Abstract
The only retrovirus protein required for the budding of virus-like particles is the Gag protein; however, recent studies of Rous sarcoma virus (RSV) and human immunodeficiency virus have suggested that modification of Gag with ubiquitin (Ub) is also required. As a consequence, the release of these viruses is reduced in the presence of proteasome inhibitors, which indirectly reduce the levels of free Ub within the cell. Here we show that the budding of equine infectious anemia virus (EIAV) from infected equine cells is largely unaffected by these drugs, although use of one inhibitor (MG-132) resulted in a dramatic block to proteolytic processing of Gag. This lack of sensitivity was also observed in transiently transfected avian cells under conditions that greatly reduce RSV budding. Moreover, insensitivity was observed when the EIAV Gag protein was expressed in the absence of all the other virus products, indicating that they are not required for this phenotype. An activity that enables EIAV to tolerate exposure to proteasome inhibitors was mapped to the C-terminal p9 sequence, as demonstrated by the ability of an RSV Gag-p9 chimera to bud in the presence of the drugs. Intriguingly, the p9 sequence contains a short sequence motif that is similar to a surface-exposed helix of Ub, suggesting that EIAV Gag may have captured a function that allows it to bypass the need for ubiquitination. Thus, the mechanism of EIAV budding may not be substantially different from that of other retroviruses, even though it behaves differently in the presence of proteasome inhibitors.
Retroviruses are enveloped and obtain their lipid bilayer by budding through the plasma membrane of the host cell. Release of the nascent particle requires membrane fusion at the base of the bud, an event commonly referred to as “pinching off.” Although the mechanism of virus-cell separation is unknown, it is well established that the Gag protein is the only viral product required for budding (27).
Gag proteins are made on free ribosomes and subsequently bind to the plasma membrane by means of the M domain. Roughly 1,500 Gag molecules come together to make a virus particle (29), and the primary interactions among these proteins are provided by the I domain. As a result of the M and I functions, nascent buds rise up from the surface of the cell, but these are not released unless the L (late) domain is also present. The most striking properties of L domains are their small size (four or five amino acids) and their positional independence, both within a given Gag protein and between distantly related viruses (3, 7-9, 11, 18, 21, 26, 31-35). The L domain likely serves to recruit host machinery that mediates the pinching off step (6), but little is known about the specific host factors involved.
Numerous lines of evidence have accumulated to suggest that ubiquitin (Ub) plays an important role in virus budding. All examined retroviruses have been found to contain roughly 100 copies of Ub, and, with the exception of those in Rous sarcoma virus (RSV), about one-third of these molecules have been found to be individually conjugated to Gag at positions near the L domain (16, 17, 23). Moreover, L domains have been shown to recruit Ub ligase activity to facilitate virus release (26), and components of the ubiquitination machinery have been identified in searches for the potential binding partners of L domains (12, 28). Proteasome inhibitors, which deplete the intracellular levels of free Ub, dramatically reduce budding, resulting in the accumulation of virus particles on the surfaces of infected cells (19, 24). Overexpression of Ub stimulates particle release in the presence of the inhibitors, and a Gag chimera that has Ub fused to its C terminus is insensitive to the drugs (19). The specific role of Ub in budding is unknown.
To further explore the requirements of Ub in retrovirus budding, we decided to test the sensitivity of equine infectious anemia virus (EIAV) to proteasome inhibitors. This study was of interest because EIAV has an L domain sequence (Y-P-D-L) that is highly divergent from the proline-rich motifs found in other retroviruses (for example, P-P-P-P-Y in RSV and P-T-A-P in human immunodeficiency virus [HIV]) and its binding partner is not a component of the ubiquitination machinery but instead is the well-known clathrin adapter protein, AP-2 (21, 22). Our results indicate that EIAV has acquired a novel function that enables it to escape the effects of proteasome inhibitors (see also the accompanying paper by Ott et al. [17]). This property maps to the p9 sequence present at the C terminus of Gag. Intriguingly, p9 contains a motif that bears striking similarity to a surface-exposed helix of Ub, suggesting that the mechanism of EIAV budding may not be different from that of other retroviruses, even though it behaves differently in the presence of proteasome inhibitors.
MATERIALS AND METHODS
Cell lines.
