Abstract
The expression of human papillomavirus type 16 late genes encoding virus capsid proteins L1 and L2 is restricted to terminally differentiated epithelial cells in the superficial layers of the squamous epithelium. We wish to understand the molecular mechanisms that determine the levels of expression of the human papillomavirus type 16 late genes. We have previously shown that the L1 coding region contains inhibitory sequences. Here we extend previous findings to show that the 5′ end of the L1 gene contains strong inhibitory sequences but that the 3′ end does not. We show that the first 514 nucleotides of the L1 coding region contain multiple inhibitory elements that act independently of one another and that the major inhibitory element is located within the first 129 nucleotides of the L1 gene. Introduction of point mutations in the inhibitory elements in the 5′ end of the L1 gene which altered the RNA sequence without affecting the protein sequence specifically inactivated the inhibitory elements and resulted in production of high levels of human papillomavirus type 16 L1 mRNA and protein in human epithelial cells. Furthermore, we show that inhibitory sequences are present in the L1 coding regions of multiple human papillomavirus types, demonstrating that these elements are conserved among the human papillomaviruses, and suggest that they have an important function in the viral life cycle.
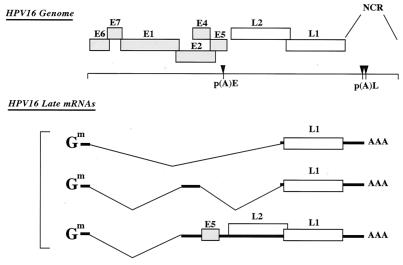
Human papillomaviruses (HPVs) are a group of small, double-stranded DNA tumor viruses (22, 36). There are more than 70 different types of HPVs identified, which can be classified into groups of viruses which infect either the cutaneous or mucosal epithelium. The HPVs can be further classified according to their low- and high-risk status (58). Low-risk types are the major cause of benign tumors and warts, while high-risk types, including HPV type 16 (HPV-16) and HPV-18, are the primary HPV types responsible for anogenital cancers. The HPV genome is approximately 8 kb in length and can be divided into early (E) and late (L) regions (Fig. 1) (22). Among the eight open reading frames (ORFs), the early genes code for proteins which primarily control viral DNA replication, transcription, and transformation of the host cell. The late region codes for the two structural proteins, L1 and L2, which are the major and minor capsid proteins, respectively. Together they form the icosahedral capsid that contains the viral genomic DNA.
FIG. 1.
Schematic representation of the genomic organization of HPV-16. The major late mRNAs are diagrammed. The cap and the poly(A) tail of the mRNAs are indicated. p(A)E, early poly(A) signal; p(A)L, late poly(A) signal; NCR, noncoding region; Gm, methylated cap structure.
The HPV life cycle is tightly linked to the differentiation stage of the infected cell (22). Upon entry into the basal epithelial cells, papillomaviruses establish and maintain their genomes extrachromosomally at approximately 20 to 50 episomes per cell. At this stage, only early genes are expressed. As the cell progresses toward terminal differentiation, induction of viral DNA replication occurs at high levels. This is followed by activation of viral late-gene expression and assembly of infectious viral particles at the uppermost layers of the epithelium. As differentiation of the infected cell occurs, it is believed that late-gene expression from a differentiation-dependent promoter begins (21, 29). L1 and L2 expression is also regulated at the posttranscriptional level by RNA elements in the 3′ untranslated region (UTR) and in the coding regions of the mRNAs (1, 31, 32, 34).
The strong association between late-gene expression and terminal differentiation of the epithelium has prevented the production of virus in a tissue culture system and therefore hindered the study of the roles of individual protein functions in the viral life cycle. However, propagation of papillomavirus has been successful in a xenograft model and in the organotypic (raft) culture systems, where the use of HPV-containing cell lines leads to the complete differentiation-dependent life cycle of HPVs in vitro (14, 28, 45). More recently, the transfection of HPV-11, -16, -18, and -31 genomic DNA sequences into primary human keratinocytes, which are subsequently grown in organotypic cultures, has allowed the study of various mutations in the HPV genome and a detailed analysis of their effects (16-18, 28, 49). Furthermore, production of infectious bovine papillomavirus type 1 (BPV-1) virions was achieved by an organotypic raft/xenograft technique (27). These types of experiments also confirmed the differentiation-dependent expression of the papillomavirus L1 and L2 late genes, which was previously observed in vivo. L1 and L2 proteins were detected only after culturing transfected cells in raft cultures and then only in the superficial layers of the terminally differentiated cells. In contrast, L1 and L2 were never seen in the proliferating transfected keratinocytes.
We are interested in the regulation of late-gene expression, and, in our attempt to decipher the absence of L1 and L2 production in proliferating cells, we identified inhibitory sequences in late HPV mRNAs (31, 32, 34). Sequences with inhibitory function have been reported for many cellular and viral mRNAs and have been shown to be important determinants in gene expression. In general, inhibitory sequences have been located in 3′ UTR sequences and less frequently in the protein-coding regions. For example, both c-fos and c-myc mRNAs contain negative regulatory sequences in the 3′ UTR and in the protein-coding regions (8, 30). Interestingly, papillomavirus late mRNAs contain inhibitory sequences, both in the late 3′ UTR and in the late-gene protein-coding regions (31, 32, 34). Sequences with inhibitory function have been identified in the late-gene 3′ UTRs of HPV-1, HPV-16, and BPV-1. A 57-nucleotide AU-rich inhibitory element containing two AUUUA and three UUUUU motifs has been identified in the late-gene 3′ UTR of HPV-1 (43, 48), a 53-nucleotide inhibitory sequence has been identified in the 3′ UTR of BPV-1 (19), and a 79-nucleotide negative element has been identified in the 3′ UTR of HPV16 (23, 24). The HPV-1 AU-rich element reduces mRNA stability and inhibits translation (43, 46), the BPV-1 element acts by inhibiting polyadenylation (20), and the HPV-16 element appears to reduce mRNA stability in vitro (24). We have previously shown that the HPV-1 AU-rich element interacts specifically with cellular factors (52, 53), primarily HuR (40) and heterogeneous nuclear ribonucleoprotin (hnRNP) C (41, 43). Analysis of the activity of the HPV-1 AU-rich element in a panel of epithelial cell lines revealed that there was an inverse correlation between the inhibitory activity and the levels of the shuttling HuR in the cytoplasm, suggesting that high levels of HuR in the cytoplasm have a stimulatory effect on HPV-1 late-gene expression (6). The BPV-1 element interacts with U1 snRNP, which inhibits polyadenylation at the late poly(A) signal (20). The HPV-16 3′ UTR element appears to interact with multiple cellular factors including U2AF65, HuR, CstF64, and U1snRNP (12, 20, 25). Interestingly, the inhibitory effect of all the 3′ UTR elements could be overcome by the human immunodeficiency virus type 1 (HIV-1) nuclear RNA export factors Rev and the Rev-responsive element (RRE) (3, 46, 48) and the simian retrovirus type 1 constitutive transport element (CTE) (46, 48), demonstrating that nuclear events contribute to the inhibition of late-gene expression by these elements.
Previous work led to the identification of negative elements within the HPV-16 L1 and L2 coding regions (31, 32, 34). We have previously shown that an inhibitory sequence present in the 5′ end of the HPV-16 L2 coding region acts by reducing the stability of the L2 mRNAs (10, 42), whereas another element, located in the 3′ end of the L2 gene, acts by reducing L2 protein production (42). It was shown that the RNA binding protein hnRNP K and poly(rC) binding proteins (PCBP-1 and PCBP-2) interact with the 3′ end inhibitory element and cause reduced translation of L2 mRNA in vitro (10). Our previous work on HPV-16 L1 expression demonstrated the presence of cis-acting negative elements in the L1 coding region (46). The HPV-16 L1 coding region was shown to act in cis to inhibit chloramphenicol acetyltransferase (CAT) protein production in an orientation-dependent manner when placed downstream of the CAT reporter gene (46). The presence on the mRNA of the L1 coding sequence resulted in reduced mRNA levels.
In this study, we sought to further this work on HPV-16 the L1 gene by mapping and characterizing its inhibitory elements. We found that multiple, independently acting negative elements are located in the first 514 nucleotides of the L1 gene, but no inhibitory sequences are found between nucleotide positions 514 and 1518 in the L1 gene open reading frame. The shortest element which retains full inhibitory activity was mapped to the first 129 nucleotides of the L1 gene coding sequence. We successfully inactivated the inhibitory elements by mutagenesis and generated a synthetic HPV16 L1 gene, which produced high levels of L1 mRNA and L1 protein in proliferating, human epithelial cells. Furthermore, we show that inhibitory sequences are present in the L1 gene coding regions of multiple HPV types, suggesting that these elements are conserved among the HPVs and serve an important function in the viral life cycle.
MATERIALS AND METHODS
Plasmid constructions. (i) L1-EIAV hybrids.
To generate pCL130EIAV, the equine infectious anemia virus (EIAV) coding sequence was amplified by PCR with oligonucleotides L1startEIAV (5′-GTCGACCAGCGCGCCAAGATGTCTCTTTGGCTGCCTGGAGCAAGGCGCTCAAGAAGTTAGAGAAGG-3′),which introduced 30 nucleotides the of HPV-16 L1 gene in frame with EIAV, and EIAVstop (5′-TCTAGACTCGAGTTACTCCCACAAACTGTCC-3′). Restriction sites are underlined. To generate pCEIAV30L1, the L1 coding sequence was amplified by PCR with oligonucleotide EIAVstartL1 (5′-CAGCGCGCCAAGATGGGAGACCCTTTGACATGGAGCAAGGCGAGTGAGGCCACTGTC-3′), which introduced 30 nucleotides of EIAV in frame with the L1 gene, and L1stop (5′-CTCGAGCTTACAGCTTA-3′). pCL125EIAV75 was generated by PCR amplification of nucleotides 1 to 367 of the HPV-16 L1 gene with oligonucleotides L1STARTSalI (5′-GTCGACCAGCGCGCCAAGATGTC-3′) and L15′25AS (5′-CCAATTTACGCGTTAAAGG-3′) and amplification of nucleotides 358 to 1461 of EIAV gag using oligonucleotides EIAV3′75S (5′-CAGTCTGAGACGCGTGAAGAATATCC-3′) and EIAVstop. pCL150EIAV50 was generated by PCR amplification of nucleotides 1 to 747 of the HPV-16 L1 gene using oligonucleotides L1STARTSalI and L15′50AS (5′-CTTTCGTAAACGCGTAAATAAGCT-3′), followed by PCR amplification of nucleotides 751 to 1461 of EIAV gag using oligonucleotides EIAV3′50S (5′-CAGTTTAGGACGCGTTATAGACAATGG-3′) and EIAVstop. pCL175EIAV25 was generated by PCR amplification of nucleotides 1 to 1156 of L1 gag using oligonucleotides L1STARTSalI and L15′75AS (5′-CGTCTGCAGTACGCGTTATTTTGCA-3′), followed by PCR amplification of nucleotides 1123 to 1461 of EIAV gag using oligonucleotides EIAV3′25S (5′-TTGAAAGGAACGCGTCTAAAGGCAGC-3′) and EIAVstop. pCEIAV25L175 was generated by PCR amplification of nucleotides 1 to 358 of EIAV gag using oligonucleotides EIAVstart (5′-CAGCGCGCCAAGATGGGAGACCCTTTGACATGG-3′) and EIAV 5′25AS (5′-GGATATTCTTCACGCGTCTCAGACTG-3′), followed by PCR amplification of nucleotides 367 to 1518 of the HPV-16 L1 gene using oligonucleotides L13′75S (5′-CCTTTAACGCGTAAATTGG-3′) and L1stop. pCEIAV75L125 was generated by PCR amplification of nucleotides 1 to 1123 of EIAV gag using oligonucleotides EIAVstart and EIAV5′75AS (5′-GCTGCCTTTAGACGCGTTCCTTTCAA-3′, followed by PCR amplification of nucleotides 1156 to 1518 using oligonucleotides L13′25S (5′-TGCAAAATAACGCGTACTGCAGACG-3′) and L1stop. All PCR fragments were subcloned into an EcoRV-digested pBluescript KS(−). Ligation of the L1 gene and EIAV PCR fragments was facilitated by use of the MluI restriction sites present in the oligonucleotides and allowed the generation of pBluescript KS(−) intermediates pKSL125EIAV75, pKSL150EIAV50, pKSL175EIAV25, pKSEIAV25L175, and pKSEIAV75L125. Finally, the various hybrid ORFs were excised using BssHII and XhoI and ligated into the pCL086 vector (Fig. 2A).
