclin expression does not play a significant role in regulating serotype 3 reovirus-induced G1-to-S progression. Alterations in histone synthesis and alterations in the relative levels of histone proteins could alter G1-to-S progression, but no clear model has been elucidated (19). Collectively, these findings indicate that serotype 3 reovirus-induced alterations in gene expression are not consistent with G1-to-S arrest in HEK293 cells.
G2-to-M checkpoint: reovirus-induced inhibition of cdc2 by phosphorylation.
Serotype 3 reovirus-induced G2/M phase cell cycle arrest results from a reduction in cdc2 kinase activity due to inhibitory phosphorylation, which has been demonstrated in infection of multiple cell lines, including HEK293, MDCK, C127, and HeLa cells (62). An increase in hyperphosphorylated cdc2 could be due either to increases in cdc2-specific kinase activity or to decreases in cdc2-specific phosphatase activity, or to both. The kinases wee1 and myt1 and the phosphatase cdc25C regulate the phosphorylation state of cdc2 (reviewed in reference 38). Following serotype 3 reovirus infection, levels of wee1 transcripts are elevated, suggesting that this kinase may play a role in inhibiting cdc2 (Fig. 2). However, an elevation in wee1 transcript levels is not sufficient for serotype 3 reovirus-induced inhibitory phosphorylation of cdc2, since the transcript encoding wee1 kinase is similarly upregulated following T1L infection. The activity of the cdc2-specific phosphatase cdc25C is regulated by phosphorylation. Hyperphosphorylation of cdc25C results in active phosphatase activity capable of removing inhibitory phosphates from cdc2 kinase (reviewed in reference 38). Following reovirus infection, cdc25C is inhibited by dephosphorylation (62). It is suggested that the HIV Vpr protein can inhibit cdc2 kinase activity (30, 64) through mechanisms requiring wee1 and involving the inactivation of cdc25C (35, 48).
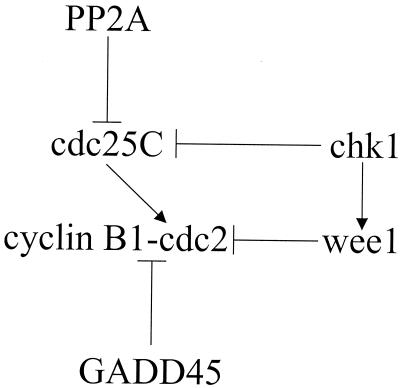
FIG. 2.
Proposed model for serotype 3 reovirus-induced G2-to-M transition arrest. cdc2 kinase activity is required for entry into mitosis. Active cdc2 kinase is complexed with cyclin B and dephosphorylated at Thr14/Tyr15. Following reovirus infection, cdc2 is inhibited, in part, by phosphorylation. The increase in phosphorylated cdc2 may be due to upregulation of the kinases chk1 and wee1 and/or localization to the nucleus of the phosphatase PP2A, which can inhibit the cdc2-activating phosphatase cdc25C. cdc2 kinase activity may also be inhibited by dissociation of cyclin B by GADD45. Arrows indicate activation; blunted lines indicate inhibition.
PP2A or PP2A-like activity is capable of negatively regulating cdc25C by removing activating phosphates (12, 42, 45). HIV Vpr is reported to inactivate cdc25C by physically targeting PP2A to the nucleus and enhancing the recruitment and dephosphorylation of cdc25C though association with the PP2A-B55 regulatory subunit (35). Following serotype 3 reovirus infection, we found selective decreased expression of the PP2A-B56-β regulatory subunit. Since the B56-β regulatory subunit is known to localize PP2A to the cytoplasm (50), downregulation of B56-β production may lead to increased nuclear localization of PP2A following reovirus infection (Fig. 2). Furthermore, upregulation of S/G2 nuclear antigen (SG2NA) may target PP2A to the nucleus following serotype 3 reovirus infection, since SG2NA is known both to localize to the nucleus (52) and to interact with the C subunit of PP2A (51). However, an elevation in SG2NA transcript levels is not sufficient for serotype 3 reovirus-induced dephosphorylation of cdc25C, since the transcript encoding SG2NA is similarly upregulated following T1L infection. Nonetheless, it is possible that reovirus-induced nuclear localization of PP2A could inhibit cdc25C, which is consistent with the cdc2 hyperphosphorylation and G2/M phase cell cycle arrest found following serotype 3 reovirus infection.
