Abstract
Some retroviruses contain monoubiquitinated Gag and do not bud efficiently from cells treated with proteasome inhibitors, suggesting an interaction between the ubiquitin-proteasome system and retrovirus assembly. We examined equine infectious anemia virus (EIAV) particles and found that approximately 2% of the p9Gag proteins are monoubiquitinated, demonstrating that this Gag protein interacts with an ubiquitinating activity. Different types of proteasome inhibitors were used to determine if proteasome inactivation affects EIAV release from chronically infected cells. Pulse-chase immunoprecipitation and time course immunoblot analyses showed that proteasome inactivation slightly decreased virus release (at most a twofold effect), while it did not affect Gag processing. These results contrast with those obtained with other viruses which are sensitive to these inhibitors. This suggests that, although its Gag is monoubiquitinated, the requirements for EIAV release are somewhat different from those for retroviruses that are sensitive to proteasome inhibitors.
Lentiviruses and type C retroviruses assemble in association with the host plasma membrane, forming a bud that is released from the cell to produce a virion (46). The late assembly domain (L) within Gag is crucial for the efficient release of the budding virus from the plasma membrane (42). Three different sequences have been shown to possess L domain function: PPPY, found in Rous sarcoma virus (RSV) (51, 52), murine leukemia virus (MuLV), (54), and Mason-Pfizer monkey virus (53); PTAP, found in human immunodeficiency virus type 1 (HIV-1) (presumably P[T/S]AP for HIV-2 and simian immunodeficiency virus [SIV]) (11, 18); and YPDL, found in equine infectious anemia virus (EIAV) (34). Deletion or replacement of these sequences causes virions to mostly remain attached to the plasma membrane by a thin tether and to fail to separate from the cell. These L domain sequences can interact directly with cellular proteins (8, 9, 12, 13, 19, 35, 45), suggesting potential cellular partners for virus budding. Despite these findings, the pathway(s) used by retroviruses for budding is mostly unknown, though recent results suggest that components of the vacuolar protein sorting pathway might be used by HIV-1 (9). Experiments with several retroviruses have shown that Gag interacts with the ubiquitination pathway and that efficient budding requires active proteasomes (47). Here we examine EIAV for interactions with the ubiquitin (Ub)-proteasome system.
EIAV particles contain free Ub and Ub-Gag conjugates.
For several retroviruses, the mature protein within Gag that contains the L domain, p6Gag in HIV-1 and SIV and p12Gag in MuLV, is also monoubiquitinated (27). HIV-1 Pr55Gag can be monoubiquitinated within the p6Gag region, consistent with Gag being modified during assembly (26). The significance of Gag monoubiquitination is not clear. The best-known role for Ub is as the basic monomer in the formation of polyubiquitin, where Ub itself is ubiquitinated to form a polymeric chain. Ubiquitination can be a rapidly reversible process that is regulated by a complex pathway of ubiquitinating and deubiquitinating enzymes (5, 49, 50). A chain at least four molecules long is sufficient as a signal for degradation of the conjugated protein by the 26S proteasome (15, 17, 20, 21, 43). In contrast, it appears that monoubiquitination is mostly involved in cellular processes other than degradation, including endocytosis and histone-mediated transcriptional regulation (16).
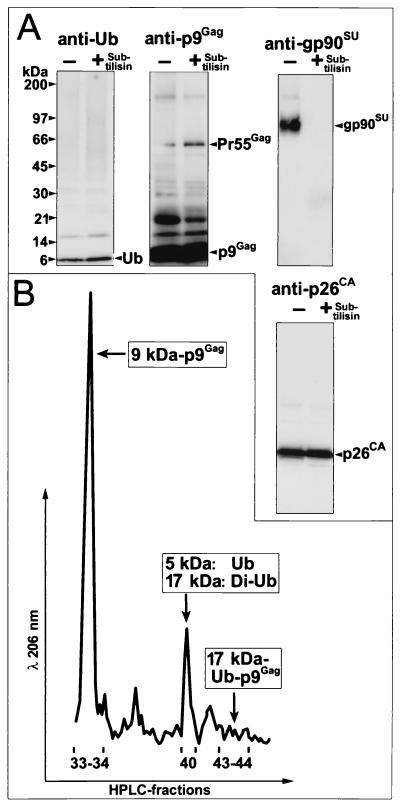
To better understand the interaction of lentiviruses with the ubiquitination system, we examined the proteins inside particles of EIAV, another member of this genus, for the presence of free Ub and Ub-Gag conjugates. Since even highly purified virus preparations can contain microvesicles, i.e., protein-containing membranous particles (2, 10), we digested a sucrose density-purified preparation of EIAV (produced from EIAVWyoming-infected Cf2th cells) with subtilisin as previously described (29). This protease treatment removes proteins outside the virus, including greater than 95% of the contaminating proteins that are associated with microvesicles. Removal of these proteins allows for the detection and characterization of the proteins that are inside the virions (28-30). Immunoblot detection of proteins was carried out as previously described (27) with a Ub monoclonal antibody, clone 2C5 (PanVera Corporation, Madison, Wis.); EIAV p15MA- and p26CA-reactive goat serum (AIDS Vaccine Program, National Cancer Institute [NCI]-Frederick); or EIAV p9Gag and gp90SU rabbit antiserum (Advanced Biosciences, Basic Research Program, NCI-Frederick). A Ub immunoblot of 20 μg (determined by the Lowry method [24]) of a purified EIAV virus preparation digested either with or without subtilisin showed that the majority of the free Ub (present as a 5-kDa band) remained in the virion samples after subtilisin digestion (Fig. 1A), thus protected from the protease. Immunoblotting the samples with EIAV gp90SU antiserum demonstrated that this exterior protein was removed by the subtilisin treatment as expected (Fig. 1A), confirming that the proteins on the surface of the virus were removed. The amounts of p9Gag and p26CA were not altered by the digestion procedure, as revealed by immunoblot analysis, showing that the treatment did not digest the interior virion proteins (Fig. 1A). Together, these results show that free Ub is present inside EIAV particles.