Uninfected and EIAVuk-infected equine dermal cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and 0.1% penicillin-streptomycin. Canine (Cf2th) cells infected with EIAV (kindly provided by David Ott) were cultured in the same medium. Uninfected and RSV-infected avian (QT6) cells were grown in F-10 medium supplemented with 8.5% tryptose phosphate broth, 5.1% fetal bovine serum, 1.0% chicken serum, and 0.1% penicillin-streptomycin.
EIAV expression plasmids.
To express all of the structural genes of EIAV in avian cells, pCMV.EIAVuk was used (2). This plasmid contains a recombinant proviral genome in which the retrovirus promoter has been replaced with the cytomegalovirus (CMV) immediate-early promoter. To prevent Gag and Gag-Pol processing, a derivative of this plasmid, pCMV.EIAVuk.pro-, was made; in the derivative, the codon for the Asp residue in the catalytic site of the viral protease was changed to an Ala codon.
To express only the Gag protein of EIAV in avian cells, pEG.PRE was constructed to enable expression in the absence of the regulatory proteins of EIAV. Four steps were required. First, pTR-UF5 (13, 36) (a gift from N. Muzyczka) was digested with SalI to excise the neomycin resistance gene and the remaining DNA was recircularized to create pTRU5Δneo. Second, pLG.TRU5Δneo was created by ligating a PstI fragment of pTRU5Δneo into the PstI site of a derivative of pLG338-30 (4) whose SalI site had been destroyed by treatments with SalI, T4 DNA polymerase, and ligase. The transferred PstI fragment contains the 145-bp terminal repeat (TR) sequences from adeno-associated virus, between which reside the CMV immediate-early promoter, simian virus 40 (SV40) splice donor and splice acceptor sequences, the green fluorescence protein (GFP)-coding sequence (flanked by NotI sites), and two polyadenylation signals (the first from SV40 and the second from the bovine growth hormone gene) separated by the unique SalI site created in step one. Third, the GFP-coding sequence of pLG.TRU5Δneo was removed by digestion with NotI and replaced with the gag sequence of EIAVuk. The gag gene was amplified by PCR using the following upstream and downstream oligonucleotides (respectively) containing NotI restriction sites (underlined) for cloning: 5′-ATAAGGAATGCGGCCGCCACCATGGGAGACCCTTTGACATGG-3′ and 5′-ATAAGGAATGCGGCCGCTTACTCCCACAAACTGTCCAGGTT-3′. The resulting plasmid was named pLG.Gag. Fourth, to facilitate export of unspliced transcripts into the cytoplasm for optimal EIAV Gag expression, the hepatitis B virus posttranscriptional regulatory element (PRE) (5) was inserted between the termination codon of gag and the bovine growth hormone polyadenylation signal in pLG.Gag to create pGag.PRE. PRE was amplified from pDM138 (a gift from G. Smith and T. Hope) with the following upstream and downstream PCR primers (respectively): 5′-ATAAGGAATGCGGCCGCTTGCTCGGCAACGGCCTGGTC-3′ (NotI) and 5′-ATAAGGAATGTCGACACATTGCTGAAAGTCCAAGAG-3′ (SalI). The underlined sequences indicate the restriction endonuclease sites used for cloning (for the enzymes in parentheses). Following complete digestion of the amplified fragment with NotI and SalI, the product was ligated with pLG.Gag DNA, which had been completely digested with SalI and partially digested with NotI. The resulting expression plasmid was designated pGag.PRE.
RSV expression plasmids.
pCMV.Gag-GFP encodes a chimera of the RSV Gag protein (RG.GFP) in which the GFP sequence replaces the viral protease and six adjoining residues of the NC sequence (1). pCMV.Gag.Ub encodes a similar chimera (RG.Ub) except that the GFP sequence has been replaced with that of Ub (19). To fuse the EIAV p9 sequence to the end of RSV Gag (at the same position used for GFP and Ub), the p9-coding sequence was amplified by PCR using the wild-type EIAV gag gene as the template. The following upstream primer (containing the coding sequence for a flexible glycine-serine linker, indicated by brackets) and downstream primer (respectively) were utilized: 5′-CTATACGGGCCC[GGGAGTGGGAGTGGGAGTGGGAGTGGGAGT]CCGATACAACAGAAG-3′ (ApaI) and 5′-TACGACGCGGCCGCTTACTCCCACAAACTGTCCAGGTT-3′ (NotI). The underlined sequences indicate the restriction endonuclease sites used for cloning (for the enzymes in parentheses). Following amplification, the PCR product and pCMV.Gag-GFP (1) were digested with ApaI and NotI. The large fragment from the plasmid was ligated to the p9-coding sequence to produce pRG.p9.