cdc25C activity is also inhibited following phosphorylation by the kinases chk1 and chk2 (22, 69). Phosphorylation of cdc25C on Ser-216 by chk1 or chk2 leads to 14-3-3 protein binding, which results in sequestration of cdc25C into the cytoplasm (47, 61). Export to the cytoplasm physically separates cdc25C from cdc2 kinase. We have found that reovirus infection is associated with increased expression of chk1, suggesting that this kinase plays a role in reovirus-induced inhibition of cdc25C (Fig. 2). chk1 kinase activity is also suggested to regulate wee1 subcellular localization, such that cdc2 kinase is phosphorylated and inactivated (56) (Fig. 2). This suggests that serotype 3 reovirus infection may selectively increase the level of chk1 kinase activity to modulate the activities of kinases and phosphatases directly responsible for regulating cdc2 activity.
G2-to-M checkpoint: reovirus-induced inhibition of cdc2 by physical dissociation from cyclin B.
An alternative mechanism for cdc2 inhibition is by physical interaction with GADD45. GADD45 inhibits cdc2 kinase by physically dissociating cdc2 from cyclin B (83, 87). Physical dissociation of cdc2 kinase from its regulatory cyclin results in elimination of cdc2 kinase activity and G2-to-M checkpoint arrest (24, 55). Serotype 3 reovirus induces an increase in GADD45 expression, suggesting that GADD45 plays a role in reovirus-induced G2-to-M checkpoint arrest (Fig. 2). However, an elevation in GADD45 transcript levels is not sufficient for serotype 3 reovirus-induced inhibition of cdc2 kinase activity, since the transcript encoding GADD45 is similarly upregulated following T1L infection. Interestingly, it is possible that GADD45-induced inhibition of cdc2 kinase activity results in accumulation of cells in the G2/M phase of the cell cycle following infection with a herpes simplex virus (HSV) restricted to expressing the infected-cell polypeptide ICP0, since this virus induces an increase in GADD45 protein levels (34). The capacity of GADD45 to regulate cdc2 activity is a function of protein concentration (83). GADD45 transcription is regulated by p53 (41, 87) and Brca1 (28, 39). We have preliminary data suggesting that serotype 3 reovirus infection may result in increased expression of Brca1 protein. Reovirus-induced increases in Brca1 activity may result from changes in the activity of ATM or ATM-related kinases (46). Although we did not detect changes in expression of the gene encoding ATM, the expression of FRP1, a kinase with significant homology to ATM kinases (11), was selectively increased following serotype 3 reovirus infection.
The data presented in this report and our previous studies are consistent with a model in which reovirus-induced phosphorylation of cdc2 results in G2/M phase cell cycle arrest that is dependent on σ1s expression (63). σ1s is a highly basic protein (16, 17) that is present in the nuclei of serotype 3-infected cells (7, 67), where it could directly interact with cdc2 or proteins regulating cdc2 kinase activity. However, the role of σ1s in serotype 3 reovirus-induced G2/M phase cell cycle arrest has not yet been determined.
Mitotic spindle checkpoint: reovirus-induced alterations in spindle checkpoint signaling.
Reovirus-induced disruption of the microtubule-kinetochore association may contribute to G2/M arrest by activating the mitotic spindle checkpoint, which inhibits the APC and results in M phase arrest (2, 78). Selective expression of several genes encoding proteins with putative roles in regulating the mitotic spindle checkpoint were altered following serotype 3 reovirus infection. For example, Bub3 is an integral component of mitotic spindle checkpoint signaling, localizes to unattached kinetochores, and phosphorylates and complexes with Bub1 (6, 40, 66, 79). The functions of MPP11 and HZWINT1 are not completely understood but have been speculated to contribute to mitotic spindle checkpoint regulation, since they localize to mitotic structures, including kinetochores (49, 65, 77). These results suggest that deregulation of mitotic spindle checkpoint proteins may also contribute to serotype 3 reovirus-induced G2/M phase cell cycle arrest.
Other proteins functioning in G2 or M.