FIG. 1.
Analysis of EIAV virions. (A) Immunoblots of EIAV virions digested in the absence (−) or presence (+) of subtilisin. The antibody or antiserum used is indicated above each blot. Molecular mass markers are indicated at the left, and bands are identified at the right. (B) High-pressure liquid chromatography chromatogram of the region containing Ub and Ub-p9Gag proteins. Fractions are identified under the A206 tracing, with the identities of the sequenced proteins indicated next to their respective peaks.
In addition to free Ub, there was a band at 17 kDa. Stripping and reprobing this blot with p9Gag antiserum also revealed a band at 17 kDa, suggesting the presence of monoubiquitinated p9Gag (Fig. 1A). Other prominent bands present in the p9Gag blot were at 20 kDa, likely p11NC-p9Gag, and at 55 kDa, the Pr55Gag precursor.
Based on our previous experience, the search for ubiquitinated Gag proteins is complicated by the relatively small amounts of this species inside virions and the presence of multiple Ub conjugates (26, 27) (data not shown). To isolate and quantitate ubiquitinated Gag species and free Ub inside EIAV virions, 200 mg of a virus preparation (total protein by the Lowry method) was treated with subtilisin and subjected to reversed-phase high-pressure liquid chromatography as previously described (14). Samples of the fractions containing protein peaks were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with Coomassie brilliant blue staining and by immunoblotting. Fractions containing proteins of interest were blotted onto filters, and the bands were excised and subjected to protein sequence analysis on an Applied Biosystems (Foster City, Calif.) Procise model 494 microsequencer as previously described (14). The results showed that fraction 40 contained most of the free Ub in the preparation (Fig. 1B and data not shown). Based on the relative absorbance of the free-Ub peak versus those of the p9Gag and p26CA peaks, we estimate that free Ub is present in the virion at 10% of the number of Gag molecules, or 200 molecules based on 2,000 Gag proteins per virion (22, 32, 41, 48).
In addition to free Ub, we detected a 17-kDa species that reacted with both the Ub antibody and p9Gag antiserum in fractions 43 and 44 (Fig. 1B and data not shown). Protein sequencing of this isolated band produced an equimolar sequence of both p9Gag (PIQQKSQHNKSVVQE) and Ub (MQIFVKTLTGKTITL). The apparent 17-kDa molecular mass, the immunoreactivity of the protein, and the stoichiometry of the two sequences demonstrate that this band is a p9Gag-Ub conjugate. Conjugation of Ub to proteins is accomplished by the formation of an isopeptide bond between the C-terminal carboxyl group of Ub and the ɛ-amino group of a lysine in the target protein (reviewed in references 3 and 7). The presence of lysines 5 and 10 in the p9Gag sequence of this conjugate indicates that these residues are not ubiquitinated, since modified lysines are not detected by Edman degradation sequencing (27). Therefore, this protein is monoubiquitinated on one of the four C-terminal lysines (at positions 30, 31, 36, and 38) in p9Gag. These residues are too distal to be detected by N-terminal sequencing, given the small amount of protein available. Based on the relative absorbance of the peaks containing Ub-p9Gag and other Gag peaks, approximately 2% of the p9Gag proteins (40 molecules) are monoubiquitinated, similar to the numbers of modified Gag proteins found in HIV-1, SIV, and MuLV (27). Together, these results show that EIAV Gag interacts with an ubiquitinating activity.
Effect of various proteasome inhibitors on EIAV.