FIG. 2.
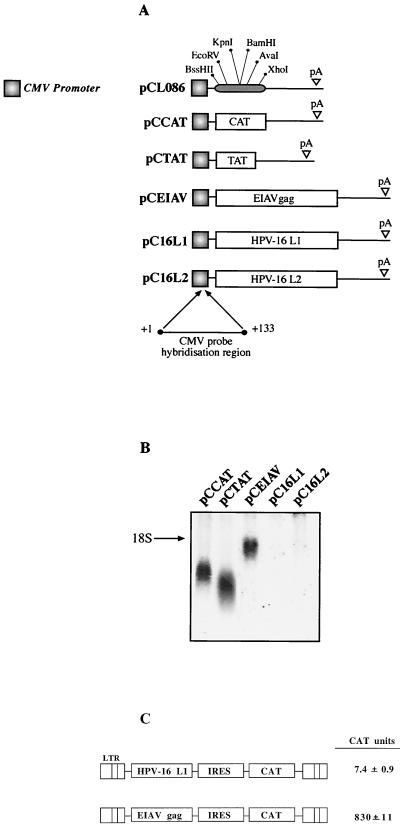
Assay for identification of inhibitory sequences in protein-coding regions. (A) Structures of the indicated expression plasmids. PCL086 contains the human CMV immediate-early promoter followed by a polylinker with the indicated restriction enzyme sites and the HPV-16 late poly(A) (pA) signal from nucleotide 7282 to 7453 (35). The bacterial CAT gene, HIV-1 tat cDNA 1.4.7 (TAT) (33), the EIAV p55gag (EIAV), and the HPV-16 L1 and L2 ORFs were inserted into pCL086 to produce the indicated plasmids. The region to which the probe used in the Northern blots hybridizes is indicated. (B) Northern blot of total cytoplasmic RNA harvested from HeLa cells transfected with the plasmids shown in panel A. (C) Structures of the two plasmids. The plasmids were transfected into HLtat cells (33), and the levels of CAT were determined at 20 h posttransfection using a CAT ELISA as described in Materials and Methods. LTR, HIV-1 long terminal repeat promoter; IRES, poliovirus internal ribosome entry site.
pCEIAV75L125s was generated by PCR amplification of nucleotides +4 to +367 of the HPV-16 L1 gene using oligonucleotides L15′25start (5′-ACGCGTTCTCTTTGGCTGCCTAGTGAGGCC-3′) and L15′25stop (5′-CTCGAGTTACCAATTTATTTAATAAAGG-3′). PCR fragments were subcloned into EcoRV-digested pBluescript KS(−), generating pKSL125S. This plasmid was digested with MluI and XhoI and was exchanged for the L1 gene fragment present in pCEIAV75L125, resulting in pCEIAV75L125s.
(ii) L1 gene deletions.
pCL1(−10)EIAV was generated by PCR amplification of nucleotides 130 to 367 of the HPV-16 L1 gene using oligonucleotides L1(−10)start (5′-CAGCGCGCCAAGATGGCAGTTGGACATCCCTATTTTCCTATTAAAAAACC-3′) and L15′25AS. pCL1(−15)EIAV was generated by PCR amplification of nucleotides 151 to 367 of the HPV-16 L1 gene using oligonucleotides L1(−15)start (5′-GCGCGCATGCCTAACAATAACAAAATATTAGTTCC-3′) and L15′25AS. pC16L1-129WT was generated by PCR amplification of nucleotides 1 to 129 of the HPV-16 L1 gene using oligonucleotides L1STARTSalI and L15AS129NT (5′-ACGCGTAAGTAGTCTGGATGTTCCTGCATG-3′). pC16L1-129MUT was generated by PCR amplification of nucleotides 1 to 129 of the HPV-16 L1 gene using oligonucleotides L15′-MUTANT#1(5′-CCGTCGACGCGCGCCAAGATGAGCCTGTGGCTGCCCAGCG-3′)and L1M129AS (5′-CCACGCGTAGCAGGCGGCTGGTGCCGGCGTGG-3′). To generate pCL1-129MUTCAT and pCL1-129WTCAT, the CAT gene was first amplified by PCR with oligonucleotides CAT-MluI (5′-ACGCGTATGGAGAAAAAAATCACTGGATATACC-3′) and CATXHO (5′-CTCGAGTTACGCCCCGCCCTGCCAC-3′), inserted into the pCR-TOPO cloning vector (Invitrogen), and then transferred into pC16L1-129MUT and pC16L1-129WT by using MluI and XhoI, generating pCL1-129MUTCAT and pCL1-129WTCAT. pCL1(−20)EIAV was generated by PCR amplification of nucleotides 250 to 367 of the HPV-16 L1 gene using oligonucleotides L1(−20)start (5′-GCGCGCCAAGATGGGTTTTCCTGACACCTCATTTTATAATCCAGATACACAGCGG-3′) and L15′25AS. pCL1(+10)EIAV was generated by PCR amplification of nucleotides 1 to 145 of the HPV-16 L1 gene using oligonucleotides L1STARTSalI and L1(+10)AS (5′-GGTTTTTTAATAGGACGCGTGGGATGTCCAACTGC-3′). pCL1(+20)EIAV was generated by PCR amplification of nucleotides 1 to 268 of the HPV-16 L1 gene using oligonucleotides L1STARTSalI and L1(+20)AS (5′-CCGCTGTGTATCTGGATTACGCGTTGAGGTGTCAGGAAAACC-3′). The PCR fragments were subcloned into EcoRV-digested pBluescript KS(−) or into the pCR-TOPO cloning vector (Invitrogen), before digestion with SalI and MluI and subsequent ligation to SalI- and MluI-digested pCL125EIAV75.
(iii) L1 gene mutants.
pC16L1MUT#123 was generated by dividing the HPV-16 L1 gene sequence from nucleotide +1 to +514 into three sections; then where possible nucleotides were changed without changing the protein amino acid sequence, taking care not to introduce rare codons. When this was achieved long oligonucleotides corresponding to the new L1 gene sequence were used as PCR templates for the construction of the L1 mutant gene. The first section from nucleotides +1 to +188 was generated by PCR on long oligonucleotide L1Mut#1 (5′-ATGAGCCTGTGGCTGCCCAGCGAGGCCACCGTGTACCTGCCCCCCGTGCCCGTGAGCAAGGTGGTGAGCACCGACGAGTACGTGGCCCGCACCAACATCTACTACCACGCCGGCACCAGCCGCCTGCTGGCCGTGGGCCACCCCTACTTCCCCATCAAGAAGCCTAACAACAACAAGATCCTGGTGCCCAAGG-3′) using L15′-MUTANT#1 (5′-CCGTCGACGCGCGCCAAGATGAGCCTGTGGCTGCCCAGCG-3′) and L13′-MUTANT#1 (5′-AGGCCTCCTTGGGCACCAGGATCTTG-3′). The second section from nucleotides +188 to +356 was generated by PCR on long oligonucleotide L1Mut#2 (5′-CCCAAGGTGAGCGGCCTGCAGTACCGCGTGTTCCGCATCCACCTGCCCGACCCCAACAAGTTCGGCTTCCCCGACACCAGCTTCTACAACCCCGACACCCAGCGCCTGGTGTGGGCCTGCGTGGGCGTGGAGGTGGGCCGCGGCCAGCCCCTGGGCGTGGGCATCTCTGGCCACCGGATCCGTCGAC-3′) using L15′-MUTANT#2 (5′-CCCAAGGTGAGCGGCCTGCAGTACCGC-3′) and L13′-MUTANT#2 (5′-GTCGACGGATCCGGTGGCCAGAGATGCCCACGCCCAGGGG-3′). The third section from nucleotides +356 to +514 was generated by PCR on long oligonucleotideL1Mut#3 (5′-AGCGGCCACCCCCTGCTGAACAAGCTGGACGACACCGAGAACGCCAGCGCCTACGCCGCCAACGCCGGCGTGGACAACCGCGAGTGCATCAGCATGGACTACAAGCAGACCCAGCTGTGCCTGATCGGCTGCAAGCCCCCCATCGGCGAGCACTGGGGCAAGGGATCCCTCGAG-3′) using L15′-MUTANT#3 (5′-AGTGGCCACCCCCTGCTGAACAAGCTGG-3′) and L13′-MUTANT#3 (5′-CTCGAGGGATCCCTTGCCCCAGTGCTCGCC-3′). The PCR fragments were subcloned and transferred into the pC16L1:8 expression plasmid by using BssHII and BamHI, generating pC16L1MUT#123. pT7L1 was generated by PCR on pC16L1:8 using oligonucleotides T7CMV (5′-GGGAGCTCTAATACGACTCACTATAGGGTCAGATCGCCTGGAGACGCC-3′) and pAsignal(AS) (5′-TCTAGAACGCGTGGGATGCATAGGTGTTGAAACAATAAGTTTATTTAT-3′), and pT7L1MUT was generated by PCR on pC16L1MUTANT#123 using oligonucleotides T7CMV and pAsignal(AS). The fragments were subcloned and transferred to pUC19 using SacI and XbaI, generating pT7L1 and pT7L1MUT.