Several genes encoding proteins that influence both the G2-to-M and mitotic spindle checkpoints were selectively differentially regulated following serotype 3 reovirus infection. For example, polo-like kinase activity is important in regulating G2-to-M transition events by activating cdc25C and in regulating M phase events including APC regulation, centrosome maturation, and bipolar spindle formation (25, 54). SAK, a polo-like kinase (36), was found to be upregulated following serotype 3 reovirus infection; however, the role of SAK in cell cycle regulation is not clear. Interestingly, expression of murine SAK-a is cell cycle regulated, with levels peaking in M phase, which is consistent with a role in the latter portion of the cell cycle (21). This suggests that serotype 3 reovirus may perturb both the G2-to-M transition and the mitotic spindle checkpoint to ensure host cell cycle arrest in the G2/M phase.
Serotype 3 reovirus infection also selectively altered the expression of genes encoding proteins with cdc2-related serine/threonine kinase activity, including C-2k (5) and PCTAIRE-2 (33). This suggests that serotype 3 reovirus may require cdc2-like kinase activity to posttranslationally modify a viral protein in a manner similar to the phosphorylation of varicella-zoster virus glycoprotein gI (84), Epstein-Barr virus EBNA-LP (43), hepatitis E virus ORF3 (86), and herpes simplex virus ICP0 (1) by cdc2. Alternatively, cdc2-like kinase activity may be responsible for alterations in cytoskeletal structure following serotype 3 reovirus infection. As discussed earlier, reovirus infection leads to disruption and reorganization of vimentin filaments into viral inclusions. Vimentin organization is regulated by cdc2-related kinase activity (10). PCTAIRE-2 kinase activity may regulate cytoskeletal protein distribution (33). Thus, alterations in PCTAIRE-2 and Trap (tudor repeat associator with PCTAIRE-2 [32]) following reovirus infection may lead to cytoskeletal changes necessary for viral inclusion formation. These results suggest that the reovirus life cycle may require modulation of cdc2-like kinase activity.
The present experiments shed light on the kinetics of key cellular processes that may directly influence reovirus-induced G2/M arrest. The lack of differential expression of key transcripts prior to 12 h postinfection is consistent with prior findings, which showed no alteration in the activities of several key regulators of cell cycle progression (including cdc2 kinase) at 6 h postinfection (62). It is important to note that the lack of detectable altered gene expression at 6 h postinfection in these experiments does not preclude either early activation of cell cycle regulators at the protein level or altered expression of cell cycle-regulatory genes not represented on the U95A microarray. It is interesting that UV-inactivated (replication-incompetent) virus induced small but detectable changes in gene expression in the direction of those seen with infectious virus. This suggests that while viral replication may not be required, it substantially augments the expression of cellular regulators of cell cycle progression. Since the reovirus replication cycle is approximately 24 h in HEK293 cells, the requirement for viral replication for maximal alteration in gene expression may account for the lack of detectable alterations at the earliest time point postinfection.
In conclusion, reovirus infection alters a limited subset of transcripts encoding proteins that function to regulate cell cycle progression. Transcripts encoding proteins that function at the G1-to-S, G2-to-M, and mitotic spindle checkpoints were altered following serotype 3 reovirus infection. Of this group, genes encoding proteins regulating the G2-to-M cell cycle transition were among the earliest to show changes in expression. This is consistent with the findings of our previous studies indicating that serotype 3 reovirus infection of HEK293 cells induces a G2/M phase cell cycle arrest (63). We recently showed that G2/M arrest results from inhibition of the key G2-to-M transition kinase cdc2 (62). Consistent with these findings, we found changes in expression of kinases, phosphatases, and GADD proteins that directly and indirectly influence cdc2 kinase activity. These results have enabled us to provide a hypothetical model for modulation of cdc2 kinase activity in serotype 3 reovirus-infected cells.
Acknowledgments
G. J. Poggioli and R. L. DeBiasi contributed equally to this work.
This work was supported by Public Health Service award AG14071 from the National Institute on Aging, Merit and REAP grants from the Department of Veterans Affairs, a U.S. Army Medical Research Acquisition Activity Grant (DAMD17-98-1-8614) (to K.L.T.), and an Infectious Diseases Society of America Young Investigator Award (to R.L.D.).
