Abstract
Transcription represents a crucial step in the life cycle of human immunodeficiency virus (HIV) and is highly regulated. Here we show that the strength of the viral long terminal repeat (LTR) promoter is optimized for efficient replication. Artificially increasing the rate of LTR-driven transcription was strongly detrimental for viral fitness, and HIV was able to regain replication capacity by selecting for variants with a weaker LTR. Strikingly, the strength of the evolved promoter was equivalent to that of the wild-type LTR.
The long terminal repeat (LTR) promoter of human immunodeficiency virus (HIV) represents a most interesting example of molecular adaptation of a virus to the host cellular environment. The activity of the promoter is dependent on the viral transactivator Tat (3, 8) and a variety of cellular transcription factors that bind the LTR in a cell-specific and cell activation-dependent manner (7, 12). The overall architecture of the promoter is remarkably conserved among different viral isolates, but some degree of variation in the number and type of binding sites is evident in the different HIV subtypes (11). These variations, although small, modulate virus replication in a cell type-specific manner (9). The transcriptional strength of the LTR could be easily increased by duplication of binding sites, yet such viruses are generally not observed. However, defective viruses can regain replication competence by duplication of existing binding sites (4). This body of evidence implies that transcription of the HIV LTR is optimized for the needs of viral replication and that increased transcription rates are not beneficial, unless in the context of a replication-impaired mutant.
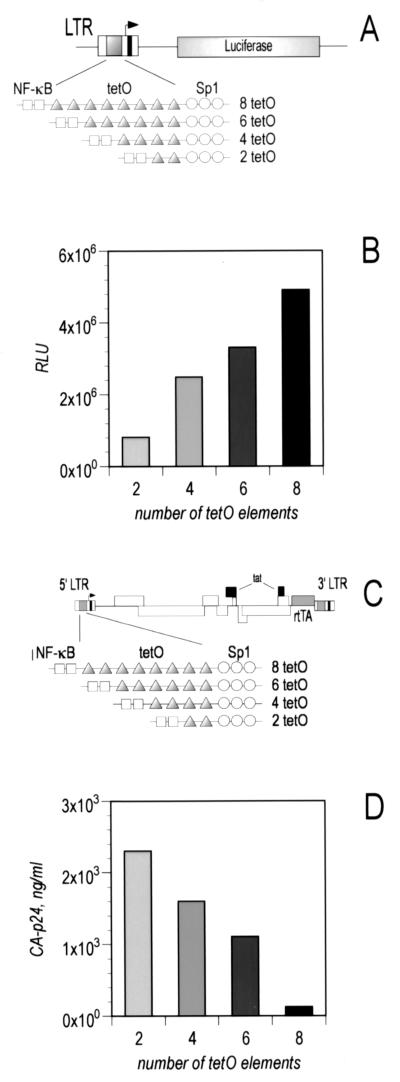
In order to determine the relationship between optimal HIV replication and transcription efficiency, we constructed a panel of LTR promoters with graded transcriptional activities. This was achieved by inserting two, four, six, or eight binding sites for the rtTA protein derived from the Tet system (2) in the LTR. RtTA is a chimeric protein containing an Escherichia coli-derived DNA binding domain and a VP16-derived transactivating domain. RtTA was chosen for its ability to activate viral transcription independently of the cellular environment and in a strictly doxycycline (dox)-dependent fashion. Dox is necessary to induce DNA binding of rtTA and was added in all the experiments described below at the optimal concentration (1,000 ng/ml) (Sigma). The binding sites for rtTA, termed tetO elements, were cloned in between the NF-κB and the SP1 sites, as described in detail in reference 17. The resulting LTRs were placed in front of a luciferase reporter gene (Fig. 1A) and tested in transient-transfection assays. After electroporation (1) of SupT1 T cells with the indicated reporter plasmids (5 μg) and the pCMV-rtTA expression vector (1 μg), we found that the transcription rate of the LTRs was directly proportional to the number of the rtTA binding sites. The following ranking order was observed: LTR of eight tetO (designated 8 tetO) > 6 tetO > 4 tetO > 2 tetO (Fig. 1B). This was not entirely unexpected, as in many other experimental settings multiple binding sites for a transcriptional activator are commonly used when stronger transcription is required (2). These results were not dependent on the amount of rtTA protein available, as we observed an identical pattern of luciferase activity when we cotransfected much larger or smaller amounts of rtTA-expression vector (results not shown).