To make pREI.T10c, pCMV.Gag-GFP (1) and pSV.REI.T10c (18) were digested with BsrG1 and BssHII, respectively. The DNA fragments were treated with the Klenow fragment of DNA polymerase I and then digested with SstI. The large fragment from pCMV.Gag-GFP and the small fragment from pSV.REI.T10c were gel purified and ligated.
Proteasome inhibitors and Ub immunoblots.
MG-132 (also known as zLLL), MG-262, lactacystin, and epoxomicin were obtained from Calbiochem. All of the inhibitors were dissolved in dimethyl sulfoxide (DMSO) and used within 2 weeks at a final concentration of 10 μM, unless otherwise indicated. To measure changes in free-Ub levels following MG-132 treatment, standard immunoblotting techniques using rabbit anti-Ub serum (kindly provided by Caroline E. Shamu) were employed.
Budding assay, metabolic labeling, and immunoprecipitation.
Infected cells and cells transfected by the calcium phosphate precipitation method were pretreated with DMSO or proteasome inhibitors for 90 min immediately prior to labeling with [35S]methionine for 2.5 h, also in the presence of the drug (19). The virus particles in the growth medium and the cell monolayers were separated from one another and disrupted with detergents. The Gag proteins present in each sample were immunoprecipitated with polyclonal rabbit serum against whole RSV or EIAV using previously described methods (30). Proteins were separated by electrophoresis in sodium dodecyl sulfate (SDS)-12% polyacrylamide gels, which were then fixed and dried. The labeled proteins were detected by autoradiography using Kodak X-Omat AR5 film at −80°C. The effects of proteasome inhibitors on budding were quantitated using a phosphorimager.
Electron microscopy.
EIAV-infected equine dermal cells on Permanox dishes were treated for 3 h with DMSO (negative control) or 80 μM MG-132 prior to the analysis. Cells were fixed, dehydrated, embedded, and sectioned for electron microscopy using standard methods.