REFERENCES
- 1.Advani, S. J., R. R. Weichselbaum, and B. Roizman. 2000. The role of cdc2 in the expression of herpes simplex virus genes. Proc. Natl. Acad. Sci. USA 97:10996-11001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amon, A. 1999. The spindle checkpoint. Curr. Opin. Genet. Dev. 9:69-75. [DOI] [PubMed] [Google Scholar]
- 3.Babiss, L. E., R. B. Luftig, J. A. Weatherbee, R. R. Weihing, U. R. Ray, and B. N. Fields. 1979. Reovirus serotypes 1 and 3 differ in their in vitro association with microtubules. J. Virol. 30:863-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barton, E. S., J. C. Forrest, J. L. Connolly, J. D. Chappell, Y. Liu, F. J. Schnell, A. Nusrat, C. A. Parkos, and T. S. Dermody. 2001. Junction adhesion molecule is a receptor for reovirus. Cell 104:441-451. [DOI] [PubMed] [Google Scholar]
- 5.Best, J. L., D. H. Presky, R. A. Swerlick, D. K. Burns, and W. Chu. 1995. Cloning of a full-length cDNA sequence encoding a cdc2-related protein kinase from human endothelial cells. Biochem. Biophys. Res. Commun. 208:562-568. [DOI] [PubMed] [Google Scholar]
- 6.Brady, D. M., and K. G. Hardwick. 2000. Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr. Biol. 10:675-678. [DOI] [PubMed] [Google Scholar]
- 7.Ceruzzi, M., and A. J. Shatkin. 1986. Expression of reovirus p14 in bacteria and identification in the cytoplasm of infected mouse L cells. Virology 153:35-45. [DOI] [PubMed] [Google Scholar]
- 8.Chen, P. L., Y. C. Ueng, T. Durfee, K. C. Chen, T. Yang-Feng, and W. H. Lee. 1995. Identification of a human homologue of yeast nuc2 which interacts with the retinoblastoma protein in a specific manner. Cell Growth Differ. 6:199-210. [PubMed] [Google Scholar]
- 9.Cheng, A., P. Kaldis, and M. J. Solomon. 2000. Dephosphorylation of human cyclin-dependent kinases by protein phosphatase type 2Cα and β2 isoforms. J. Biol. Chem. 275:34744-34749. [DOI] [PubMed] [Google Scholar]
- 10.Chou, Y. H., J. R. Bischoff, D. Beach, and R. D. Goldman. 1990. Intermediate filament reorganization during mitosis is mediated by p34cdc2 phosphorylation of vimentin. Cell 62:1063-1071. [DOI] [PubMed] [Google Scholar]
- 11.Cimprich, K. A., T. B. Shin, C. T. Keith, and S. L. Schreiber. 1996. cDNA cloning and gene mapping of a candidate human cell cycle checkpoint protein. Proc. Natl. Acad. Sci. USA 93:2850-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke, P. R., I. Hoffmann, G. Draetta, and E. Karsenti. 1993. Dephosphorylation of cdc25-C by a type-2A protein phosphatase: specific regulation during the cell cycle in Xenopus egg extracts. Mol. Biol. Cell 4:397-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Connolly, J. L., S. E. Rodgers, P. Clarke, D. W. Ballard, L. D. Kerr, K. L. Tyler, and T. S. Dermody. 2000. Reovirus-induced apoptosis requires activation of transcription factor NF-κB. J. Virol. 74:2981-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox, D. C., and J. E. Shaw. 1974. Inhibition of the initiation of cellular DNA synthesis after reovirus infection. J. Virol. 13:760-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dales, S. 1963. Association between the spindle apparatus and reovirus. Proc. Natl. Acad. Sci. USA 50:268-275. [DOI] [PMC free article] [PubMed]
- 16.Dermody, T. S., M. L. Nibert, R. Bassel-Duby, and B. N. Fields. 1990. Sequence diversity in S1 genes and S1 translation products of 11 serotype 3 reovirus strains. J. Virol. 64:4842-4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duncan, R., D. Horne, L. W. Cashdollar, W. K. Joklik, and P. W. Lee. 1990. Identification of conserved domains in the cell attachment proteins of the three serotypes of reovirus. Virology 174:399-409. [DOI] [PubMed] [Google Scholar]