(iv) L1 gene mutant hybrids.
pCL1H:1 was generated by PCR of the HPV-16 L1 gene using oligonucleotides L1STARTSalI and L1(AS)StyI (5′-CCTGATACCTTGGGAACTAAT-3′) followed by subcloning, digestion with BssHII and StyI, and transfer into pC16L1MUTANT#123. pCL1H:II was generated by PCR using oligonucleotides L1(S)StyI (5′-ATTAGTTCCCAAGGTATCAGG-3′) and L1(AS)MscI (5′-GGATGGCCACTAATGCCC-3′) followed by subcloning, digestion with StyI and MscI, and transfer to pC16L1MUTANT#123. pCL1H:III was generated by PCR using oligonucleotide L15′-MUTANT#1 and L1(AS)MscI followed by subcloning and transfer to pC16L1MUTANT#123 with BssHII and MscI. pCL1H:IV was generated by PCR using oligonucleotides L1STARTSalI and L1(AS)MscI, followed by subcloning and transfer into pC16L1MUTANT#123 with BssHII and MscI. pCL1H:V was generated by PCR using oligonucleotides L1(S)StyI and L1(AS)BamHI (5′-GGATCCTTTGCCCCAGTGTTCC-3′) followed by subcloning and transfer into pC16L1MUTANT#123 with StyI and BamHI.
L1 genes from various HPV types.
pC5L1, pC6bL1, pC18L1, pC31L1, pC45L1, and pC56L1 were generated by PCR amplification from the L1 gene start codon to the stop codon from each individual coding sequence using the following oligonucleotides: HPV5L1start (5′-CCGTCGACGCGCGCGAAATGGCAGTGTGGCACTCGG-3′) and HPV5L1stop (5′-CCCTCGAGTCAATTTTTACGTTTGCGTTTTG-3′), HPV6bL1start (5′-CCGTCGACGCGCGCGAAATGTGGCGGCCTAGCGAC-3′) and HPV6bL1stop (5′-CCCTCGAGTTACCTTTTGGTTTTGGCGCG-3′), HPV18L1start (5′-CCGTCGACGCGCGCGAAATGGCTTTGTGGCGGCCTAGTG-3′) and HPV18L1stop (5′-CCCTCGAGTTACTTCCTGGCACGTACAC-3′), HPV31L1start (5′-CCGTCGACGCGCGCGAAATGTCTCTGTGGCGGCCTAG-3′) and HPV31L1stop (5′-CCCTCGAGTTACTTTTTAGTTTTTTTACGTTTTGC-3′), HPV45L1start (5′-CCGTCGACGCGCGCGAAATGGCTTTGTGGCGGCCTAG-3′) and HPV45L1stop (5′-CCCTCGAGTTATTTCTTACTACGTATACGTAC-3′), HPV56L1start (5′-CCGTCGACGCGCGCGAAATGGCGACGTGGCGGCC-3′) and HPV56L1stop (5′-CCCTCGAGCTACCGCCTTTTACGTTTTTGC-3′). The PCR fragments were subcloned into the pCR-TOPO cloning vector (Invitrogen) before excision with BssHII and XhoI and transfer to the pCL086 vector (Fig. 2A).
To generate pC16L1:8, the HPV-16 L1 gene was transferred from pT7-16L1 (46) by digestion with BssHII and XhoI and inserted into pCH16pA (46). The following plasmids have been described previously, pCE55 (47), pC16L1UTR (46), pCCAT as pΔKXb (53), and pCTAT as pHCMVtat (42).
Cells and transfections.
Transfections were performed using the HeLa or HeLa-tat cells maintained in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal calf serum. Transfections were performed according to the Fugene 6 method (Roche Molecular Biochemicals). Briefly, 1 μg of DNA was transfected in combination with 3 μl of Fugene 6 and added in 200-μl aliquots consisting of DNA, Fugene 6, and medium to 60-mm-diameter plates containing subconfluent HeLa or HeLa-tat cells. All transfection mixtures contained 1 μg of pCTAT or pCCAT as an internal control for measuring transfection efficiency.
RNA extraction and Northern blotting.
Total RNA extraction was performed 24 h posttransfection according to the RNeasy Mini protocol (Qiagen) for the isolation of total RNA from animal cells. Cytoplasmic RNA extraction was performed as previously described (46) and according to the RNeasy Mini protocol for the isolation of cytoplasmic RNA from animal cells (Qiagen). Nuclear RNA fractionation was performed by separating the nucleus and cytoplasm according to the RNeasy Mini protocol, followed by added washing steps before continuing with the protocol for total RNA extraction. Northern blot analysis was performed by the separation of 10 μg of total or cytoplasmic RNA on 1% agarose gels containing 2.2 M formaldehyde, followed by transfer to a nitrocellulose filter and hybridization. Random priming of the DNA probe was performed with a Decaprime kit (Ambion) according to the manufacturer's instructions. The DNA probe used was generated by digestion of the pCL086 vector with SacI and SalI, thereby releasing the 133-nucleotide cytomegalovirus (CMV) leader sequence (Fig. 2A). Alternatively, end-labeled oligonucleotides 28S AS (5′-CCCGCGCCCCGCGGGGCGGGGATTCGGCGCTGGG-3′) and 47S AS (5′-CCGGGCGCCCGCAGCGGAGAGCGCACGGGGCACGGTGGCC-3′) were used to detect 28S and 47S rRNA.
CAT ELISA, in vitro translation, Western blotting, and indirect immunofluorescence.
Production of the CAT protein was quantitated by a CAT antigen capture enzyme-linked immunosorbent assay (ELISA; Roche Molecular Biochemicals) according to the manufacturer's protocol. In vitro translation was performed in the coupled transcription/translation rabbit reticulocyte lysate system (Promega) according to the manufacturer's instructions. Western blot analysis was performed as described previously (48). The rabbit anti-L1 peptide serum was generously provided by J. Dillner and was used at a dilution of 1:5,000 in immunoblots and 1:50 in indirect immunofluorescence. Indirect immunofluorescence was performed on formaldehyde-fixed transfected cells.
In vitro RNA synthesis and RNA transfection.
Plasmids pT7L1 and pT7L1MUT were linearized with XbaI and transcribed in transcription buffer (40 mM Tris [pH 8], 8 mM MgCl2, 2 mM spermidine, 25 mM NaCl) containing cap analogue 5′7MeGpppG5′ (1.0 mM), ribonucleotide mixture (1 mM [each] CTP, ATP, and UTP and 0.3 mM GTP), RNA guard (1 U/μl), and bacteriophage T7 RNA polymerase (2 U/μl) in a total volume of 50 μl at 37°C for 1 h. Samples were than treated with RNase-free DNase I (1 U/μl) at 37°C for 1 h followed by purification of the RNAs on RNeasy minispin columns according to the manufacturer's instructions (Qiagen). The RNAs were then polyadenylated in vitro by incubating the RNA in a mixture containing 40 mM Tris, pH 8, 10 mM MgCl2, 2.5 mM MnCl2, 250 mM NaCl, and 0.25 mM ATP (Amersham Pharmacia Biotech Inc.) in the presence of 12 U of poly(A) polymerase (Life Technologies) for 30 min at 37°C. The mRNAs were purified on RNeasy minispin columns, and the concentrations were determined.
Six micrograms of mRNA was mixed with 10 million cells in 1× phosphate-buffered saline and subjected to electroporation in a Bio-Rad Gene Pulser electroporator under the following conditions: 380 V, 960 μF, 950 V/cm. The cells were harvested at 20 h posttransfection. Capped and polyadenylated CAT mRNA was included as an internal control. The levels of CAT were determined in CAT capture ELISAs according to the manufacturer's instructions (Roche Molecular Biochemicals).
RESULTS
Detection of negative elements in HPV-16 L1 and L2 protein coding regions.
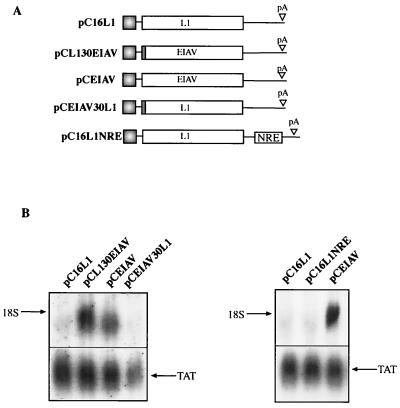
Here we have used a transfection system to monitor mRNA levels produced from any gene placed between the human CMV immediate-early promoter and the HPV-16 late pA signal (Fig. 1) in the pCL086 vector (Fig. 2A). The system involves the use of a DNA probe that is complementary to the leader sequence transcribed from the CMV promoter, which is present on all mRNAs produced from pCL086-derived plasmids independent of the insert (Fig. 2A). As can be seen in Fig. 2B, sequences with negative elements can be immediately detected when analyzed in tandem with genes which are known to contain no such elements. pCTAT (42), expressing HIV-1 Tat cDNA 1.4.7 (33), pCCAT, expressing bacterial CAT, and pCEIAV (47), expressing EIAV p55gag (Fig. 2A), produced detectable mRNA levels when analyzed with the same probe (Fig. 2B). In contrast, the mRNA levels produced from the HPV-16 L1- and L2-encoding plasmids pC16L1 and pC16L2 are undetectable (Fig. 2B), confirming our earlier findings that inhibitory sequences are present within their respective coding regions. Insertion of an internal ribosome entry site-driven reporter gene on the L1 and EIAV mRNAs confirmed the large difference in RNA levels between the two mRNAs (Fig. 2C). Similar results were obtained with different promoters, e.g., the HIV-1 long terminal repeat promoter and the human RNA polymerase I promoter, and different cell lines, including cells of nonhuman and nonepithelial origin (data not shown). We can exclude the possibility that interactions between the inhibitory sequences in the L1 gene and early ORF products are required for inhibition mediated by the L1 gene sequences. Since the probe hybridized to the 133 nucleotides of the leader sequence from the CMV promoter (Fig. 2A), we wished to exclude the possibility that the first nucleotides of the HPV sequences interfered with the ability of the probe to hybridize to its target sequence. We therefore transferred 30 nucleotides of the immediate 5′ end of the L1 coding region to EIAV (which produced high mRNA levels), generating pCL130EIAV (Fig. 3A). In addition, 30 nucleotides of the 5′ end of the EIAV coding sequence were fused to the L1 coding sequence in pC16L1, generating pCEIAV30L1 (Fig. 3A). The presence of the L1 gene 5′ 30 nucleotides had no effect on the EIAV mRNA levels, and the presence of the EIAV 5′ 30 nucleotides on the L1 gene did not alleviate the inhibitory activity in the L1 gene (Fig. 3B). The pCTAT plasmid served as an internal control and produced similar mRNA levels in all transfections (Fig. 3B).
FIG. 3.
Lack of inhibitory sequences in the first 30 nucleotides of the HPV-16 L1 gene. (A) Structures of the indicated expression plasmids. pCL130EIAV contains the first 30 nucleotides of the HPV-16 L1 gene fused in frame with the EIAV p55gag coding sequence, and pCEIAV30L1 contains the first 30 nucleotides of the EIAV p55gag fused in frame with the HPV-16 L1 coding sequence. (B) Northern blot of total cytoplasmic RNA harvested from HeLa cells transfected with the plasmids shown in panel A. Plasmid pCTAT (42), expressing the HIV-1 tat cDNA (33), was included as an internal control for transfection efficiency. L1, HPV-16 L1 gene; EIAV, the EIAV p55gag coding sequence; NRE, negative regulatory element in the HPV-16 late 3′ UTR from nucleotide 7282 to 7453 (35).