FIG. 1.
Transcriptional activity driven by a set of dox-dependent HIV LTR promoters. (A) Schematic of the LTR-luc vectors used in the experiment. The inactivated TAR sequence is indicated by a black rectangle. (B) SupT1 T cells were transfected with the indicated molecular clones and the rtTA expression vector. Luciferase activity of the SupT1 cell lysates was measured on day 3 posttransfection. RLU, relative luciferase units. (C) Schematic of the viruses used in the experiment. Indicated in gray are the components of the Tet-system (tetO elements and rtTA) and in black the inactivated TAR and Tat elements. (D) SupT1 cells were infected with the indicated viruses, and viral replication was monitored by measuring production of CA-p24 protein in the supernatant by enzyme-linked immunosorbent assay (day 3 posttransfection).
We have recently demonstrated that such a dox-dependent LTR can support HIV replication when inserted in the viral genome together with the coding sequence for rtTA (17). However, we also observed that the virus with eight tetO elements underwent a drastic promoter rearrangement upon prolonged culture. The evolved virus was characterized by the loss of six of the original eight binding sites, and often the spacer region between the remaining two tetO motifs showed a deletion, which we termed 2Δ tetO (10). We have since repeated this evolution experiment several times with virus constructs containing eight and six tetO motifs. The severe truncation to two or three tetO motifs is consistently observed, without the appearance of intermediate forms (results not shown). These virus replication results are surprising, as the 8 tetO promoter configuration showed greater transcriptional strength than did its shorter counterparts (Fig. 1B). We therefore decided to study the activities of these promoters in the context of the replicating virus.
We used a set of HIVLAI-derived infectious molecular clones containing each of the dox-dependent LTRs, as previously described (10). These viruses differ only in the number of tetO elements present in the 3′ LTR, which will be inherited in the 5′ LTR after reverse transcription. RtTA was inserted in the viral genome in place of the nef gene, and both Tat and its RNA binding site, TAR, were inactivated (Fig. 1C). It was recently demonstrated that these viruses are replication competent and that replication is completely dox dependent (10). Infectious supernatants of each molecular clone in C33A cells were prepared as previously described (5). SupT1 T cells were infected (50 ng of CA-p24) and subsequently washed to remove unbound virus. We monitored HIV-rtTA gene expression by measuring the production of the CA-p24 protein in the supernatant (9). In striking contrast to the luciferase data, the virus with the highest number of tetO elements replicated the worst, and the apparent fitness ranking observed in Fig. 1B was reversed, with HIV-rtTA 2 tetO producing the highest values of CA-p24 protein (Fig. 1D).
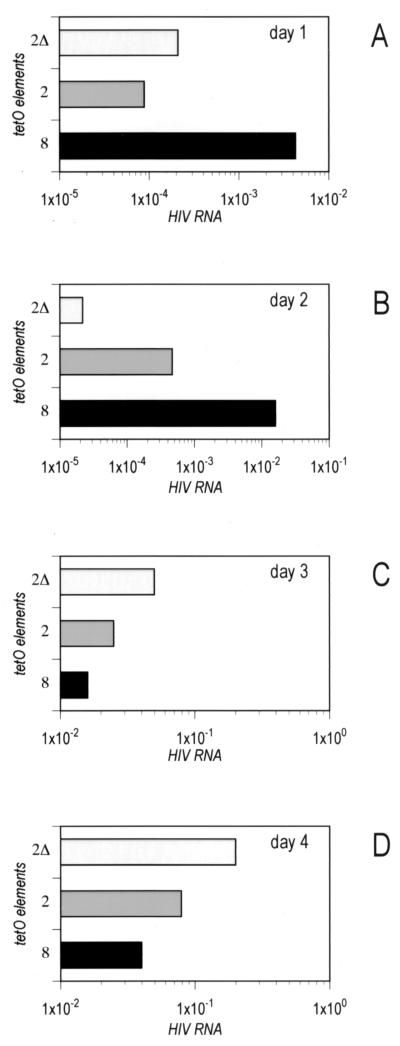
In order to explain the apparent paradox of the strongest promoter resulting in the lowest level of gene expression in the context of the whole viral genome, we set out to measure directly the amount of HIV RNA produced by the LTR in infected T cells. This experimental model also allowed us to study the promoter in its natural configuration, where it is integrated into the host cell genome. We infected SupT1 cells with viruses containing the 8, 2, and evolved 2Δ tetO LTR (50 ng of CA-p24). We extracted RNA from cell samples that were taken at regular intervals, and the total amount of HIV RNA present in each sample was determined by quantitative nucleic acid sequence-based amplification (NASBA) as previously described (6). Values were standardized for extraction efficiency and amount of input RNA by using the cellular snRNP U1A mRNA as an internal standard. To