- 18.Endl, E., and J. Gerdes. 2000. The Ki-67 protein: fascinating forms and an unknown function. Exp. Cell Res. 257:231-237. [DOI] [PubMed] [Google Scholar]
- 19.Ewen, M. E. 2000. Where the cell cycle and histones meet. Genes Dev. 14:2265-2270. [DOI] [PubMed] [Google Scholar]
- 20.Fajardo, E., and A. J. Shatkin. 1990. Expression of the two reovirus S1 gene products in transfected mammalian cells. Virology 178:223-231. [DOI] [PubMed] [Google Scholar]
- 21.Fode, C., C. Binkert, and J. W. Dennis. 1996. Constitutive expression of murine Sak-a suppresses cell growth and induces multinucleation. Mol. Cell. Biol. 16:4665-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furnari, B., N. Rhind, and P. Russell. 1997. Cdc25 mitotic inducer targeted by chk1 DNA damage checkpoint kinase. Science 277:1495-1497. [DOI] [PubMed] [Google Scholar]
- 23.Gaulton, G. N., and M. I. Greene. 1989. Inhibition of cellular DNA synthesis by reovirus occurs through a receptor-linked signaling pathway that is mimicked by antiidiotypic, antireceptor antibody. J. Exp. Med. 169:197-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gautier, J., J. Minshull, M. Lohka, M. Glotzer, T. Hunt, and J. L. Maller. 1990. Cyclin is a component of maturation-promoting factor from Xenopus. Cell 60:487-494. [DOI] [PubMed] [Google Scholar]
- 25.Glover, D. M., I. M. Hagan, and A. A. Tavares. 1998. Polo-like kinases: a team that plays throughout mitosis. Genes Dev. 12:3777-3787. [DOI] [PubMed] [Google Scholar]
- 26.Grana, X., J. Garriga, and X. Mayol. 1998. Role of the retinoblastoma protein family, pRB, p107 and p130 in the negative control of cell growth. Oncogene 17:3365-3383. [DOI] [PubMed] [Google Scholar]
- 27.Hand, R., and I. Tamm. 1974. Initiation of DNA replication in mammalian cells and its inhibition by reovirus infection. J. Mol. Biol. 82:175-183. [DOI] [PubMed] [Google Scholar]
- 28.Harkin, D. P., J. M. Bean, D. Miklos, Y. H. Song, V. B. Truong, C. Englert, F. C. Christians, L. W. Ellisen, S. Maheswaran, J. D. Oliner, and D. A. Haber. 1999. Induction of GADD45 and JNK/SAPK-dependent apoptosis following inducible expression of BRCA1. Cell 97:575-586. [DOI] [PubMed] [Google Scholar]
- 29.Harper, J. W. 1997. Cyclin dependent kinase inhibitors. Cancer Surv. 29:91-107. [PubMed] [Google Scholar]
- 30.He, J., S. Choe, R. Walker, P. Di Marzio, D. O. Morgan, and N. R. Landau. 1995. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 69:6705-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herwig, S., and M. Strauss. 1997. The retinoblastoma protein: a master regulator of cell cycle, differentiation and apoptosis. Eur. J. Biochem. 246:581-601. [DOI] [PubMed] [Google Scholar]
- 32.Hirose, T., M. Kawabuchi, T. Tamaru, N. Okumura, K. Nagai, and M. Okada. 2000. Identification of tudor repeat associator with PCTAIRE 2 (Trap). A novel protein that interacts with the N-terminal domain of PCTAIRE 2 in rat brain. Eur. J. Biochem. 267:2113-2121. [DOI] [PubMed] [Google Scholar]
- 33.Hirose, T., T. Tamaru, N. Okumura, K. Nagai, and M. Okada. 1997. PCTAIRE 2, a Cdc2-related serine/threonine kinase, is predominantly expressed in terminally differentiated neurons. Eur. J. Biochem. 249:481-488. [DOI] [PubMed] [Google Scholar]
- 34.Hobbs, W. E., and N. A. DeLuca. 1999. Perturbation of cell cycle progression and cellular gene expression as a function of herpes simplex virus ICP0. J. Virol. 73:8245-8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hrimech, M., X. J. Yao, P. E. Branton, and E. A. Cohen. 2000. Human immunodeficiency virus type 1 vpr-mediated G2 cell cycle arrest: vpr interferes with cell cycle signaling cascades by interacting with the B subunit of serine/threonine protein phosphatase 2A. EMBO J. 19:3956-3967. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Hudson, J. W., L. Chen, C. Fode, C. Binkert, and J. W. Dennis. 2000. Sak kinase gene structure and transcriptional regulation. Gene 241:65-73. [DOI] [PubMed] [Google Scholar]