It has been shown previously that the HPV-16 late 3′ UTR contains a negative element termed NRE (23, 24). However, the absence or presence of the late 3′ UTR does not affect the expression of L1 mRNA from the plasmids used here (Fig. 3A and B), demonstrating that the inhibitory sequences in the HPV-16 L1 ORF act independently of the 3′ UTR NRE.
Mapping of the L1 gene negative elements to the 5′ end of the L1 gene.
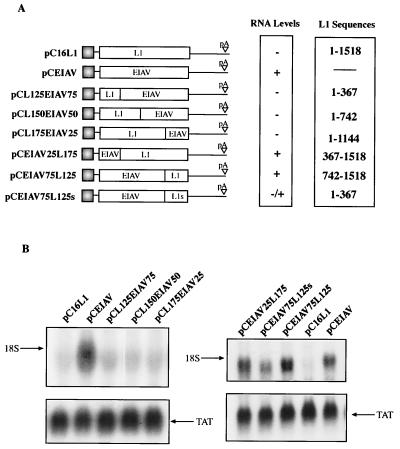
To locate the negative element in the HPV-16 L1 coding region, we designed hybrid constructs which consisted of sequences from the L1 and EIAV ORFs fused in frame. We selected EIAV gag as a fusion partner since EIAV gag and the HPV-16 L1 gene both encode viral capsid proteins of approximately 55 kDa. The L1 and EIAV coding sequences were divided into four parts and fused to each other (Fig. 4A). These hybrid plasmids were transfected into HeLa cells, and the resulting mRNA levels were monitored by Northern blotting. The results showed that the mRNA levels produced from pCL125EIAV75, pCL150EIAV50, and pCL175EIAV25 were undetectable (Fig. 4B, left), whereas high mRNA levels were produced from pCEIAV and internal control pCTAT (Fig. 4B). Therefore, the first 367 nucleotides contain sequences that reduce mRNA levels to the same extent as the full wild-type L1 gene. Interestingly, when these L1 gene sequences were fused to the 3′ end of the EIAV coding sequence, as in pCEIAV75L125s, inhibition was substantially reduced (Fig. 4B). When the first 367 nucleotides of the full wild-type L1 gene sequence are replaced with EIAV sequences, as in pCEIAV25L175, inhibition is shown to be abolished (Fig. 4B, right). As expected, plasmid pCEIAV75L125 (Fig. 4A) produced high mRNA levels (Fig. 4B). These results demonstrated that the major inhibitory sequences are located in the 5′ end of the L1 gene and also suggest that no significant negative elements are present in the 3′ end of the L1 gene.
FIG. 4.
Mapping of the inhibitory sequences in the HPV-16 L1 gene. (A) Structures of the indicated expression plasmids. The HPV-16 L1 gene sequences were fused in frame with the EIAV p55gag coding sequence to produce the diagrammed plasmids. Nucleotide position 1 refers to the first purine in the translational start codon of the L1 coding sequence. (B) Northern blot of total cytoplasmic RNA harvested from HeLa cells transfected with the plasmids shown in panel A. Plasmid pCTAT (42), expressing the HIV-1 tat cDNA (33), was included as an internal control for transfection efficiency. L1, HPV-16 L1 gene; EIAV, EIAV p55gag coding sequence.
Mutational inactivation of the negative elements in the HPV-16 L1 gene.
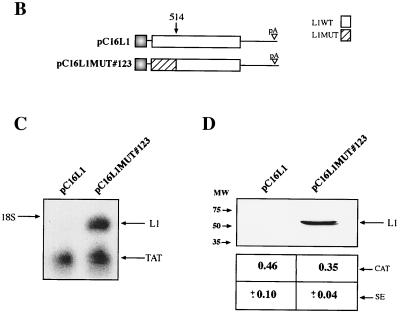
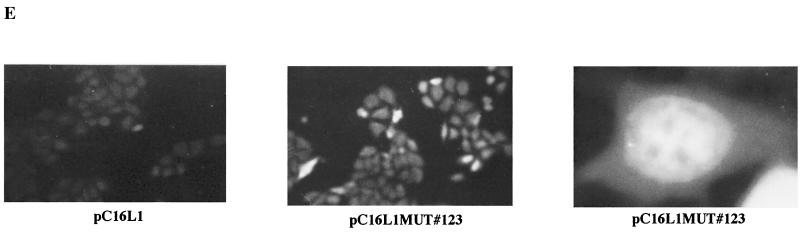
To provide further evidence for the existence of an inhibitory element in the 5′ end of the L1 gene, we reasoned that alterations in the RNA sequence which would not affect the protein sequence of L1 might destroy the inhibitory element and induce high production of L1. Since the negative elements appeared to be so extensive within the 5′ end of the L1 gene, at least up to nucleotide 367, we decided to mutate sequences between nucleotides 1 and 514 in the L1 coding sequence. Primarily wobbling nucleotide positions were changed so as not to alter the protein sequence of L1 (Fig. 5A). Care was taken not to introduce rare codons. This resulted in plasmid pC16L1MUT#123 (Fig. 5B). After transfection, total RNA extractions and protein extractions were performed, followed by Northern blot analysis and Western blot analysis on the respective samples. The results displayed in Fig. 5C and D show that the introduced mutations inactivated the inhibitory sequences, which resulted in the production of high L1 mRNA and protein levels. In contrast, L1 mRNA and protein levels produced from the wild-type L1 gene sequence were undetectable (Fig. 5C and D), whereas Tat or CAT levels produced from internal control plasmids pCTAT and pCCAT in all transfections were similar. The mRNA levels produced from pC16L1MUT#123 were comparable to or higher than those produced from the EIAV gag-containing plasmid, demonstrating that all negative elements in the L1 gene had been inactivated by these point mutations. We also performed immunofluorescence on the respective samples. For the wild-type L1 gene sequence, no staining was observed after immunofluorescence (Fig. 5E). In contrast, cells transfected with the L1 gene mutant showed specific staining, with the localization of the L1 protein in the nucleus but not in the nucleolus (Fig. 5E). In conclusion, these experiments proved the existence of inhibitory elements within the first 514 nucleotides in the 5′ end of the L1 gene and demonstrated that the L1 gene sequence downstream of position 514 lacks inhibitory sequences.
FIG. 5.
Mutational analysis of the inhibitory sequences in the HPV-16 L1 coding sequence. (A) The sequences of the wild-type (WT) and mutant (MUT) HPV-16 L1 gene sequences from position 1 to 514 is shown. Numbering starts at the first purine in the translational start codon of the L1 gene coding sequence. Altered nucleotides are in boldface italics. The StyI, MscI, and BamHI sites at nucleotides 188, 356, and 514, respectively, are indicated. Arrowhead, nucleotide 129. (B) Structures of the plasmids expressing the wild-type and the mutant HPV-16 L1 genes. (C) Northern blot of total cytoplasmic RNA harvested from HeLa cells transfected with the two plasmids shown in panel B. Plasmid pCTAT (42), expressing the HIV-1 tat cDNA (33), was included as an internal control for transfection efficiency. (D) Western blot of extracts from HeLa cells transfected with the two plasmids shown in panel B. The L1 protein is indicated. The CAT protein levels produced from internal control plasmid pCCAT and standard errors from triplicate experiments are shown. (E) Indirect immunofluorescence on HeLa cells transfected with the two plasmids shown in panel B. The cells were stained with an anti-HPV-16 L1 peptide antiserum (13).
Mapping of the inhibitory elements through the use of hybrids between the mutant and the wild-type HPV-16 L1 gene sequence.
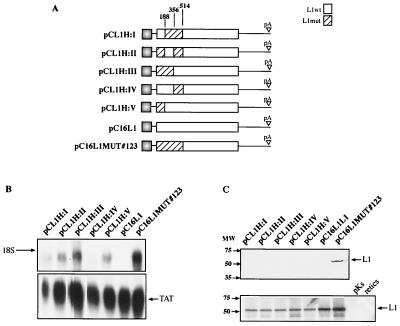
To determine the exact location of the inhibitory elements in the 5′ end of the L1 gene, we first constructed hybrids between the wild-type L1 and mutant L1 gene sequences (Fig. 6A). The L1 gene sequence between positions 1 and 514 was divided into three segments, each flanked by hinge restriction sites at positions 188, 356, and 514 (Fig. 5A and 6A). The different segments could be exchanged to generate the plasmids shown in Fig. 6A. All plasmids contained an uninterrupted L1 coding sequence. Transfections and Northern blot analysis revealed the presence of a strong negative element within the first 188 nucleotides (Fig. 6B). L1 gene hybrid constructs containing this sequence showed no detectable L1 mRNA levels (Fig. 6B, lanes 1, 4, and 6). In respect to other negative elements within nucleotides 1 to 514, it appears that another weaker element lies in the region of nucleotides 188 to 356 (Fig. 6B) and that there may be one element between nucleotides 356 and 514 (Fig. 6B). Normalization of the L1 mRNA levels to the internal control mRNA does not alter the conclusion of the results. These elements are clearly significantly weaker than the major inhibitory sequence located within the first 188 nucleotides of the HPV-16 L1 gene coding sequence. However, although the different fragments differ in the efficiency of inhibition, they act independently of each other.
FIG. 6.
Mapping of inhibitory sequences in the HPV-16 L1 gene using hybrids between wild-type (wt) and mutant (mut) HPV-16 L1 gene sequences. (A) Structures of the plasmids expressing the wt and the mutant HPV-16 L1 genes. Numbering starts at the first purine in the translational start codon of the L1 coding sequence. The StyI, MscI, and BamHI sites at nucleotides 188, 356, and 514, respectively, which were used to generate the hybrids, are indicated. pA, poly(A). (B) Northern blot of total cytoplasmic RNA harvested from HeLa cells transfected with the plasmids shown in panel A. Plasmid pCTAT (42), expressing the HIV-1 tat cDNA (33), was included as an internal control for transfection efficiency. (C) (Top) Western blot of extracts from HeLa cells transfected with the plasmids shown in panel A. The filter was incubated with an anti-HPV-16 L1 peptide antiserum (13). (Bottom) In vitro translation of the various L1 coding sequences in the plasmids shown in panel A after insertion downstream of the T7 promoter in pBluescript. The radiolabeled products were analyzed by autoradiography. pKS, in vitro translation of empty pBluescript vector; retics, in vitro translation reaction in the absence of plasmid DNA.
Analysis of protein levels produced by the L1 wild-type gene and the hybrids by Western blotting showed that all hybrids were negative compared to the L1 gene mutant carried by pC16L1MUT#123 (Fig. 6C, top). These results may be due to the relative insensitivity of the Western blotting. Therefore, we would not expect to detect the same level of variability that was observed among the mRNA levels (Fig. 6B). In vitro translation of all hybrids showed that all hybrid ORFs were translatable in a rabbit reticulocyte lysate system (Fig. 6C, bottom). From these findings, combined with the data shown in Fig. 6B, we concluded that multiple independent inhibitory sequences are located between nucleotides 1 and 514 in the 5′ end of the L1 coding sequence and that the major negative element lies between nucleotides 1 and 188 of the L1 gene.