Recently, efficient release of HIV-1 and -2, SIV, and RSV from cells has been shown to require proteasome activity, i.e., treatment of virus-producing cells with proteasome inhibitors reduces viral budding (31, 39, 40). For HIV-1 and -2, proteasome inhibition also decreased the viral protease-mediated processing of Gag into its mature proteins (39), a step required for the production of an infectious virus (42). However, this decrease in Gag processing was not a result of a direct inhibition of HIV-1 protease activity by the proteasome inhibitors themselves (39). The mechanism for the effects of proteasome inhibitors on lentiviruses has not been determined.
Proteasome inhibitors can affect many cellular processes, potentially making experimental results difficult to interpret. We have previously optimized the treatment of cells with proteasome inhibitors (10 μM zLLL [N-carbobenzoxyl-l-leucinyl-l-leucinyl-leucinal] and lactacystin) to avoid unwanted effects on translation and maintain their integrity during these experiments while completely inhibiting proteasome activity in different cell types from different species (37, 38; U. Schubert, unpublished data). Therefore, we chose these established conditions to examine the effect of proteasome inhibition on EIAV budding and Gag processing by using the same pulse-chase procedure as before (38, 39) with various types of inhibitors that block proteasome activity by different mechanisms: lactacystin (Calbiochem-Novabiochem Corp, San Diego, Calif.), PS-341 (Millennium Pharmaceuticals, Cambridge, Mass.), epoxomicin (Affiniti Research Products Ltd., Mamhead, Exeter, Devon, United Kingdom), and zLLL (MG-132; Calbiochem-Novabiochem Corp.). Lactacystin is a highly specific and potent inhibitor, as its metabolic product, β-lactone, irreversibly alkylates the catalytic threonine of the 26S proteasome β subunit (4, 6). PS-341, a dipeptide boronic acid derivative, is a reversible inhibitor that tightly interacts with the hydroxyl threonine in position 1 of the catalytic β subunit of the proteasome (Ki = 0.6 nM) (1). We also tested epoxomicin, a natural epoxy-ketone-containing peptoid that is a specific, irreversible inhibitor of the chymotrypsin-like activity of the proteasome in various cell types with a molar efficiency comparable to that of lactacystin (25, 33) and that has been shown to block HIV-1 assembly (39). The least-specific proteasome inhibitor used was zLLL, which reversibly inhibits proteasomes (23). Whereas this compound forms a transient hemiacetal with the catalytic threonine of the proteasome, it also inhibits other cellular enzymes. As a negative control for this nonspecific inhibitor, we also used zLL, a dipeptide analogue of zLLL that inhibits a similar spectrum of thiol proteases but that affects neither the proteasome (44) nor HIV assembly (39).
Pulse-chase experiments were carried out as follows. Parallel inhibitor-treated and untreated cultures of EIAV-infected Cf2th cells were methionine starved for 30 min before being metabolically labeled during a 30-min pulse of [35S]methionine (2 mCi/ml; Du Pont Inc., Boston, Mass.), which was followed by up to an 8-h chase with an excess of methionine. The chase started with the removal of the labeling medium followed by a single wash with Dulbecco's phosphate-buffered saline (PBS; Life Technologies Inc., Gaithersburg, Md.); then the cells were divided into equal aliquots with 1 ml of prewarmed RPMI 1640 with fetal bovine serum containing 1 mM methionine. Proteasome inhibitors were added during the methionine starvation period, and this treatment was continued throughout the experiment. Cells were lysed in a buffer containing 50 mM Tris-hydrochloride (pH 8.0), 5 mM EDTA, 100 mM NaCl, 0.5% (wt/vol) CHAPS {[3-(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, and 0.2% (wt/vol) deoxycholate. At various times during the chase, samples of these cells, virus fractions (material pelleted from cell-free supernatants in a refrigerated Eppendorf Microfuge [4°C, 100 min, 16,000 × g] and lysed in a buffer containing 300 mM NaCl, 50 mM Tris-hydrochloride [pH 7.4], and 0.1% [vol/vol] Triton X-100 by shaking at room temperature for 20 min), and clarified supernatant fractions were subjected to immunoprecipitation with EIAV-specific antisera (a cocktail of goat 15MA and p26CA antisera and rabbit p9Gag antiserum preadsorbed to protein G-Sepharose) and analyzed by SDS-PAGE and fluorography.