- 37.Iijima, M., Y. Kano, T. Nohno, and M. Namba. 1996. Cloning of cDNA with possible transcription factor activity at the G1-S phase transition in human fibroblast cell lines. Acta Med. Okayama 50:73-77. [DOI] [PubMed] [Google Scholar]
- 38.Jackman, M. R., and J. N. Pines. 1997. Cyclins and the G2/M transition. Cancer Surv. 29:47-73. [PubMed] [Google Scholar]
- 39.Jin, S., H. Zhao, F. Fan, P. Blanck, W. Fan, A. B. Colchagie, A. J. Fornace, and Q. Zhan. 2000. BRCA1 activation of the GADD45 promoter. Oncogene 19:4050-4057. [DOI] [PubMed] [Google Scholar]
- 40.Kalitsis, P., E. Earle, K. J. Fowler, and K. H. Choo. 2000. Bub3 gene disruption in mice reveals essential mitotic spindle checkpoint function during early embryogenesis. Genes Dev. 14:2277-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kastan, M. B., Q. Zhan, W. S. el Deiry, F. Carrier, T. Jacks, W. V. Walsh, B. S. Plunkett, B. Vogelstein, and A. J. Fornace. 1992. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia telangiectasia. Cell 71:587-597. [DOI] [PubMed] [Google Scholar]
- 42.Kinoshita, N., H. Ohkura, and M. Yanagida. 1990. Distinct, essential roles of type 1 and 2A protein phosphatases in the control of the fission yeast cell division cycle. Cell 63:405-415. [DOI] [PubMed] [Google Scholar]
- 43.Kitay, M. K., and D. T. Rowe. 1996. Cell cycle stage-specific phosphorylation of the Epstein-Barr virus immortalization protein EBNA-LP. J. Virol. 70:7885-7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumada, K., S. Su, M. Yanagida, and T. Toda. 1995. Fission yeast TPR-family protein nuc2 is required for G1-arrest upon nitrogen starvation and is an inhibitor of septum formation. J. Cell Sci. 108:895-905. [DOI] [PubMed] [Google Scholar]
- 45.Lee, T. H., M. J. Solomon, M. C. Mumby, and M. W. Kirschner. 1991. INH, a negative regulator of MPF, is a form of protein phosphatase 2A. Cell 64:415-423. [DOI] [PubMed] [Google Scholar]
- 46.Li, S., N. S. Ting, L. Zheng, P. L. Chen, Y. Ziv, Y. Shiloh, E. Y. Lee, and W. H. Lee. 2000. Functional link of BRCA1 and ataxia telangiectasia gene product in DNA damage response. Nature 406:210-215. [DOI] [PubMed] [Google Scholar]
- 47.Lopez-Girona, A., B. Furnari, O. Mondesert, and P. Russell. 1999. Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature 397:172-175. [DOI] [PubMed] [Google Scholar]
- 48.Masuda, M., Y. Nagai, N. Oshima, K. Tanaka, H. Murakami, H. Igarashi, and H. Okayama. 2000. Genetic studies with the fission yeast Schizosaccharomyces pombe suggest involvement of wee1, ppa2, and rad24 in induction of cell cycle arrest by human immunodeficiency virus type 1 Vpr. J. Virol. 74:2636-2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsumoto-Taniura, N., F. Pirollet, R. Monroe, L. Gerace, and J. M. Westendorf. 1996. Identification of novel M phase phosphoproteins by expression cloning. Mol. Biol. Cell 7:1455-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCright, B., A. M. Rivers, S. Audlin, and D. M. Virshup. 2089. 1996. The B56 family of protein phosphatase 2A (PP2A) regulatory subunits encodes differentiation-induced phosphoproteins that target PP2A to both nucleus and cytoplasm. J. Biol. Chem. 271:22081-22082. [DOI] [PubMed] [Google Scholar]
- 51.Moreno, C. S., S. Park, K. Nelson, D. Ashby, F. Hubalek, W. S. Lane, and D. C. Pallas. 2000. WD40 repeat proteins striatin and S/G2 nuclear autoantigen are members of a novel family of calmodulin-binding proteins that associate with protein phosphatase 2A. J. Biol. Chem. 275:5257-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muro, Y., E. K. Chan, G. Landberg, and E. M. Tan. 1995. A cell-cycle nuclear autoantigen containing WD-40 motifs expressed mainly in S and G2 phase cells. Biochem. Biophys. Res. Commun. 207:1029-1037. [DOI] [PubMed] [Google Scholar]