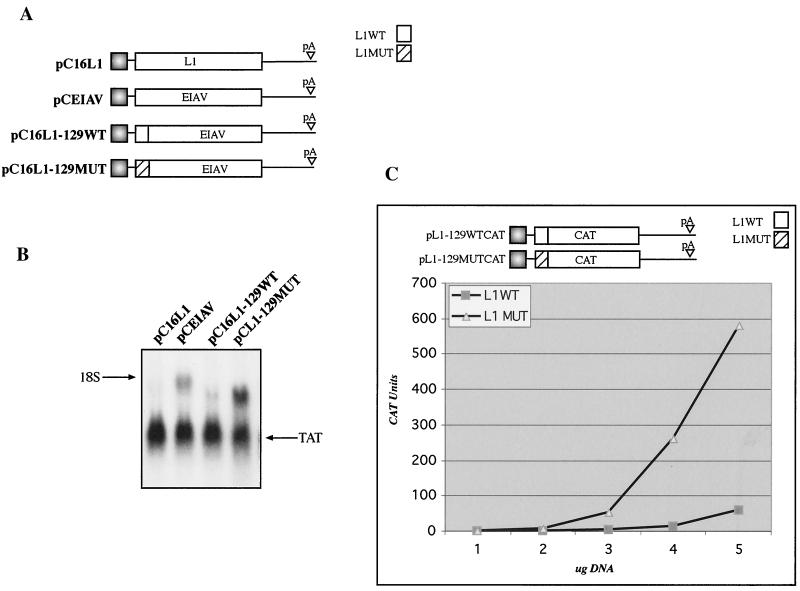
The major negative element is located within the first 129 nucleotides of the HPV-16 L1 gene.
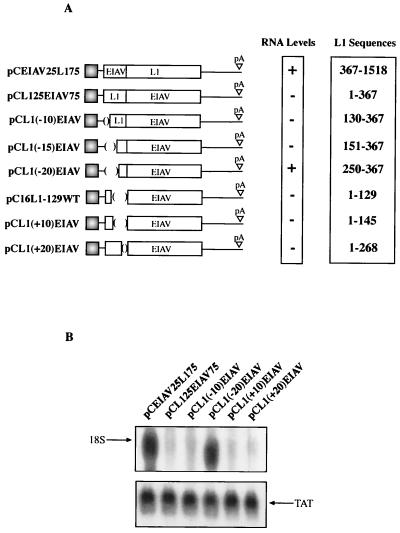
To map the major inhibitory sequences of the L1 gene in greater detail, deletion mutants were constructed from the pC16L125EIAV75 expression plasmid (Fig. 7A). This plasmid contains the L1 gene (nucleotides 1 to 367) fused to EIAV gag. The deletion constructs are displayed in Fig. 7A. All L1 gene deletion mutants contain a translational start codon and are fused in frame with EIAV. After transfection and total RNA extraction, samples were analyzed by Northern blotting. The results showed that deletion mutants that lack L1 gene nucleotides 1 to 129 [pCL1(−10)EIAV] or 1 to 150 [pCL1(−15)EIAV] remained inhibitory (Fig. 7B and data not shown), whereas a deletion mutant that lacks nucleotides 1 to 249 [pCL1(−20)EIAV] also lacks inhibitory activity (Fig. 7B). These results established that inhibitory sequences are present upstream of nucleotide 250 in the L1 gene. To map the major inhibitory element located between nucleotides 1 and 188 further, 3′ deletions were introduced into the 367-nucleotide L1 gene sequence present in pC16L125EIAV75 (Fig. 7A). When L1 gene sequences encompassing nucleotides 1 to 129, 1 to 145, and 1 to 268 were fused in frame with EIAV, as in pC16L1-129WT, pCL1(+10)EIAV, and pCL1(+20)EIAV, respectively, inhibition was observed (Fig. 7B and data not shown). From these data one may deduce that a negative element is located between nucleotides 151 and 367 and that the major negative element resides within the L1 gene fragment comprising nucleotides 1 to 129. This is in agreement with the results shown in Fig. 6. To confirm that the major negative element is located in the immediate 5′ end of the L1 gene, L1 gene nucleotides 1 to 129 in pC16L1-129WT were replaced by the corresponding 1 to 129 nucleotides from the mutant L1 coding sequence, generating pC16L1-129MUT (Fig. 8A). As expected, high mRNA levels were obtained using pC16L1-129MUT (Fig. 8B), whereas mRNA levels were substantially lower when using pC16L1-129WT (Fig. 8B). To exclude the possibility that the inhibitory activity of the L1 sequence was seen only when the L1 gene was fused to EIAV, we also fused the 5′ 129 nucleotides of wild-type and mutant sequences in frame with the CAT reporter gene, generating pL1-129WTCAT and pL1-129MUTCAT (Fig. 8C), and monitored CAT production in transfected cells. As can be seen from the results, CAT protein levels produced from the plasmid containing the L1 gene wild-type sequence were approximately 5- to 10-fold lower than those produced with the plasmid containing the mutant L1 gene sequence (Fig. 8C), confirming that the first 129 nucleotides of the HPV-16 L1 gene encode strong inhibitory elements. The half-life of an mRNA containing the major inhibitory element in the HPV-16 L1 gene was determined and compared to the half-life of an mRNA containing the noninhibitory mutant L1 gene sequence. Cells were transfected with the two plasmids shown in Fig. 8C, followed by actinomycin D treatment and harvesting of RNA at different time points. The results revealed that mRNAs containing the wild-type HPV-16 L1 gene sequence were less stable than those containing the mutant L1 gene sequence, demonstrating that the HPV-16 L1 gene region contains instability elements (Fig. 8D).
FIG. 7.
Fine mapping of the inhibitory sequences in the HPV-16 L1 gene. (A) Structures of the indicated expression plasmids. The HPV-16 L1 gene sequences were fused in frame with the EIAV p55gag coding sequence as indicated. Nucleotide 1 refers to the first purine in the translational start codon of the L1 coding sequence. pA, poly(A). (B) Northern blot of total cytoplasmic RNA harvested from HeLa cells transfected with the plasmids shown in panel A. Plasmid pCTAT (42), expressing HIV-1 tat cDNA (33), was included as an internal control for transfection efficiency. L1, HPV-16 L1 gene; EIAV, EIAV p55gag coding sequence.
FIG. 8.
The first 129 nucleotides of the HPV-16 L1 gene contain a major inhibitory element. (A) Structures of the indicated expression plasmids. The first 129 nucleotides of the HPV-16 L1 gene wild-type (WT) and mutant (MUT) sequences were fused in frame with the EIAV gag coding sequence. (B) Northern blot of total cytoplasmic RNA harvested from HeLa cells transfected with the plasmids shown in panel A. Plasmid pCTAT (42), expressing the HIV-1 tat cDNA (33), was included as an internal control for transfection efficiency. (C) Structures of the expression plasmids which contained the first 129 nucleotides of the HPV-16 L1 wild-type or mutant sequence fused in frame with the CAT ORF. Different amounts of the two CAT plasmids were transfected into HeLa cells, and the levels of CAT were monitored in a CAT capture ELISA. CAT units, arbitrary CAT units; L1, HPV-16 L1 gene; EIAV, the EIAV p55gag coding sequence. (D) Plasmids pL1-129WTCAT and pL1-129MUTCAT were transfected into HeLa cells that were treated with actinomycin D at 20 h posttransfection. The RNA levels were analyzed by Northern blotting, and the RNA blots were quantitated by phosphorimager and plotted against time after actinomycin D addition. lg, logarithmic value.
The inhibitory sequences in the HPV-16 L1 gene coding region act in the nucleus.
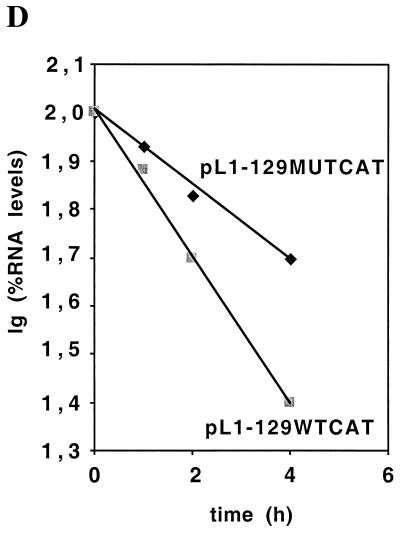
We have previously shown that HIV-1 nuclear export protein Rev and its target sequence, RRE, can at least partially overcome the inhibition exerted by the L1 gene elements and induce production of the HPV-16 L1 protein in transient transfections of HeLa cells (46), suggesting that the HPV-16 L1 mRNAs are defective and are retained in the nucleus. To compare nuclear and cytoplasmic mRNA levels produced from the wild-type and mutant L1 genes, plasmids pC16L1MUT#123 and pC16L1 were separately transfected into HeLa cells and nuclear and cytoplasmic mRNAs were extracted and analyzed by Northern blotting. The results revealed that high levels of L1 mutant mRNA are present in both the nucleus and the cytoplasm, whereas the wild-type L1 mRNA was undetectable both in the nuclear and in the cytoplasmic fractions (Fig. 9). The results demonstrated that the inhibitory sequences in the HPV-16 L1 coding region act in the nucleus.
FIG. 9.
The inhibitory sequences in the HPV-16 L1 gene act in cis by reducing the mRNA levels in the nucleus. HeLa cells were transfected with plasmid pC16L1 or pC16L1MUT#123. The transfected cells were fractionated into nuclear and cytoplasmic fractions, and RNA was extracted and analyzed by Northern blotting to detect the L1 mRNAs (A) and 28S and 47S rRNAs (B), as described in Materials and Methods. N, nuclear RNA; C, cytoplasmic RNA.
The inhibitory sequences in the HPV-16 L1 gene are not active in the cytoplasm.
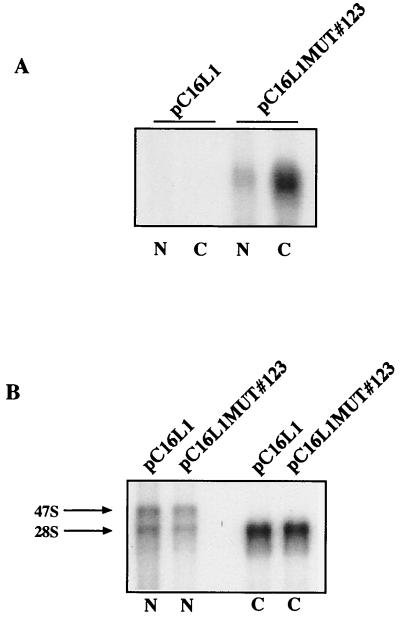
To investigate if the L1 gene inhibitory sequences acted also in the cytoplasm, we generated plasmids from which mRNAs with the same sequences as those produced from expression plasmids pC16L1 and pC16L1MUT#123 in transfected cells could be produced in vitro, capped and polyadenylated, and transfected into the cytoplasm of cells by electroporation. The resulting plasmids, pT7L1 and pT7L1MUT, are shown in Fig. 10A. Western blot analysis of extracts from RNA-transfected HeLa cells revealed that the two mRNAs produced similar levels of L1 protein when transfected into cells (Fig. 10B). In contrast, parallel DNA transfections of pC16L1 and pC16L1MUT#123 gave rise to L1 protein production only from the L1 gene mutant, not from the wild-type L1 gene sequence (Fig. 10B), as expected. Therefore, we concluded that the inhibitory elements in the HPV-16 L1 gene were not active in the cytoplasm, at least not in the absence of a nuclear experience of the mRNA.