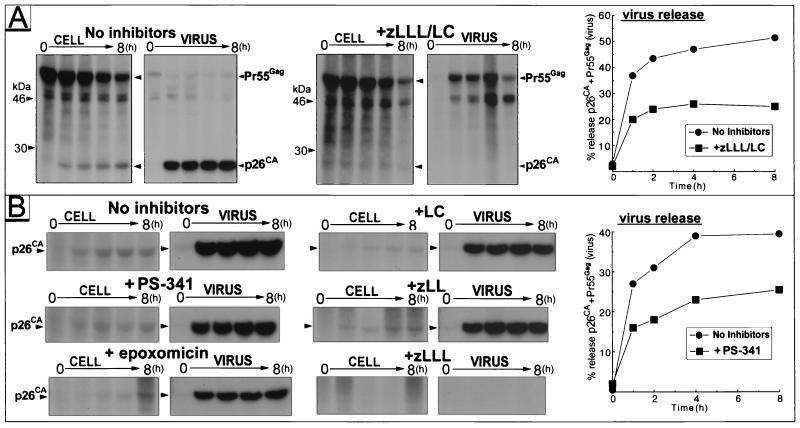
Since we previously have shown that the budding and processing of HIV-1 and HIV-2 are efficiently blocked by a combination of zLLL and lactacystin in various cell types (38, 39), we examined the effect of this treatment on EIAV produced from chronically infected Cf2th by pulse-chase analysis (Fig. 2A). Comparable amounts of radiolabeled Pr55Gag were recovered from treated-cell and control cell lysates at the end of pulse-labeling (the zero time point in Fig. 2A), confirming that the 60-min zLLL-lactacystin treatment during the starvation and pulse-labeling steps did not affect protein synthesis in the EIAV-expressing cells, similar to our previous results with other viruses and cell lines (37-39). The relative amounts of Gag precursor Pr55Gag and its major processing product p26CA were quantitated with a PhosphorImager and used to calculate the percentage of particle-associated Gag proteins (i.e., virus released) in the total number of Gag proteins (i.e., that in the cell, virus, and supernatant fractions) at the various time points during the chase (Fig. 2B). We observed a subtle, approximately twofold reduction of EIAV release in the presence of zLLL-lactacystin during the 8-h chase. Most strikingly, this treatment nearly eliminated the processing of Pr55Gag into p26CA as evidenced by an up-to-32-fold-lower ratio of p26CA to Pr55Gag observed in virions released from treated culture at the end of the chase period (Fig. 2A). Partial Gag processing did occur, as a 46-kDa processing intermediate accumulated in virion samples from the treated cells. This severe inhibition of Gag processing and the subtle decrease in virus release for EIAV are quite different from the effect of proteasome inhibition on HIV-1 and -2, where we observed a three- to sixfold decrease in both virus release and Gag processing under nearly identical experimental conditions.
FIG. 2.
Effect of proteasome inhibitors on EIAV release and Gag processing. (A) (Left) Fluorograms of SDS-PAGE gels of immunoprecipitates from cell or virus lysates produced from EIAV-infected cells cultured in the absence or presence of zLLL and lactacystin (LC) (10 μM each). The treatments used and sample identities are indicated. Sample time points: 0, 1, 2, 4, and 8 h. (Right) Graph of virus released in the presence or absence of zLLL and lactacystin. (B) (Left) Fluorograms of SDS-PAGE gels of immunoprecipitates from cell and virus lysates from EIAV-infected cells cultured in the absence or presence of PS-341 (5 μM), epoxomicin (10 μM), lactacystin (20 μM), zLL (20 μM), and zLLL (20 μM). Labeling is as in panel A. (Right) Graph of virus released in the presence or absence of PS-341.
To investigate this effect further, parallel pulse-chase experiments were conducted with EIAV-infected cells that were treated individually with various proteasome inhibitors. The pulse-chase experiments and PhosphorImager analysis revealed that treatment of EIAV-infected cells with 5 μM PS-341 (a highly specific inhibitor) had a subtle, approximately twofold effect on particle release (Fig. 2B). Similar release kinetics, a decrease of at most twofold in virus release, were produced in parallel experiments with 10 μM epoxomicin and 20 μM lactacystin (Fig. 2B and data not shown). For these inhibitors, no processing defect was observed, unlike our previous results with HIV-1 and -2 (39) and the strong processing block of EIAV Pr55Gag with the zLLL-lactacystin combination (Fig. 2B). In contrast, the presence of 20 μM zLLL strongly inhibited Pr55Gag processing and resulted in the accumulation of Pr55Gag (data not shown) and the absence of p26CA in both the cell and virus samples (Fig. 2B), similar to the data from the zLLL-lactacystin combination experiments (Fig. 2A). Therefore, only treatments that contained zLLL blocked Gag processing, suggesting that zLLL itself, and not proteasome inactivation per se, is responsible for this decrease in EAIV Gag processing. However, treatment with zLLL still produced the same subtle effect on virus release (data not shown) as that observed for the other single proteasome inhibitors and for the zLLL-lactacystin combination (Fig. 2A). Treatment with the control inhibitor, zLL, had little if any effect on virus release and processing (Fig. 2B and data not shown), showing that the effect on virus processing is specific for zLLL.