- 53.Nibert, M. L., L. A. Schiff, and B. N. Fields. 1996. Reoviruses and their replication, p. 1557-1596. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 54.Nigg, E. A. 1998. Polo-like kinases: positive regulators of cell division from start to finish. Curr. Opin. Cell Biol. 10:776-783. [DOI] [PubMed] [Google Scholar]
- 55.Nurse, P. 1990. Universal control mechanism regulating onset of M-phase. Nature 344:503-508. [DOI] [PubMed] [Google Scholar]
- 56.O'Connell, M. J., J. M. Raleigh, H. M. Verkade, and P. Nurse. 1997. Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J. 16:545-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O'Connor, P. M. 1997. Mammalian G1 and G2 phase checkpoints. Cancer Surv. 29:151-182. [PubMed] [Google Scholar]
- 58.Op, D. B., and P. Caillet-Fauquet. 1997. Viruses and the cell cycle. Prog. Cell Cycle Res. 3:1-19. [DOI] [PubMed] [Google Scholar]
- 59.Page, A. M., and P. Hieter. 1997. The anaphase promoting complex. Cancer Surv. 29:133-150. [PubMed] [Google Scholar]
- 60.Parker, L. L., S. Atherton-Fessler, and H. Piwnica-Worms. 1992. p107wee1 is a dual-specificity kinase that phosphorylates p34cdc2 on tyrosine 15. Proc. Natl. Acad. Sci. USA 89:2917-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peng, C. Y., P. R. Graves, R. S. Thoma, Z. Wu, A. S. Shaw, and H. Piwnica-Worms. 1997. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science 277:1501-1505. [DOI] [PubMed] [Google Scholar]
- 62.Poggioli, G. J., T. S. Dermody, and K. L. Tyler. 2001. Reovirus-induced σ1s-dependent G2/M phase cell cycle arrest is associated with inhibition of p34cdc2. J. Virol. 75:7429-7434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poggioli, G. J., C. Keefer, J. L. Connolly, T. S. Dermody, and K. L. Tyler. 2000. Reovirus-induced G2/M cell cycle arrest requires σ1s and occurs in the absence of apoptosis. J. Virol. 74:9562-9570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Re, F., D. Braaten, E. K. Franke, and J. Luban. 1995. Human immunodeficiency virus type 1 Vpr arrests the cell cycle in G2 by inhibiting the activation of p34cdc2-cyclin B. J. Virol. 69:6859-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Resto, V. A., O. L. Caballero, M. R. Buta, W. H. Westra, L. Wu, J. M. Westendorf, J. Jen, P. Hieter, and D. Sidransky. 2000. A putative oncogenic role for MPP11 in head and neck squamous cell cancer. Cancer Res. 60:5529-5535. [PubMed] [Google Scholar]
- 66.Roberts, B. T., K. A. Farr, and M. A. Hoyt. 1994. The Saccharomyces cerevisiae checkpoint gene BUB1 encodes a novel protein kinase. Mol. Cell. Biol. 14:8282-8291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rodgers, S. E., J. L. Connolly, J. D. Chappell, and T. S. Dermody. 1998. Reovirus growth in cell culture does not require the full complement of viral proteins: identification of a σ1s-null mutant. J. Virol. 72:8597-8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roner, M. R., and D. C. Cox. 1985. Cellular integrity is required for inhibition of initiation of cellular DNA synthesis by reovirus type 3. J. Virol. 53:350-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sanchez, Y., C. Wong, R. S. Thoma, R. Richman, Z. Wu, H. Piwnica-Worms, and S. J. Elledge. 1997. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science 277:1497-1501. [DOI] [PubMed] [Google Scholar]
- 70.Saragovi, H. U., A. Bhandoola, M. M. Lemercier, G. K. Akbar, and M. I. Greene. 1995. A receptor that subserves reovirus binding can inhibit lymphocyte proliferation triggered by mitogenic signals. DNA Cell Biol. 14:653-664. [DOI] [PubMed] [Google Scholar]