focus on the differences between the virus constructs, we plotted the results for each sampling day in a separate graph, each with its own scale to highlight these differences. The results for the sample taken on day 1 (Fig. 2A) reflected those we had observed in the luciferase assay (Fig. 1B). The promoter with eight tetO was the most transcriptionally active, resulting in a 10-fold higher production of viral RNA than that observed with the 2 tetO promoter. The following ranking order was observed: 8 tetO > 2Δ tetO > 2 tetO. At day 2, the 8 tetO construct remained the most active transcriptional promoter (Fig. 2B). However, the picture changed drastically at a later time point, when the RNA values in the samples were mainly determined by the contribution of virus replication. On day 3 (Fig. 2C), the virus with the 2Δ tetO promoter showed the best replication capacity, closely followed by 2 tetO, while the virus with the 8 tetO promoter resulted in the lowest production of viral RNA. The same results were observed on day 4 (Fig. 2D), with severalfold-higher values due to virus spread in the cultures. These results suggested that higher transcription levels are detrimental for the virus, which over time gets rid of excessive transcriptional power by reducing the number of operators in the promoter from 8 to 2.
FIG. 2.
RNA production versus virus replication. SupT1 cells were infected with the indicated HIV-rtTA viruses containing either eight or two tetO elements, or the evolved 2Δ tetO version. RNA was extracted at day 1 (A), day 2 (B), day 3 (C), and day 4 (D), and HIV RNA was quantitated by NASBA. All values were standardized using the cellular snRNP U1A mRNA as an internal control.
It seems theoretically possible that the observed promoter shortening is due to an RNA packaging problem of the lengthy RNA genome of the HIV-rtTA 8 tetO virus construct. Alternatively, extended genomes may put an extra burden on the viral replication cycle in terms of the time that is required to complete the process of reverse transcription. The RNA genome of HIV rtTA 8 tetO is 9,875 nucleotides (nt), compared to 9,229 nt for the wild-type HIV-1LAI isolate. The loss of six tetO motifs results in a shortening of the genome by 6 × 42 = 252 nt, which is only 2.6% of the genome length. We think that this relatively minor reverse transcription burden is not responsible for the observed promoter trimming because the 8 tetO-to-2 tetO evolution takes place within one or two viral passages. The packaging problem is also less likely to be responsible for the observed evolution because studies with the Moloney murine leukemia virus indicate that RNA genomes with an extension of up to 11 kb can be packaged and that there is no strict upper limit for the genome size (15). Furthermore, the subsequent evolution event that we observed is the deletion of just 14 nt in the 2Δ tetO spacer region, which does not significantly change the genome size. We therefore think that it is unlikely that matters related to the viral genome size played a major role in the observed virus evolution. Consistent with this idea, we have in fact observed further evolution and lengthening of the 2Δ promoter towards a 3Δ configuration in several long-term infections (results not shown).
It could also be argued that the reduction in number of tetO motifs is beneficial for the virus because it down-modulates the expression of the exogenous and potentially toxic rtTA protein. To investigate this issue, we performed several fluorescence-activated cell sorter analyses on SupT1 cells that were infected by the different HIV-rtTA viruses. We were not able to detect any difference in terms of cell proliferation, cell cycle progression, or apoptosis (results not shown). Thus, it seems that the tetO adjustment is key for virus replication but its benefits are not due to avoiding rtTA-mediated toxicity problems.