RESULTS
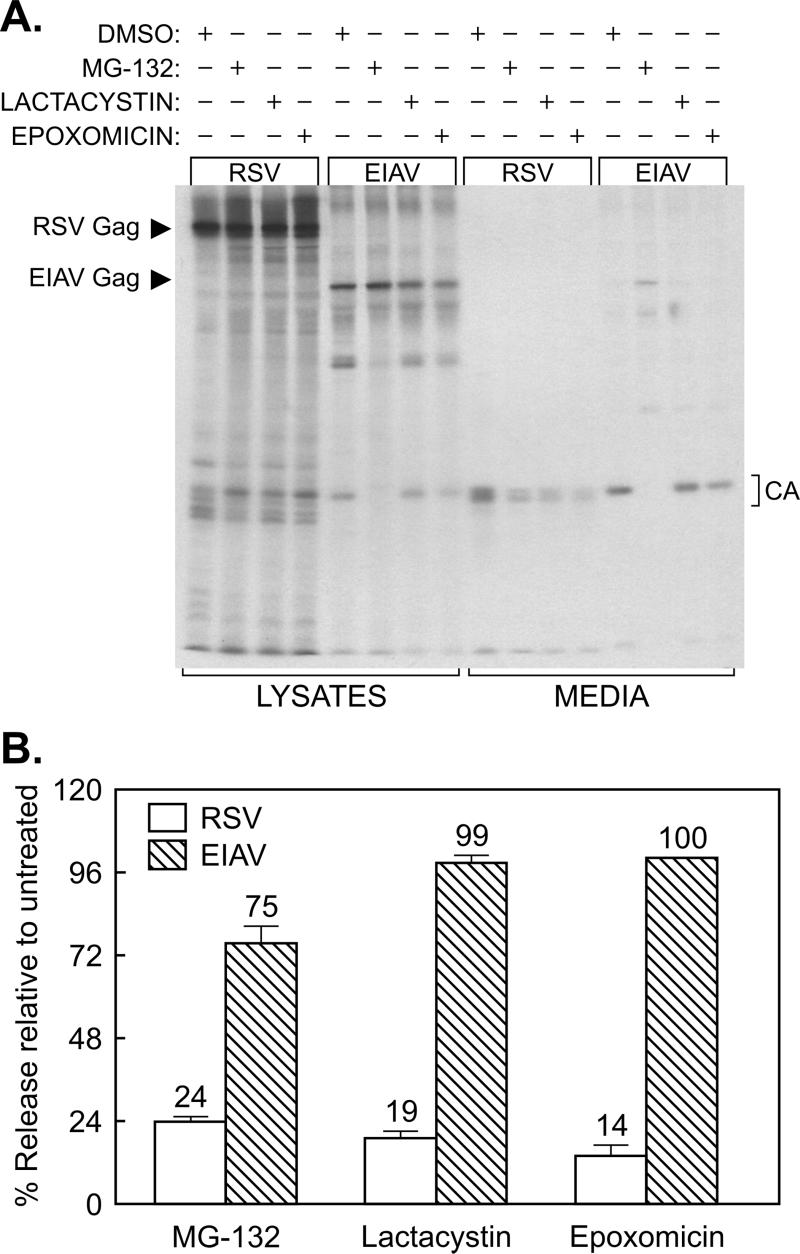
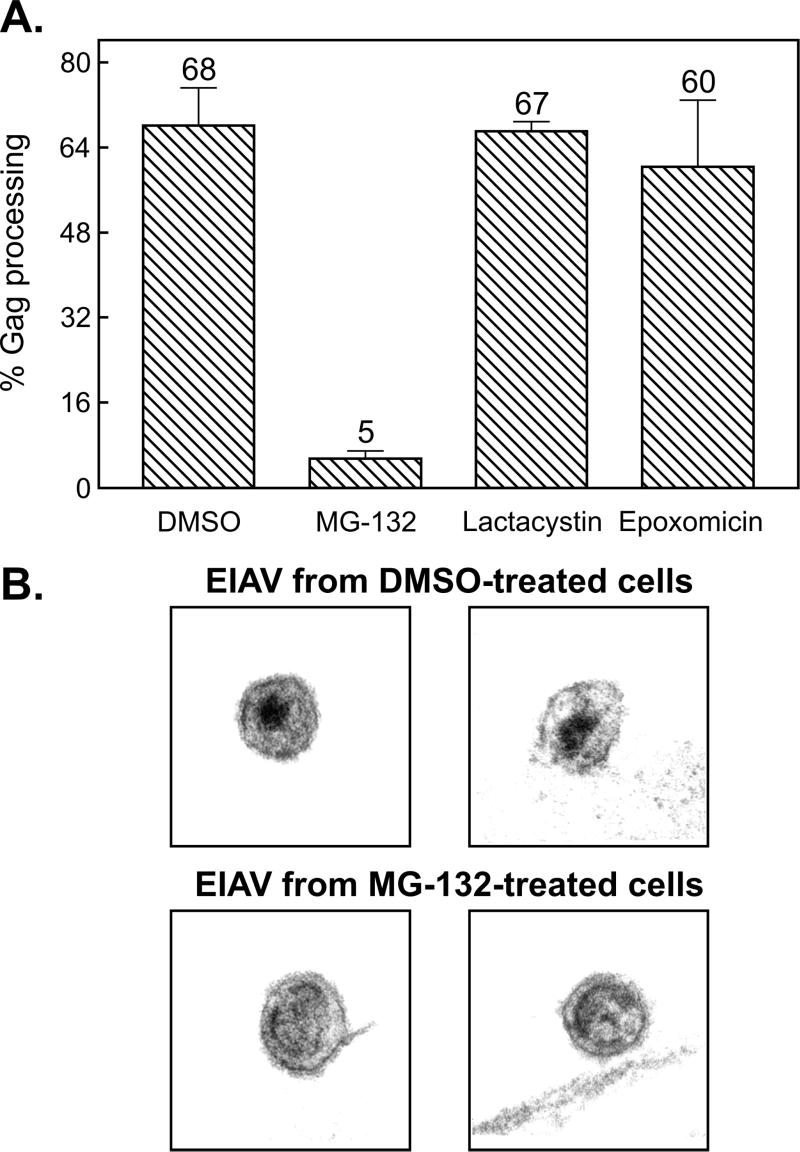
To determine whether the release of EIAV is reduced by exposure to proteasome inhibitors, EIAV-infected equine cells and RSV-infected avian cells were treated with a panel of three different inhibitors (or with DMSO as the vehicle control) while being metabolically labeled with [35S]methionine for 2.5 h. Labeled Gag proteins present in the cells and growth media were immunoprecipitated, subjected to SDS-polyacrylamide gel electrophoresis, and quantitated by phosphorimager analysis (Fig. 1). The activity of the inhibitors was demonstrated by their ability to reduce RSV budding to levels similar to those previously reported (19). However, in spite of their potency, none had a large effect on the release of EIAV (Fig. 1). The most obvious effect was seen with MG-132. This inhibitor had only a mild effect on particle release (Fig. 1B) but produced a powerful block to EIAV (but not RSV) Gag processing, as shown by the absence of CA products in the medium (Fig. 1A and 2A) and the appearance of immature virions on the surfaces of the infected cells (Fig. 2B). MG-132 treatment of a canine (Cf2th) cell line infected with EIAV yielded identical results (data not shown). In contrast, lactacystin and epoxomicin had essentially no effect on EIAV processing (Fig. 1A and 2A). Moreover, a derivative of MG-132, MG-262, which differs by the presence of an additional boronic acid group, showed no effect on EIAV protease activity and budding (data not shown). These results provided the first suggestion that EIAV might be very different from RSV and HIV in its insensitivity to proteasome inhibitors.
FIG. 1.