- 71.Saragovi, H. U., N. Rebai, G. M. Di Guglielmo, R. Macleod, J. Sheng, D. H. Rubin, and M. I. Greene. 1999. A G1 cell cycle arrest induced by ligands of the reovirus type 3 receptor is secondary to inactivation of p21ras and mitogen-activated protein kinase. DNA Cell Biol. 18:763-770. [DOI] [PubMed] [Google Scholar]
- 72.Saragovi, H. U., N. Rebai, E. Roux, M. Gagnon, X. Zhang, B. Robaire, J. Bromberg, and M. I. Greene. 1998. Signal transduction and antiproliferative function of the mammalian receptor for type 3 reovirus. Curr. Top. Microbiol. Immunol. 233:155-166. [DOI] [PubMed] [Google Scholar]
- 73.Scholzen, T., and J. Gerdes. 2000. The Ki-67 protein: from the known and the unknown. J. Cell. Physiol. 182:311-322. [DOI] [PubMed] [Google Scholar]
- 74.Sharpe, A. H., L. B. Chen, and B. N. Fields. 1982. The interaction of mammalian reoviruses with the cytoskeleton of monkey kidney CV-1 cells. Virology 120:399-411. [DOI] [PubMed] [Google Scholar]
- 75.Sharpe, A. H., and B. N. Fields. 1981. Reovirus inhibition of cellular DNA synthesis: role of the S1 gene. J. Virol. 38:389-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Solomon, M. J., M. Glotzer, T. H. Lee, M. Philippe, and M. W. Kirschner. 1990. Cyclin activation of p34cdc2. Cell 63:1013-1024. [DOI] [PubMed] [Google Scholar]
- 77.Starr, D. A., R. Saffery, Z. Li, A. E. Simpson, K. H. Choo, T. J. Yen, and M. L. Goldberg. 2000. HZwint-1, a novel human kinetochore component that interacts with HZW10. J. Cell Sci. 113:1939-1950. [DOI] [PubMed] [Google Scholar]
- 78.Straight, A. F. 1997. Cell cycle: checkpoint proteins and kinetochores. Curr. Biol. 7:R613-R616. [DOI] [PubMed] [Google Scholar]
- 79.Taylor, S. S., E. Ha, and F. McKeon. 1998. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol. 142:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tyler, K. L., and B. N. Fields. 1996. Reoviruses, p. 1597-1623. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 81.Tyler, K. L., D. A. McPhee, and B. N. Fields. 1986. Distinct pathways of viral spread in the host determined by reovirus S1 gene segment. Science 233:770-774. [DOI] [PubMed] [Google Scholar]
- 82.Tyler, K. L., M. K. Squier, A. L. Brown, B. Pike, D. Willis, S. M. Oberhaus, T. S. Dermody, and J. J. Cohen. 1996. Linkage between reovirus-induced apoptosis and inhibition of cellular DNA synthesis: role of the S1 and M2 genes. J. Virol. 70:7984-7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang, X. W., Q. Zhan, J. D. Coursen, M. A. Khan, H. U. Kontny, L. Yu, M. C. Hollander, P. M. O'Connor, A. J. Fornace, Jr., and C. C. Harris. 1999. GADD45 induction of a G2/M cell cycle checkpoint. Proc. Natl. Acad. Sci. USA 96:3706-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ye, M., K. M. Duus, J. Peng, D. H. Price, and C. Grose. 1999. Varicella-zoster virus Fc receptor component gI is phosphorylated on its endodomain by a cyclin-dependent kinase. J. Virol. 73:1320-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yoshida, Y., S. Matsuda, N. Ikematsu, J. Kawamura-Tsuzuku, J. Inazawa, H. Umemori, and T. Yamamoto. 1998. ANA, a novel member of the Tob/BTG1 family, is expressed in the ventricular zone of the developing central nervous system. Oncogene 16:2687-2693. [DOI] [PubMed] [Google Scholar]
- 86.Zafrullah, M., M. H. Ozdener, S. K. Panda, and S. Jameel. 1997. The ORF3 protein of hepatitis E virus is a phosphoprotein that associates with the cytoskeleton. J. Virol. 71:9045-9053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhan, Q., M. J. Antinore, X. W. Wang, F. Carrier, M. L. Smith, C. C. Harris, and A. J. Fornace, Jr. 1999. Association with cdc2 and inhibition of cdc2/cyclin B1 kinase activity by the p53-regulated protein GADD45. Oncogene 18:2892-2900. [DOI] [PubMed] [Google Scholar]