To confirm these pulse-chase results, we used time course immunoblots as an alternate method of examining virus release and Gag processing. For time course immunoblot analysis, identical flasks of EIAV-infected cells were washed three times with PBS and then the experimental medium was added. Proteasome inhibitors were used at the following concentrations: 20 μM zLLL and lactacystin (both from Biomol, Plymouth Meeting, Pa.), 1 μM PS-341, and 5 μM epoxomicin. At various time points, supernatants and cell samples were removed, prepared, and analyzed by immunoblotting. Virions were isolated by pelleting cell-free supernatant through 20% (wt/vol) sucrose in PBS in an SW41Ti rotor (Beckman-Coulter, Inc., Fullerton, Calif.) at 37,000 rpm and at 4°C for 1 h. Cytoplasmic extracts were prepared by lysing cells in 1% (vol/vol) Triton X-100 in 10 mM Tris, pH 7.5-100 mM NaCl, and the nuclei were removed by centrifugation at 2,500 × g for 5 min at 4°C.
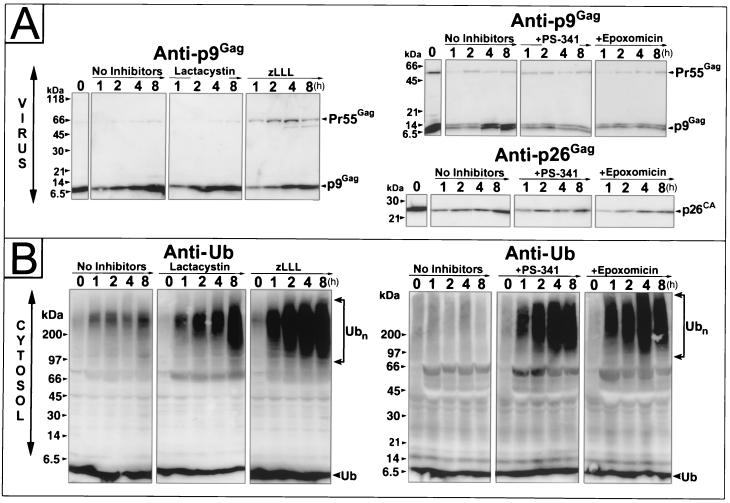
The results of these time course experiments on EIAV-infected cells were consistent with our pulse-chase results (Fig. 3A): lactacystin, PS-341, and epoxomicin treatment of cells had little discernible effect on virus release, and virions produced from treated cells showed no defect in Pr55Gag processing. As seen before, treatment of cells with zLLL alone interfered with Gag processing (Fig. 3A); similar results were observed with the zLLL-lactacystin combination (data not shown).
FIG. 3.
Time course analysis of virus release and cellular Ub levels. (A) Immunoblots of virion samples produced by EIAV-infected cells that were untreated or treated with 20 μM lactacystin, 20 μM zLLL, 1 μM PS-341, or 10 μM epoxomicin. (B) Ub immunoblots of the cell lysates that correspond to the virion samples. The antiserum and antibody used are indicated above each blot, and the samples are labeled above the respective lanes.
The efficacy of proteasome treatment was determined by observing the status of Ub inside the cells during the time course experiments presented above using our monoclonal antibody (the same 2C5 clones used previously) that readily detects free and conjugated Ub (26). Immunoblots of cell lysates (from equal numbers of cells) demonstrated that high-molecular-weight Ub complexes, readily detectable after 1 h of treatment, accumulated rapidly for all of the proteasome inhibitors tested (Fig. 3B), indicating that the inhibitors efficiently blocked proteasome function. However, the levels of free Ub were not dramatically decreased for any of the inhibitors. Experiments with the zLLL-lactacystin combination on these cells and other cell types have shown that proteasome inhibition decreased free-Ub levels by approximately twofold after 8 h (data not shown), reproducing the results presented in Fig. 3B.
EIAV and the Ub-proteasome pathway.
Our data demonstrate that EIAV Gag interacts with the ubiquitination system since it is monoubiquitinated, similar to HIV-1. However, unlike the findings for HIV-1, proteasome inhibition has only a small effect on budding and produces no detectable decrease in Gag processing. The accompanying paper by Patnaik et al. (31a) also notes a similar insensitivity of EIAV budding and Gag processing to proteasome inhibitors in cell types different from ours. Therefore, this particular interaction between EIAV Gag and the ubiquitinating machinery is not reflected in a marked reduction in virus budding by proteasome inhibitors, implying that monoubiquitination of p9Gag is not required for virus release. This is consistent with our previous results demonstrating that the ubiquitination of HIV-1 p6Gag and MuLV p12Gag is not required for replication in cell culture (26). However, those data could not exclude a necessary and reversible ubiquitination in another region of Gag. The finding that RSV uses Ub as part of its budding machinery provides a clear example of a link between the Ub-proteasome pathway and RSV budding (31). However, a requirement for the monoubiquitination of EIAV, HIV-1, SIV, and MuLV Gag in the viral life cycle remains to be elucidated. An alternative explanation is that, for these four viruses, the monoubiquitinated Gag found inside the virions is simply a consequence of Gag interacting with a protein or protein complex that has a ubiquitinating activity during the assembly and budding process (26).