FIG. 10.
The inhibitory sequences in the HPV-16 L1 gene are not active in the cytoplasm. (A) Structures of the indicated expression plasmids. In plasmids pT7L1 and pT7L1MUT the CMV promoter is replaced by the T7 promoter, which was inserted immediately 5′ of position +1 in the CMV promoter, and the polyadenylation signal is flanked by the XbaI restriction site. XbaI-restricted plasmids were used for in vitro synthesis of capped mRNAs followed by in vitro polyadenylation. The sequences of these mRNA are therefore the same as the sequences of the mRNAs produced in the nuclei of cells transfected with corresponding eukaryotic expression plasmids pC16L1 and pC16L1MUT#123 (Fig. 5B), respectively. T7, bacteriophage T7 promoter; C, nucleotide +1 to +133 leader sequence from the CMV promoter. (B) Western blot analysis of extracts from cells transfected with pC16L1 or pC16L1MUT#123 plasmid DNA or capped and polyadenylated mRNAs produced in vitro from pT7L1 and pT7L1MUT. (C) In vitro translation of various amounts of pT7L1 or pT7L1MUT plasmid DNA in a coupled reticulocyte lysate in vitro translation system (Promega). The radiolabeled products were separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels and then subjected to autoradiography. The HPV-16 L1 protein is indicated.
It has been shown previously that BPV-1 L1 mRNAs are inefficiently translated in the TNT rabbit reticulocyte lysate translation system (Promega) and that replacing rare codons in the BPV-1 L1 gene sequence with those preferred in humans resulted in increased translation efficiency in the same system (57). We therefore wished to investigate if the wild-type HPV-16 L1 mRNA was inefficiently translated compared with the mutant L1 RNA. We used the plasmids shown in Fig. 10A, which contain the wild-type L1 ORF and the mutant L1 ORF downstream of the bacteriophage T7 promoter. These plasmids were transcribed and translated in the coupled rabbit reticulocyte lysate system. A dilution series of the wild-type and mutant L1-encoding plasmids was performed and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autoradiography. As seen in Fig. 10C, the levels of the L1 protein produced from the wild-type L1 gene sequence and the mutant L1 gene sequence in all dilution steps were similar, demonstrating that both mRNAs were translated with similar efficiencies in vitro. We concluded that the HPV-16 L1 gene inhibitory sequences do not inhibit translation in vitro.
The presence of negative RNA elements in the L1 ORF is a conserved property among HPVs.
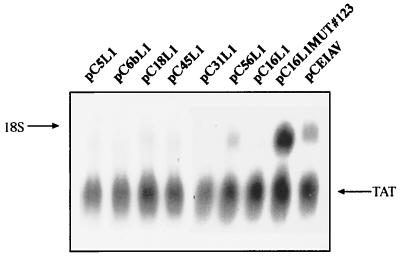
Since the negative elements appeared to be present in abundance in both L1 and L2 genes of HPV-16, we wished to analyze the L1 coding sequences from different HPV types. We selected a number of distantly related HPV types (HPV-5, -6b, -18, -31, -45, and -56), amplified the L1 coding sequences by PCR, and inserted the sequences in the pCL086 assay vector (Fig. 2A). The different resulting plasmids were transfected into HeLa cells, and cytoplasmic mRNA levels were monitored by Northern blotting. The results shown in Fig. 11 demonstrate that negative elements are present in the majority of the HPV types tested since L1 mRNA was undetectable in most transfections. However, HPV-56 produced detectable L1 mRNA levels, indicating that HPV-56 contained weaker inhibitory sequences. In contrast, the EIAV expression plasmid produced high mRNA levels as expected and the pCTAT internal control produced high levels of Tat mRNA in all transfections (Fig. 11). The conservation of the negative elements present in the coding regions of HPV L1 ORFs suggests that they have an important function in the HPV replication cycle.
FIG. 11.
Inhibitory sequences in the majority of the HPV L1 protein-coding sequences. Northern blot of total cytoplasmic RNA harvested from HeLa cells transfected with expression plasmids containing the L1 coding sequences of the various HPV types. The L1 coding sequences from the indicated HPV types were amplified by PCR and subcloned into pCL086 (Fig. 2A). The expression levels were compared to those of EIAV gag- and HPV-16 L1 gene-expressing plasmids pCEIAV and pC16L1, respectively. Plasmid pCTAT (42), expressing the HIV-1 tat cDNA (33), was included as an internal control for transfection efficiency.
DISCUSSION
Mapping experiments using L1 and EIAV hybrid genes demonstrated that multiple inhibitory sequences were located in the first 514 nucleotides of the L1 coding region. Analysis of the first 514 nucleotides of the wild-type L1 gene sequence showed that the sequence has a 51% purine content and a 59% AU content. This is similar to the contents of purine (51%) and AU (62%) of the entire L1 coding sequence, suggesting that the inhibitory activity of that region as a whole does not correlate with a certain content of purines or AU nucleotides. Inactivation of the negative elements through point mutations resulted in 31% of nucleotides being mutated, while not affecting the coding sequence. However, this caused only a modest change in the purine content to 48% but significantly reduced the AU content to 33%. It remains to be investigated if the alleviation in inhibition is a direct result of the alteration of the AU content of the first 514 nucleotides of the L1 coding sequence.
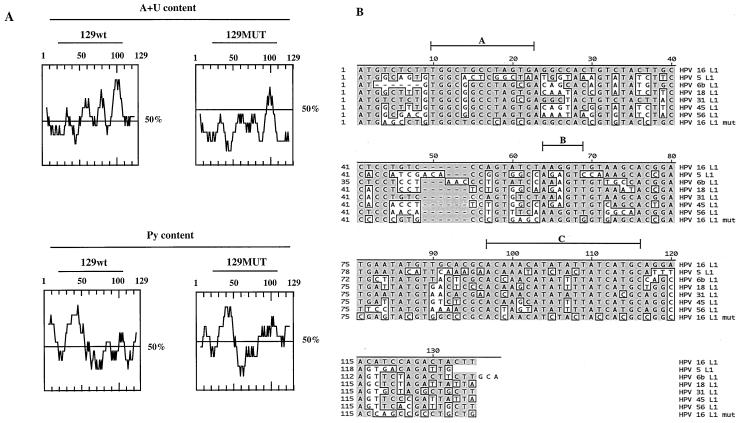
Using fine deletion mapping we showed that the strongest inhibitory element was located within the first 129 nucleotides. A look at the distribution of A and U and purines revealed that the 129-nucleotide L1 gene sequence can be divided into a pyrimidine-rich 5′ half and an AU-rich 3′ half (Fig. 12A). Overall, the 129-nucleotide sequence contains 55% A and U and 46% purines; these contents are slightly lower than the AU and purine contents of the full 520-nucleotide sequence (59% A and U and 51% purines) and the entire L1 coding sequence (62% A and U and 51% purine). In this case, mutagenesis resulted in a sequence with a significantly lowered purine content (37%) and AU content (32%) (Fig. 12A). In the mutant, the pyrimidine content in the 5′ end is slightly altered whereas the AU content in the AU-rich 3′ end has been dramatically reduced. One or both of these regions may encode the inhibitory elements. Interestingly, one of the more conserved regions is located in the AU-rich 3′ end of the first 129 nucleotides of the L1 gene and is an AU-rich sequence between nucleotides 97 and 116 with the consensus AC(A/C)APuPyAUAU(A/U)PyUAUCAPyGC (sequence C in Fig. 12B) and an AU content of 75%. It remains to be determined if this sequence is involved in the regulation of HPV-16 late-gene expression.
FIG. 12.
(A) (Top) Pyrimidine content of the first 129 nucleotides of the wild-type (wt) and mutant (MUT) HPV-16 L1 gene sequences. (Bottom) AU contents of the first 129 nucleotides of the wild-type and mutant HPV-16 L1 gene sequences. (B) Sequence alignments of the first 129 nucleotides of the L1 gene coding sequences of HPV-5, HPV-6b, HPV-16, HPV-18, HPV-31, HPV-45, and HPV-56 and the mutant HPV-16 L1 gene sequence. Regions A, B, and C are discussed in Discussion.
Some mRNAs contain sequences in the coding regions that inhibit gene expression by reducing the mRNA half-life. RNA instability elements in protein-coding regions have been found to be present in HPV-16 L2 (42), c-myc (51), c-fos (39), urokinase receptor (uPAR) (37), vascular endothelial growth factor (VEGF) (11), and plasminogen activator type 2 (PAI2) (50) mRNA. Furthermore 10-nucleotide sequences (GATAAAATCC in PAI2) in the coding regions of the PAI2, Mat α1, c-myc, uPAR, VEGF, and TFIIA mRNAs have homology and have been proposed as candidate RNA instability motif coding regions (50). The consensus motif is not found in the first 514 nucleotides of the HPV-16 L1 coding sequence. However, other RNA instability elements that do not match previously known motifs may be present in the L1 gene sequence. The c-fos coding region RNA instability determinant is very purine rich (9), whereas the 129-nucleotide HPV-16 L1 gene sequence is relatively poor in purine content (Fig. 12A). Notably, the c-fos coding region RNA instability determinant contains multiple GAA motifs (9) and has very high homology to purine-rich splicing-enhancer sequences (4). We are currently employing other techniques that will allow us to detect the L1 mRNAs in transfected cells and to determine their stability.
One of the major splice acceptors in the HPV-16 genome is located immediately upstream of the HPV-16 L1 gene start codon. This 3′ splice site, in combination with a 5′ splice site located in the E2 ORF, is presumably utilized to generate the mRNA producing the L1 protein in HPV-16-infected cells (15). In essence, this splicing event deletes the upstream L2 ORF, thereby allowing access of the L1 ORF to the translation machinery. This arrangement requires that splicing at these sites occur with suboptimal efficiency since efficient splicing would presumably result in too low or no production of functional L2 mRNA. In contrast, inefficient splicing would cause a lack of functional L1 mRNAs. In HIV-1 suboptimal splicing is required in order to generate unspliced gag/pol mRNA and partially spliced vif, vpr, vpu, and env mRNAs, in addition to the fully spliced tat, rev, and nef mRNAs (26). It is reasonable to speculate that HPV-16 late-mRNA splice signals are flanked by sequences determining the efficiency of the splice sites, the so-called splicing silencers or suppressers (4). In addition, inefficient splice sites have been shown to require splicing enhancers in order to function, resulting in a combination of silencers and enhancers in the vicinity of a “slow” or regulated splice site. These sequences often have an exonic location in relation to a 3′ splice site, which in many cases has a suboptimal branch point and polypyrimidine tract. Visual inspection of the sequence upstream of the splice acceptor at the L1 gene start codon revealed that a pyrimidine tract is present. However, there is no pyrimidine stretch that is uninterrupted for more than nine nucleotides, which indicates that the polypyrimidine tract deviates from the consensus of an optimal polypyrimidine tract located upstream of most 3′ splice sites. In addition, a putative branch point located upstream of the 3′ splice site does not match the consensus. Therefore, this splice site may be regulated and may contain RNA elements in the L1 ORF that regulate the usage of the 3′ splice site. Potentially, this is relevant to the results shown here since interaction of splicing factors with unutilized splice sites may lead to entrapment of mRNAs in the nucleus followed by degradation (7), although this is definitely not universally true. For example, tat cDNA 1.4.7, which is used here in all transfections and which produces high mRNA levels, contains at least four unutilized 3′ splice sites (but no 5′ splice, and the mRNA is not spliced) and at least two exonic splicing repressors (31, 38, 45, 46). Although there is no strict consensus sequence for exonic splicing enhancers (ESE), most are purine rich and interact with SR proteins (4). For example, the sequence of the fibronectin ESE is (GAAGAAGA) (5). We searched the first 129 nucleotides of the HPV-16 L1 coding sequence for sequences with homology to the various exonic splicing suppressor (ESS) motifs, but we were unable to identify sequences with substantial homology to any of the above motifs.