Insensitivity of EIAV release to proteasome inhibitors. (A) Four plates each of RSV-infected avian cells and EIAV-infected equine cells were seeded, and, the next day, one from each set was pretreated for 90 min with DMSO (vehicle control), 10 μM MG-132, 10 μM lactacystin, or 10 μM epoxomicin. The cells were then metabolically labeled for 2.5 h with [35S]methionine, also in the presence of drugs. The cell monolayers and the particles in the growth medium were lysed with detergent, and the Gag proteins were immunoprecipitated and electrophoresed in an SDS-12% polyacrylamide gel prior to detection by autoradiography. Left, positions of the uncleaved Gag proteins; right, positions of the mature CA products. The majority of methionine residues in EIAV Gag (10 of 12) are found in CA, and hence it is the only cleavage product visible. (B) The impact of proteasome inhibitors on RSV and EIAV budding was quantitated with a phosphorimager. The budding efficiency in each culture was calculated as the total amount of Gag protein (Gag precursor plus intermediate cleavage products plus mature cleavage products) in the medium divided by the total amount in the culture (lysates and medium). The effects of proteasome inhibitors on budding (percent release relative to untreated) were then determined by computing the ratio of budding efficiency in the DMSO-treated cells to that in drug-treated cells. Averages from two experiments are shown, with error bars indicating the deviation of each measurement from the mean.
FIG. 2.
Inhibitory effects of MG-132 on EIAV protease activity. (A) EIAV-infected cells were labeled with [35S]methionine as indicated for Fig. 1. The impact of the proteasome inhibitor on protease activity is expressed as the ratio of mature cleavage products (CA) in the medium to the total Gag signal (Gag precursor plus intermediate cleavage products plus mature cleavage products) in the medium. Averages from two experiments are shown along with the deviation of each measurement from the mean. (B) Transmission electron microscopy of the surfaces of EIAV-infected equine cells treated with either DMSO (top) or 80 μM MG-132 (bottom).