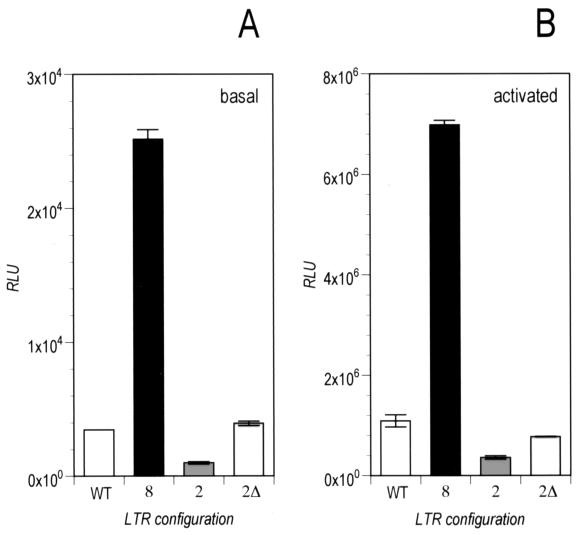
It remained to be explained why 2 tetO and especially 2Δ tetO are optimal for virus replication. By means of transient-transfection assays in SupT1 cells, we compared the transcription rates of the wild-type HIV-1 LTR with those of the original 8 tetO construct and the in vitro-selected 2 tetO and 2Δ tetO promoters (5 μg each). As shown in Fig. 3, the 8 tetO LTR was the transcriptionally strongest configuration and its strength exceeded by far that of the wild-type LTR. This was true for the basal (Fig. 3A) as well as activated transcription upon cotransfection (2 μg) of the Tat and rtTA expression vectors (Fig. 3B). Transcription driven by the 2 tetO promoter appeared to be very much reduced. Furthermore, luciferase activity generated by the evolved 2Δ tetO promoter was consistently higher than that of 2 tetO and remarkably similar to that of wild-type HIV. These results are also in good agreement with the RNA data obtained from the day 1 sample (Fig. 2 A). We are currently investigating why the observed deletion in the spacer between the remaining two tetO motifs improves transcription. This deletion appeared also in a HIV-rtTA virus with a NF-κB-less promoter (results not shown), thus excluding the possibility that the observed increase in transcription may result from a more favorable positioning of the NF-κB binding sites in the LTR. Possible explanations include the removal of a negative element (13), the requirements for cooperative DNA binding of the rtTA protein, or constraints on the positioning of the VP-16 activation domain for interaction with the cellular transcriptional machinery (14).
FIG. 3.
Basal and activated promoter activity: fine-tuning of the LTR transcriptional rate. Sup-T1 cells were transfected with a reporter plasmid containing the luciferase gene downstream of the wild-type(WT) LTR or the artificial LTRs with the indicated number of tetO elements. Luciferase activity was measured on day 3 posttransfection, and data represent the averages of three independent experiments. (A) Basal transcription. (B) Transcription activated by rtTA (2Δ tetO, 2 tetO, 8 tetO LTR: pCMV-rtTA) or Tat (WT LTR: pcDNA3Tat).
Our results indicate that HIV requires specific and fine-tuned levels of transcription and that the strength of the wild-type LTR promoter represents an evolutionary optimum in terms of viral fitness. This may also explain the difficulty in other attempts to generate replication-competent retroviruses dependent on the Tet system (16, 18). Although able to generate high transcription levels driven by LTR promoters with seven tetO elements, these viruses showed a pronounced replication defect. On the contrary, our optimized virus replicates well even in primary cells (10). Our observations argue that transcription is part of a tightly coordinated viral gene expression program that includes mRNA splicing, nuclear export, translation in the cytoplasm, virion assembly, and budding. Too-high transcription rates may disturb this well-balanced process. Furthermore, cellular cofactors are needed at multiple stages of the virus life cycle. It is conceivable that overactivation of HIV transcription may lead to exhaustion of one or more factors present in limiting amounts, resulting in a decrease in infectivity.
Acknowledgments
We thank W. A. Paxton for critical reading of the manuscript. We are grateful to H. Bujard (University of Heidelberg) for providing several reagents of the Tet-system, including the pCMV-rtTA plasmid. We thank M. van Dooren for assistance in the NASBA assays.
G.M. and K.V. are supported by HFSP and EMBO fellowships, respectively.