Regardless of whether Ub is directly involved in virus release or is a simple bystander, the monoubiquitination of p9Gag, along with the previously observed modification of HIV-1 and SIV p6Gag and MuLV p12Gag, links all three L domain sequences (YPDL, PTAP, and PPPY, respectively) with a cellular ubiquitinating activity. This is also supported by in vitro and in vivo studies where PPPY has been shown to bind to Ub ligases, especially Nedd 4 (8, 12, 13, 19). The PTAP sequence recently has been shown to bind to Tsg101, a catalytically inactive homologue of a Ub-conjugating enzyme that is involved in the vacuolar protein sorting pathway (9, 45). Experiments with a minimal Gag construct have determined that the different L domain and L domain-like sequences that provide a release function also induce ubiquitination of this small Gag protein, demonstrating that L domains functionally interact with ubiquitinating machinery (40).
In addition to monoubiquitinated p9Gag, we have found free Ub inside the virion. Ub was first found inside avian leukosis virus (36) and was later found inside HIV-1, SIV, and MuLV (27). The amounts of Ub inside these virions are similar for all viruses examined, between 10 to 15% of the molar level of the Gag protein. The mechanism for and significance of Ub incorporation is not known. For HIV-1, the ubiquitination of p6Gag is not required for the presence of free Ub in virions (26).
Our findings, along with the accompanying paper by Patnaik et al. (31a), show that EIAV release is relatively resistant to proteasome inhibitors while Gag processing is completely unaffected, except by zLLL treatment. The effect of zLLL on EIAV Gag processing is likely due to a direct inhibition of zLLL on EIAV protease (L. V. Coren and D. E. Ott, unpublished results). Since this compound also inhibits other cellular enzymes due to its chemical reactivity with threonine residues (23), great care should be exercised when using this rather nonspecific inhibitor. The insensitivity of EIAV release is markedly different from the reduction of virus budding in the presence of proteasome inhibitors observed for HIV-1, HIV-2, SIV, and RSV. The accumulation of high-molecular-weight Ub complexes in treated cells demonstrated that our conditions clearly inhibited proteasomes as expected. Thus, the insensitivity of EIAV to proteasome inhibitors was not due to a failure to effectively perturb proteasome activity.
The twofold decrease in EIAV budding could be consequent to the accumulation of misfolded Gag proteins that are polyubiquitinated. These defective ribosomal product (DRiP) forms have been previously observed for HIV-1 Gag immediately following proteasome inhibition (38). Although preliminary evidence supports the notion that HIV-1 Gag DRiPs can interfere with the folding and processing of viral structural proteins (U. Schubert and D. E. Ott, unpublished results), it remains to be established if a similar mechanism acts on EIAV Gag.
Overall, the relative insensitivity of EIAV to proteasome inhibition suggests that it may use an assembly and budding strategy somewhat different from that of other retroviruses. One difference is described in the accompanying report by Patnaik et al. (31a), which reports that a region of p9Gag, when fused to the C terminus of RSV Gag, can overcome the effect of proteasome inhibitors on RSV Gag release. Perhaps it is this activity that causes EIAV budding to resist the effects of these inhibitors.
Proteasome activity is required for many different cellular processes. The ability of EIAV to release and process Gag in the absence of active proteasomes demonstrates that inhibitor treatment does not necessarily block retrovirus budding. Therefore, the reduction in budding and, in some cases, Gag processing, seen for inhibitor-sensitive retroviruses is specific and not simply due to the pleiotropic effects of the compounds themselves. The understanding of the differing interactions between retroviruses and the ubiquitin-proteasome pathway should help unravel the mystery of retrovirus budding.
ADDENDUM IN PROOF
While this article was in press, Martin-Serrano et al. (J. Martin-Serrano, T. Zang, and P. D. Bieniasz, Nat. Med. 7:1313-1319, 2001) reported that peptides of HIV-1 and Ebola virus that contain the PTAP sequence functionally interact with Tsg101 during budding.
Acknowledgments
We thank Steven Campbell for the bacterially expressed HIV-1 Gag; Nancy Rice for the gp90SU antiserum; Robert Gorelick, Van Hoang, and James Buckman for critical reading of the manuscript; and John Wills and Akash Patnaik for helpful discussions and sharing their results before publication.
This project was funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. NO1-CO-56000. U. Schubert was supported by grant Schu11/2-1, a Heisenberg grant from the Deutsche Forschungsgemeinschaft, and a grant from the German Human Genome Research Project.