Splicing is regulated in BPV-1 (2), and splicing enhancers and silencers have been identified downstream of two weak 3′ splice signals in the early region of the BPV-1 genome (1, 54). The upstream nucleotide 3225 3′ splice site is used preferentially in proliferating cells and is followed by an ESE and an ESS 30 nucleotides downstream of the AG dinucleotide (54). In addition, 125 nucleotides downstream of the ESS, a second ESE was identified. Similarly, the downstream nucleotide 3605 3′ splice site, which is preferentially used in differentiated cells that are permissive for virus production, is followed by an ESE and an ESS. Whereas the ESEs are purine rich (nucleotide 3225) or AC rich (CACCACCACC; nucleotide 3605) (54, 56), the ESS at the nucleotide 3225 splice site can be divided into a U-rich region, a C-rich region, and a purine-rich 3′ end that bind to U2AF and PTB, the 35- and 55-kDa SR proteins, and ASF/SF2, respectively; the 129 nucleotides of the L1 gene are not purine rich and do not contain CAC repeats (Fig. 12A). The activity of the BPV-1 ESS element at the nucleotide 3225 splice site maps to C-rich core sequence GGCUCCCCC (55), whereas the ESS at nucleotide 3605 contains a UGGU motif (56). The 5′ half of the 129-nucleotide region of the HPV-16 L1 gene is pyrimidine rich. Although neither the GGCUCCCCC motif nor the UGGU motif was found in the HPV-16 L1 gene sequence, a pyrimidine-rich sequence was found in the first 10 nucleotides of the L1 gene (GUCUCUUU; sequence A in Fig. 12B), but this sequence did not possess inhibitory activity (plasmid pCL130EIAV; Fig. 3). We concluded that sequences with homology to regulatory RNA sequences in BPV-1 do not appear to be present in the first 129 nucleotides of the HPV-16 L1 coding sequence. However, this does not exclude the possibility that the 5′ region of the HPV-16 L1 ORF contains sequences that regulate splicing.
Zhou et al. observed that the BPV-1 L1 gene contains rare translational codons (57). They therefore replaced the rare codons with more commonly used codons and analyzed expression levels of a wild-type and a codon-modified BPV-1 L1 gene in transient transfection experiments. Both produced high levels of L1 mRNA in the cytoplasm of the cells, but only the codon-modified gene produced detectable levels of L1 protein (57). In addition, translation of the two sequences in a coupled rabbit reticulocyte lysate system revealed that the codon-modified L1 gene sequence was translated more efficiently than the wild-type L1 gene sequence (57). Based on their data, these investigators proposed that the usage of rare codons in the BPV-1 L1 ORF and tRNA availability prevent L1 production in proliferating cells, thereby restricting L1 production to terminally differentiated cells (57). The results for BPV-1 L1 gene expression observed by Zhou and colleagues are different from the results that we report here for the HPV-16 L1 gene. First, we found that the inhibitory sequences in the HPV-16 L1 gene reduce the L1 mRNA in the nucleus. Although this effect may be secondary to nuclear entrapment or inefficient RNA-processing steps, we were not able to detect wild-type HPV-16 L1 mRNA in the cytoplasm. Second, translation of our highly expressed mutant HPV-16 L1 coding sequence and the wild-type L1 coding sequence in the coupled rabbit reticulocyte lysate system revealed that the two sequences were translated equally well. These results were confirmed in living cells by electroporation of the two mRNAs into HeLa cells. In conclusion, the inhibitory sequences in the HPV-16 L1 mRNAs acted by reducing the levels of the L1 mRNAs in the nucleus but did not affect the translation efficiency of the L1 mRNA. If only the inhibitory sequence in the 5′ end of the L1 gene is inactivated, the L1 mRNA is efficiently translated in spite of the presence of multiple rare codons in the HPV-16 L1 coding sequence, confirming that rare codons in the HPV-16 L1 coding sequence do not affect expression levels significantly. In conclusion, inhibition of L1 production in proliferating cells occurs by different mechanisms in BPV-1 and HPV-16. Differences in the regulation of gene expression are not without precedent among papillomaviruses. For example, the regulatory element in the BPV-1 late-gene 3′ UTR interacts with U1 snRNP and acts by inhibiting polyadenylation (19, 20), whereas the late 3′ UTR element in HPV-1 interacts with HuR and hnRNP C and is an AU-rich RNA element that acts by reducing mRNA stability and translation (40, 41, 43, 48). One may speculate that the inhibitory sequence in the HPV-16 L1 gene region acts to reduce the L1 and L2 gene expression levels in proliferating cells. Cellular factors may bind specifically to the wild-type and not to the mutant L1 gene sequence. These factors may be posttranslationally modified during the course of cell differentiation, thereby losing their affinity for L1 mRNAs, which would result in high expression of L1 and L2 genes in terminally differentiated keratinocytes. Alternatively, a positive factor is activated as the cell differentiates.
Recent findings have suggested that the process of RNA synthesis to protein production is not a group of separate manufacturing stages (38). Instead it is thought of as one continuing process whereby factors which are required for RNA processing are present in the RNA polymerase II transcription complex as the mRNA is produced (44). One may speculate that the inhibitory RNA sequences in the L1 gene interact with cellular factors that interfere with the RNA polymerase II function. In support of an effect early in the RNA processing pathway, we found that the ability of the inhibitory sequence to function efficiently was reduced when the L1 gene sequence was fused in frame with the 3′ end versus the 5′ end of the EIAV coding sequence in the reporter plasmids. To investigate the mechanism of action further, the L1 coding sequence will be expressed from RNA polymerase I or RNA polymerase III promoters in addition to other expression systems that allow the determination of the half-lives of the wild-type and mutant HPV-16 L1 mRNAs. We have previously shown that, through the use of the nuclear mRNA export factors HIV-1 Rev and RRE and simian retrovirus type 1 CTE, the inhibition of L1 production could be at least partially overcome as judged from the L1 protein levels produced from the mutant L1-encoding plasmids made here and from the RRE-containing L1-encoding plasmid in the presence of Rev (46). Either Rev and RRE cause the export of both functional and nonfunctional mRNAs or Rev competes inefficiently with cellular factors that bind to the L1 mRNA and traps the mRNA in the nucleus. Whether defective mRNAs are made or the mRNAs are trapped in the nucleus, for example by splicing factors, in both situations followed by rapid degradation is under investigation. Although we did not find binding sites for splicing factors in the L1 gene sequence, it is possible that the HPV-16 L1 gene inhibitory sequences overlap or contain sequences that regulate splicing and interact with splicing factors. We are therefore studying the role of the inhibitory sequences in a splicing environment by investigating the effect of the sequences on the splicing event that deletes the L2 ORF to generate the spliced L1 mRNA in the context of the full viral genome.
ADDENDUM IN PROOF
After this article was submitted, it was reported that alterations of the codon usage of the HPV-16 L1 gene results in efficient production of HPV-16 L1 (C. Leder, J. A. Kleinschmidt, C. Wiethe, and M. Müller, J. Virol. 75:9201-9209, 2001). Changing the codon usage in the unmutagenized part of the L1 gene in the expression plasmids used here may increase the levels of L1 protein further.
Acknowledgments
We thank L. Wiklund for help with RNA transfections, P. Buckley for help with plasmid constructions, P. Kreivi and A. Grynfeld for valuable discussion, J. Dillner for HPV-16 L1 antisera, R. Ostrow, E.-M. de Villiers, and A. Lörincz for HPV plasmids, and A. Carlsson for technical assistance.
The work was supported by grants from the Swedish Medical Research Council and the Swedish Cancer Society. B.C. was supported by a fellowship from the Swedish Foundation for Strategic Research and the Dublin Institute of Technology.