REFERENCES
- 1.Back, N. K., M. Nijhuis, W. Keulen, C. A. Boucher, B. O. Oude Essink, A. B. van Kuilenburg, A. H. van Gennip, and B. Berkhout. 1996. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 15:4040-4049. [PMC free article] [PubMed] [Google Scholar]
- 2.Baron, U., and H. Bujard. 2000. Tet repressor-based system for regulated gene expression in eukaryotic cells: principles and advances. Methods Enzymol. 327:401-421. [DOI] [PubMed] [Google Scholar]
- 3.Berkhout, B., R. H. Silverman, and K. T. Jeang. 1989. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell 59:273-282. [DOI] [PubMed] [Google Scholar]
- 4.Berkhout, B., K. Verhoef, J. L. van Wamel, and N. K. Back. 1999. Genetic instability of live, attenuated human immunodeficiency virus type 1 vaccine strains. J. Virol. 73:1138-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das, A. T., A. Land, I. Braakman, B. Klaver, and B. Berkhout. 1999. HIV-1 evolves into a nonsyncytium-inducing virus upon prolonged culture in vitro. Virology 263:55-69. [DOI] [PubMed] [Google Scholar]
- 6.de Baar, M. P., M. W. van Dooren, E. de Rooij, M. Bakker, B. van Gemen, J. Goudsmit, and A. de Ronde. 2001. Single rapid real-time monitored isothermal RNA amplification assay for quantification of human immunodeficiency virus type 1 isolates from groups M, N, and O. J. Clin. Microbiol. 39:1378-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaynor, R. 1992. Cellular transcription factors involved in the regulation of HIV-1 gene expression. AIDS 6:347-363. [DOI] [PubMed] [Google Scholar]
- 8.Jeang, K. T., H. Xiao, and E. A. Rich. 1999. Multifaceted activities of the HIV-1 transactivator of transcription, Tat. J. Biol. Chem. 274:28837-28840. [DOI] [PubMed] [Google Scholar]
- 9.Jeeninga, R. E., M. Hoogenkamp, M. Armand-Ugon, M. de Baar, K. Verhoef, and B. Berkhout. 2000. Functional differences between the long terminal repeat transcriptional promoters of human immunodeficiency virus type 1 subtypes A through G. J. Virol. 74:3740-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marzio, G., K. Verhoef, M. Vink, and B. Berkhout. 2001. In vitro evolution of a highly replicating, doxycycline-dependent HIV for applications in vaccine studies. Proc. Natl. Acad. Sci. USA 98:6342-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCutchan, F. E. 2000. Understanding the genetic diversity of HIV-1. AIDS 14:S31-S44. [PubMed] [Google Scholar]
- 12.Pereira, L. A., K. Bentley, A. Peeters, M. J. Churchill, and N. J. Deacon. 2000. A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res. 28:663-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rang, A., and H. Will. 2000. The tetracycline-responsive promoter contains functional interferon-inducible response elements. Nucleic Acids Res. 28:1120-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ross, E. D., A. M. Keating, and L. J. Maher III. 2000. DNA constraints on transcription activation in vitro. J. Mol. Biol. 297:321-334. [DOI] [PubMed] [Google Scholar]
- 15.Shin, N. H., D. Hartigan-O'Connor, J. K. Pfeiffer, and A. Telesnitsky. 2000. Replication of lengthened Moloney murine leukemia virus genomes is impaired at multiple stages. J. Virol. 74:2694-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith, S. M., M. Khoroshev, P. A. Marx, J. Orenstein, and K. T. Jeang. 2001. Constitutively-dead, conditionally-live HIV-1 genomes: ex vivo implications for a live-virus vaccine. J. Biol. Chem. 7:7.. [DOI] [PubMed] [Google Scholar]
- 17.Verhoef, K., G. Marzio, W. Hillen, H. Bujard, and B. Berkhout. 2001. Strict control of human immunodeficiency virus type 1 replication by a genetic switch: Tet for Tat. J. Virol. 75:979-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao, Y., T. Kuwata, T. Miura, M. Hayami, and H. Shida. 2000. Dox-dependent SIVmac with tetracycline-inducible promoter in the U3 promoter region. Virology 269:268-275. [DOI] [PubMed] [Google Scholar]