REFERENCES
- 1.Adams, J., M. Behnke, S. Chen, A. A. Cruickshank, L. R. Dick, L. Grenier, J. M. Klunder, Y. T. Ma, L. Plamondon, and R. L. Stein. 1998. Potent and selective inhibitors of the proteasome: dipeptidyl boronic acids. Bioorg. Med. Chem. Lett. 8:333-338. [DOI] [PubMed] [Google Scholar]
- 2.Bess, J. W., Jr., R. J. Gorelick, W. J. Bosche, L. E. Henderson, and L. O. Arthur. 1997. Microvesicles are a source of contaminating cellular proteins found in purified HIV-1 preparations. Virology 230:134-144. [DOI] [PubMed] [Google Scholar]
- 3.Ciechanover, A. 1994. The ubiquitin-proteasome proteolytic pathway. Cell 79:13-21. [DOI] [PubMed] [Google Scholar]
- 4.Craiu, A., M. Gaczynska, T. Akopian, C. F. Gramm, G. Fenteany, A. L. Goldberg, and K. L. Rock. 1997. Lactacystin and clasto-lactacystin beta-lactone modify multiple proteasome beta-subunits and inhibit intracellular protein degradation and major histocompatibility complex class I antigen presentation. J. Biol. Chem. 272:13437-13445. [DOI] [PubMed] [Google Scholar]
- 5.D'Andrea, A., and D. Pellman. 1998. Deubiquitinating enzymes: a new class of biological regulators. Crit. Rev. Biochem. Mol. Biol. 33:337-352. [DOI] [PubMed] [Google Scholar]
- 6.Fenteany, G., R. F. Standaert, W. S. Lane, S. Choi, E. J. Corey, and S. L. Schreiber. 1995. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science 268:726-731. [DOI] [PubMed] [Google Scholar]
- 7.Finley, D., and V. Chau. 1991. Ubiquitination. Annu. Rev. Cell Biol. 7:25-69. [DOI] [PubMed] [Google Scholar]
- 8.Garnier, L., J. W. Wills, M. F. Verderame, and M. Sudol. 1996. WW domains and retrovirus budding. Nature (London) 381:744-745. [DOI] [PubMed] [Google Scholar]
- 9.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 10.Gluschankof, P., I. Mondor, H. R. Gelderblom, and Q. J. Sattentau. 1997. Cell membrane vesicles are a major contaminant of gradient-enriched human immunodeficiency virus type-1 preparations. Virology 230:125-133. [DOI] [PubMed] [Google Scholar]
- 11.Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harty, R. N., J. Paragas, M. Sudol, and P. Palese. 1999. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J. Virol. 73:2921-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henderson, L. E., M. A. Bowers, R. C. Sowder II, S. Serabyn, D. G. Johnson, J. W. Bess, Jr., L. O. Arthur, D. K. Bryant, and C. Fenselau. 1992. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processings, and complete amino acid sequences. J. Virol. 66:1856-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 16.Hicke, L. 2001. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2:195-201. [DOI] [PubMed] [Google Scholar]
- 17.Hochstrasser, M. 1996. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30:405-439. [DOI] [PubMed] [Google Scholar]
- 18.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. USA 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kloetzel, P. M. 2001. Antigen processing by the proteasome. Nat. Rev. Mol. Cell Biol. 2:179-187. [DOI] [PubMed] [Google Scholar]
- 21.Laney, J. D., and M. Hochstrasser. 1999. Substrate targeting in the ubiquitin system. Cell 97:427-430. [DOI] [PubMed] [Google Scholar]
- 22.Layne, S. P., M. J. Merges, M. Dembo, J. L. Spouge, S. R. Conley, J. P. Moore, J. L. Raina, H. Renz, H. R. Gelderblom, and P. L. Nara. 1992. Factors underlying spontaneous inactivation and susceptibility to neutralization of human immunodeficiency virus. Virology 189:695-714. [DOI] [PubMed] [Google Scholar]
- 23.Lee, D. H., and A. L. Goldberg. 1998. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8:397-403. [DOI] [PubMed] [Google Scholar]
- 24.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 25.Meng, L., R. Mohan, B. H. Kwok, M. Elofsson, N. Sin, and C. M. Crews. 1999. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc. Natl. Acad. Sci. USA 96:10403-10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, and U. Schubert. 2000. Ubiquitination of HIV-1 and MuLV Gag. Virology 278:111-121. [DOI] [PubMed] [Google Scholar]
- 27.Ott, D. E., L. V. Coren, T. D. Copeland, B. P. Kane, D. G. Johnson, R. C. Sowder II, Y. Yoshinaka, S. Oroszlan, L. O. Arthur, and L. E. Henderson. 1998. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J. Virol. 72:2962-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ott, D. E., L. V. Coren, D. G. Johnson, B. P. Kane, R. C. Sowder II, Y. D. Kim, R. J. Fisher, X. Z. Zhou, K. P. Lu, and L. E. Henderson. 2000. Actin-binding cellular proteins inside human immunodeficiency virus type 1. Virology 266:42-51. [DOI] [PubMed] [Google Scholar]
- 29.Ott, D. E., L. V. Coren, D. G. Johnson, R. C. Sowder II, L. O. Arthur, and L. E. Henderson. 1995. Analysis and localization of cyclophilin A found in the virions of human immunodeficiency virus type 1 MN strain. AIDS Res. Hum. Retroviruses 11:1003-1006. [DOI] [PubMed] [Google Scholar]