REFERENCES
- 1.Baker, C. C. 1997. Posttranscriptional regulation of papillomavirus gene expression, p. 11-16. In S. R. Billakanti, C. E. Calef, A. D. Farmer, A. L. Halpern, and G. L. Myres (ed.), Human papillomaviruses: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 2.Barksdale, S., and C. C. Baker. 1995. Differentiation-specific alternative splicing of bovine papillomavirus late mRNAs. J. Virol. 69:6553-6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barksdale, S. K., and C. C. Baker. 1995. The human immunodeficiency virus type 1 Rev protein and the Rev-responsive element counteract the effect of an inhibitory 5′ splice site in a 3′ untranslated region. Mol. Cell. Biol. 15:2962-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blencowe, B. J. 2000. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 25:106-110. [DOI] [PubMed] [Google Scholar]
- 5.Caputi, M., G. Casari, S. Guenzi, R. Tagliabue, A. Sidoli, C. A. Melo, and F. E. Baralle. 1994. A novel bipartite splicing enhancer modulates the differential processing of the human fibronectin EDA exon. Nucleic Acids Res. 22:1018-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlsson, A., and S. Schwartz. 2000. Inhibitory activity of the human papillomavirus type 1 AU-rich element correlates inversely with the levels of the elav-like HuR protein in the cell cytoplasm. Arch. Virol. 145:491-503. [DOI] [PubMed] [Google Scholar]
- 7.Chang, D. D., and P. A. Sharp. 1990. Messenger RNA transport and HIV rev regulation. Science 249:614-615. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C. Y., and A. B. Shyu. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20:465-470. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C. Y. A., Y. You, and A.-B. Shyu. 1992. Two cellular proteins bind specifically to a purine-rich sequence necessary for the destabilization function of a c-fos protein-coding region determinant of mRNA instability. Mol. Cell. Biol. 12:5748-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collier, B., L. Goobar-Larsson, M. Sokolowski, and S. Schwartz. 1998. Translational inhibition in vitro of human papillomavirus type 16 L2 mRNA mediated through interaction with heterogenous ribonucleoprotein K and poly(rC)-binding proteins 1 and 2. J. Biol. Chem. 273:22648-22656. [DOI] [PubMed] [Google Scholar]
- 11.Dibbens, J. A., D. L. Miller, A. Damert, W. Risau, M. A. Vadas, and G. J. Goodall. 1999. Hypoxic regulation of vascular endothelial growth factor mRNA stability requires the cooperation of multiple RNA elements. Mol. Biol. Cell 10:907-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dietrich-Goetz, W., I. M. Kennedy, B. Levins, M. A. Stanley, and J. B. Clements. 1997. A cellular 65-kDa protein recognizes the negative regulatory element of human papillomavirus late mRNA. Proc. Natl. Acad. Sci. USA 94:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dillner, L., P. Heino, J. Moreno-Lopez, and J. Dillner. 1991. Antigenic and immunogenic epitopes shared by human papillomavirus type 16, bovine, canine, and avian papillomaviruses. J. Virol. 65:6862-6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dollard, S. C., J. L. Wilson, L. M. Demeter, W. Bonnez, R. C. Reichman, T. R. Broker, and L. T. Chow. 1992. Production of human papillomavirus and modulation of the infectious program in epithelial raft cultures. Genes Dev. 6:1131-1142. [DOI] [PubMed] [Google Scholar]
- 15.Doorbar, J., A. Parton, K. Hartley, L. Banks, T. Crook, M. Stanley, and L. Crawford. 1990. Detection of novel splicing patterns in a HPV16-containing keratinocyte cell line. Virology 178:254-262. [DOI] [PubMed] [Google Scholar]
- 16.Flores, E. R., B. L. Allen-Hoffmann, D. Lee, C. A. Sattler, and P. F. Lambert. 1999. Establishment of the human papillomavirus type 16 (HPV-16) life cycle in an immortalized human foreskin keratinocyte cell line. Virology 262:344-354. [DOI] [PubMed] [Google Scholar]
- 17.Frattini, M. G., H. B. Lim, J. Doorbar, and L. A. Laimins. 1997. Induction of human papillomavirus type 18 late gene expression and genomic amplification in organotypic cultures from transfected DNA templates. J. Virol. 71:7068-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frattini, M. G., H. B. Lim, and L. A. Laimins. 1996. In vitro synthesis of oncogenic human papillomaviruses requires episomal genomes for differentiation-dependent late expression. Proc. Natl. Acad. Sci. USA 93:3062-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furth, P. A., and C. C. Baker. 1991. An element in the bovine papillomavirus late 3′ untranslated region reduces polyadenylated cytoplasmic RNA levels. J. Virol. 65:5806-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furth, P. A., W. T. Choe, J. H. Rex, J. C. Byrne, and C. C. Baker. 1994. Sequences homologous to 5′ splice sites are required for the inhibitory activity of papillomavirus late 3′ untranslated regions. Mol. Cell. Biol. 14:5278-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grassmann, K., B. Rapp, H. Maschek, K. U. Petry, and T. Iftner. 1996. Identification of a differentiation-inducible promoter in the E7 open reading frame of human papillomavirus type 16 (HPV-16) in raft cultures of a new cell line containing high copy numbers of episomal HPV-16 DNA. J. Virol. 70:2339-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howley, P. M. 1996. Papillomavirinae: the viruses and their replication, p. 2045-2076. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa. [Google Scholar]
- 23.Kennedy, I. M., J. K. Haddow, and J. B. Clements. 1990. Analysis of human papillomavirus type 16 late mRNA 3′ processing signals in vitro and in vivo. J. Virol. 64:1825-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kennedy, I. M., J. K. Haddow, and J. B. Clements. 1991. A negative regulatory element in the human papillomavirus type 16 genome acts at the level of late mRNA stability. J. Virol. 65:2093-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koffa, M. D., S. V. Graham, Y. Takagaki, J. L. Manley, and J. B. Clements. 2000. The human papillomavirus type 16 negative regulatory RNA element interacts with three proteins that act at different posttranscriptional levels. Proc. Natl. Acad. Sci. USA 97:4677-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luciw, P. 1996. Human immunodeficiency viruses and their replication, p. 1881-1952. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa. [Google Scholar]
- 27.McBride, A. A., A. Dlugosz, and C. C. Baker. 2000. Production of infectious bovine papillomavirus from cloned viral DNA by using an organotypic raft/xenograft technique. Proc. Natl. Acad. Sci. USA 97:5534-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyers, C., T. J. Mayer, and M. A. Ozbun. 1997. Synthesis of infectious human papillomavirus type 18 in differentiating epithelium transfected with viral DNA. J. Virol. 71:7381-7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ozbun, M. A., and C. Meyers. 1998. Temporal usage of multiple promoters during the life cycle of human papillomavirus type 31b. J. Virol. 72:2715-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ross, J. 1995. mRNA stability in mammalian cells. Microbiol Rev. 59:423-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwartz, S. 1998. Cis-acting negative RNA elements on papillomavirus late mRNAs. Semin. Virol. 8:291-300. [Google Scholar]
- 32.Schwartz, S. 2000. Regulation of human papillomavirus late gene expression. Upsala J. Med. Sci. 105:171-192. [PubMed] [Google Scholar]
- 33.Schwartz, S., B. K. Felber, D. M. Benko, E. M. Fenyö, and G. N. Pavlakis. 1990. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J. Virol. 64:2519-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwartz, S., M. Sokolowski, B. Collier, A. Carlsson, and L. Goobar-Larsson. 1999. Cis-acting regulatory sequences on papillomavirus late mRNAs. Recent Res. Dev. Virol. 1:53-74. [Google Scholar]
- 35.Seedorf, K., G. Krämer, M. Dürst, S. Suhai, and W. G. Röwekamp. 1985. Human papillomavirus type 16 DNA sequence. Virology 145:181-185. [DOI] [PubMed] [Google Scholar]
- 36.Shah, K. V., and P. M. Howley. 1996. Papillomaviruses, p. 2077-2109. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa. [Google Scholar]
- 37.Shetty, S., A. Kumar, and S. Idell. 1997. Posttranscriptional regulation of urokinase receptor mRNA: identification of a novel urokinase receptor mRNA binding protein in human mesothelioma cells. Mol. Cell. Biol. 17:1075-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shilatifard, A. 1998. The RNA polymerase II general elongation complex. Biol Chem. 379:27-31. [DOI] [PubMed] [Google Scholar]
- 39.Shyu, A.-B., M. E. Greenberg, and J. G. Belasco. 1989. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 3:60-72. [DOI] [PubMed] [Google Scholar]
- 40.Sokolowski, M., H. Furneaux, and S. Schwartz. 1999. The inhibitory activity of the AU-rich RNA element in the human papillomavirus type 1 late 3′ untranslated region correlates with its affinity for the elav-like HuR protein. J. Virol. 73:1080-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sokolowski, M., and S. Schwartz. 2001. Heterogeneous nuclear ribonucleoprotein C binds exclusively to the functionally important UUUUU-motifs in the human papillomavirus type-1 AU-rich inhibitory element. Virus Res. 73:163-175. [DOI] [PubMed] [Google Scholar]
- 42.Sokolowski, M., W. Tan, M. Jellne, and S. Schwartz. 1998. mRNA instability elements in the human papillomavirus type 16 L2 coding region. J. Virol. 72:1504-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sokolowski, M., C. Zhao, W. Tan, and S. Schwartz. 1997. AU-rich mRNA instability elements on human papillomavirus type 1 late mRNAs and c-fos mRNAs interact with the same cellular factors. Oncogene 15:2303-2319. [DOI] [PubMed] [Google Scholar]
- 44.Steinmetz, E. J. 1997. Pre-mRNA processing and the CTD of RNA polymerase II: the tail that wags the dog? Cell 89:491-494. [DOI] [PubMed] [Google Scholar]
- 45.Stoler, M. H., A. Whitbeck, S. M. Wolinsky, T. R. Broker, L. T. Chow, M. K. Howett, and J. W. Kreider. 1990. Infectious cycle of human papillomavirus type 11 in human foreskin xenografts in nude mice. J. Virol. 64:3310-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan, W., B. K. Felber, A. S. Zolotukhin, G. N. Pavlakis, and S. Schwartz. 1995. Efficient expression of the human papillomavirus type 16 L1 protein in epithelial cells by using Rev and the Rev-responsive element of human immunodeficiency virus or the cis-acting transactivation element of simian retrovirus type 1. J. Virol. 69:5607-5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan, W., M. Schalling, C. Zhao, M. Luukkonen, M. Nilsson, E. M. Fenyö, G. N. Pavlakis, and S. Schwartz. 1996. Inhibitory activity of the equine infectious anemia virus major 5′ splice site in the absence of Rev. J. Virol. 70:3645-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan, W., and S. Schwartz. 1995. The Rev protein of human immunodeficiency virus type 1 counteracts the effect of an AU-rich negative element in the human papillomavirus type 1 late 3′ untranslated region. J. Virol. 69:2932-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas, J. T., S. T. Oh, S. S. Terhune, and L. A. Laimins. 2001. Cellular changes induced by low-risk human papillomavirus type 11 in keratinocytes that stably maintain viral episomes. J. Virol. 75:7564-7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tierney, M. J., and R. L. Medcalf. 2001. Plasminogen activator inhibitor type 2 contains mRNA instability elements within exon 4 of the coding region. Sequence homology to coding region instability determinants in other mRNAs. J. Biol. Chem. 276:13675-13684. [DOI] [PubMed] [Google Scholar]
- 51.Wisdom, R., and W. Lee. 1991. The protein-coding region of c-myc mRNA contains a sequence that specifies rapid mRNA turnover and induction by protein synthesis inhibitors. Genes Dev. 5:232-243. [DOI] [PubMed] [Google Scholar]
- 52.Zhao, C., M. Sokolowski, W. Tan, and S. Schwartz. 1998. Characterisation and partial purification of cellular factors interacting with a negative element on human papillomavirus type 1 late mRNAs. Virus Res. 55:1-13. [DOI] [PubMed] [Google Scholar]
- 53.Zhao, C., W. Tan, M. Sokolowski, and S. Schwartz. 1996. Identification of nuclear and cytoplasmic proteins that interact specifically with an AU-rich, cis-acting inhibitory sequence in the 3′ untranslated region of human papillomavirus type 1 late mRNAs. J. Virol. 70:3659-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng, Z. M., P. He, and C. C. Baker. 1996. Selection of the bovine papillomavirus type 1 nucleotide 3225 3′ splice site is regulated through an exonic splicing enhancer and its juxtaposed exonic splicing suppressor. J. Virol. 70:4691-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng, Z. M., M. Huynen, and C. C. Baker. 1998. A pyrimidine-rich exonic splicing suppressor binds multiple RNA splicing factors and inhibits spliceosome assembly. Proc. Natl. Acad. Sci. USA 95:14088-14093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng, Z. M., E. S. Reid, and C. C. Baker. 2000. Utilization of the bovine papillomavirus type 1 late-stage-specific nucleotide 3605 3′ splice site is modulated by a novel exonic bipartite regulator but not by an intronic purine-rich element. J. Virol. 74:10612-10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou, J., W. J. Liu, S. W. Peng, X. Y. Sun, and I. Frazer. 1999. Papillomavirus capsid protein expression level depends on the match between codon usage and tRNA availability. J. Virol. 73:4972-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.zur Hausen, H. 1999. Papillomaviruses in human cancers. Proc. Assoc. Am. Physicians 111:581-587. [DOI] [PubMed] [Google Scholar]