- 30.Ott, D. E., L. V. Coren, B. P. Kane, L. K. Busch, D. J. Johnson, R. C. Sowder II, E. N. Chertova, L. O. Arthur, and L. E. Henderson. 1996. Cytoskeletal proteins inside human immunodeficiency virus type 1 virions. J. Virol. 70:7734-7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31a.Patnaik, A., V. Chau, F. Li, R. C. Montelaro, and J. W. Wills. 2002. Budding of equine infectious anemia virus is insensitive to proteasome inhibitors. J. Virol. 76:2641-2647. [DOI] [PMC free article] [PubMed]
- 32.Piatak, M., M. S. Saag, L. C. Yang, S. J. Clark, J. C. Kappes, K.-C. Luk, B. H. Hahn, G. M. Shaw, and J. D. Lifson. 1993. High levels of HIV-1 in plasma during all stages of infection determined by competitive PCR. Science 259:1749-1754. [DOI] [PubMed] [Google Scholar]
- 33.Princiotta, M. F., U. Schubert, W. Chen, J. R. Bennink, J. Myung, C. M. Crews, and J. W. Yewdell. 2001. Cells adapted to the proteasome inhibitor 4-hydroxy-5-iodo-3-nitrophenylacetyl-Leu-Leu-leucinal-vinyl sulfone require enzymatically active proteasomes for continued survival. Proc. Natl. Acad. Sci. USA 98:513-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puffer, B. A., S. C. Watkins, and R. C. Montelaro. 1998. Equine infectious anemia virus Gag polyprotein late domain specifically recruits cellular AP-2 adapter protein complexes during virion assembly. J. Virol. 72:10218-10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Putterman, D., R. B. Pepinsky, and V. M. Vogt. 1990. Ubiquitin in avian leukosis virus particles. Virology 176:633-637. [DOI] [PubMed] [Google Scholar]
- 37.Schubert, U., L. C. Anton, I. Bacik, J. H. Cox, S. Bour, J. R. Bennink, M. Orlowski, K. Strebel, and J. W. Yewdell. 1998. CD4 glycoprotein degradation induced by human immunodeficiency virus type 1 Vpu protein requires the function of proteasomes and the ubiquitin-conjugating pathway. J. Virol. 72:2280-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schubert, U., L. C. Anton, A. Gibbs, C. C. Norbury, J. W. Yewdell, and J. R. Bennink. 2000. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404:770-774. [DOI] [PubMed] [Google Scholar]
- 39.Schubert, U., D. E. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. F. Princiotta, J. R. Bennink, H. G. Krausslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97:13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97:13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Summers, M. F., L. E. Henderson, M. R. Chance, J. W. Bess, Jr., T. L. South, P. R. Blake, I. Sagi, G. Perez-Alvarado, R. C. Sowder II, D. R. Hare, and L. O. Arthur. 1993. Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Sci. 1:563-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swanstrom, R., and J. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. Coffin, S. Hughes, and H. Varmus (ed.), Retroviruses. Cold Spring Harbor Press, Plainview, N.Y. [PubMed]
- 43.Thrower, J. S., L. Hoffman, M. Rechsteiner, and C. M. Pickart. 2000. Recognition of the polyubiquitin proteolytic signal. EMBO J. 19:94-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsubuki, S., Y. Saito, M. Tomioka, H. Ito, and S. Kawashima. 1996. Differential inhibition of calpain and proteasome activities by peptidyl aldehydes of di-leucine and tri-leucine. J. Biochem. (Tokyo) 119:572-576. [DOI] [PubMed] [Google Scholar]
- 45.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55Gag. Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vogt, V. 1997. Retroviral virions and genomes, p. 27-70. In J. Coffin, S. Hughes, and H. Varmus (ed.), Retroviruses. Cold Spring Harbor Press, Plainview, N.Y. [PubMed]
- 47.Vogt, V. M. 2000. Ubiquitin in retrovirus assembly: actor or bystander? Proc. Natl. Acad. Sci. USA 97:12945-12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vogt, V. M., and M. N. Simon. 1999. Mass determination of Rous sarcoma virus virions by scanning transmission electron microscopy. J. Virol. 73:7050-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weissman, A. M. 2001. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2:169-178. [DOI] [PubMed] [Google Scholar]
- 50.Wilkinson, K. D. 2000. Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Semin. Cell Dev. Biol. 11:141-148. [DOI] [PubMed] [Google Scholar]
- 51.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed]
- 52.Xiang, Y., C. E. Cameron, J. W. Wills, and J. Leis. 1996. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol. 70:5695-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72:4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan, B., S. Campbell, E. Bacharach, A. Rein, and S. P. Goff. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly. J. Virol. 74:7250-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]