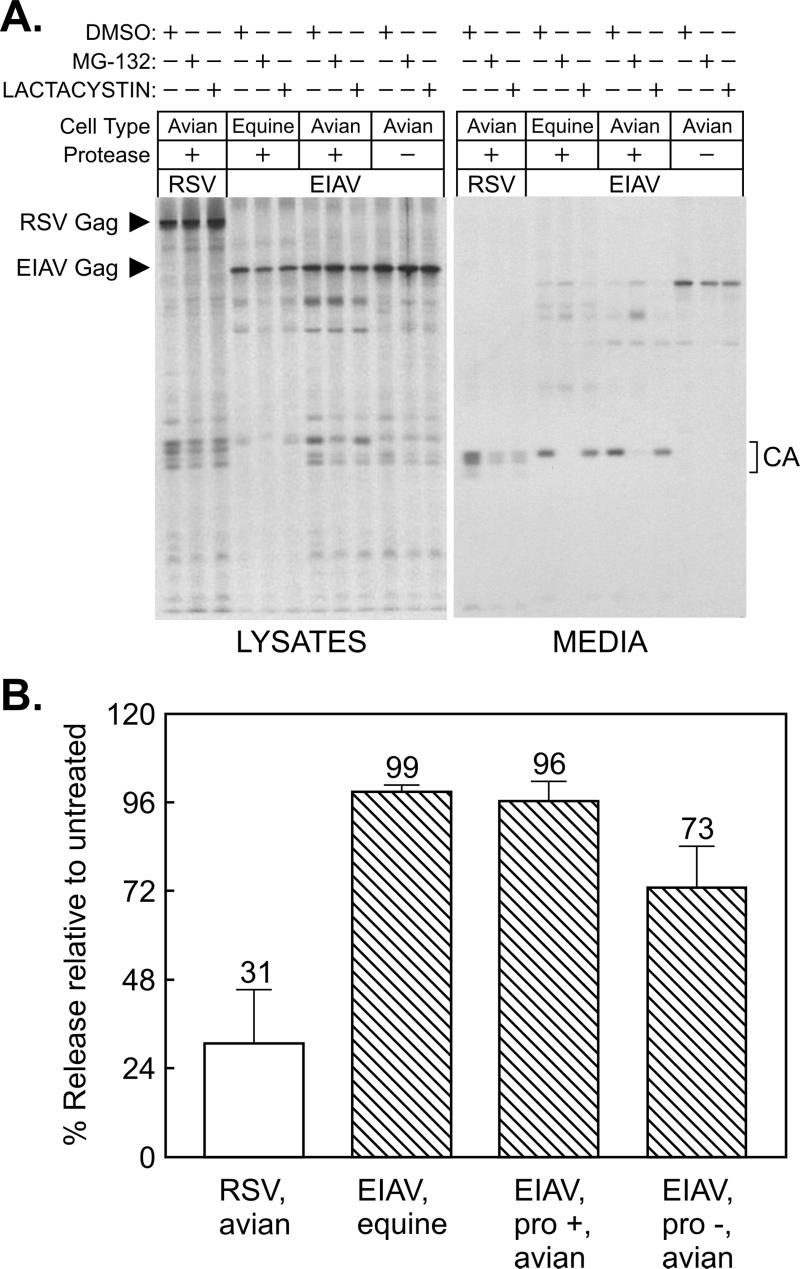
Proteasome inhibitors do not affect the budding machinery of other retroviruses in a direct manner but instead have the indirect effect of reducing the availability of free Ub (14, 24), and it is this molecule that is required for budding. If equine (or canine) cells have unusually high numbers of proteasomes and correspondingly high basal levels of free Ub compared to avian cells, then this could account for the failure of the inhibitors to reduce EIAV budding. To address this concern, EIAV proviruses were transfected into the same avian cell line (QT6) that we previously used to show that RSV budding is responsive to proteasome inhibitors (19). Positive and negative controls were provided by RSV-infected avian cells and EIAV-infected equine cells, respectively. Conditions that are inhibitory to RSV budding again did not have much effect on the release of particles when all of the EIAV structural proteins were expressed in avian cells (Fig. 3). MG-132 again resulted in a striking block to Gag processing, but lactacystin did not (Fig. 3A). A mild reduction in particle release was seen when a protease-negative provirus was expressed in lactacystin-treated avian cells, suggesting that protease activity might be required for maximum budding, but these conditions had no effect on the wild type (Fig. 3B). These results demonstrate that EIAV is indeed quite different from RSV and HIV in having the ability to resist the effects of proteasome inhibitors.
FIG. 3.
Direct comparison of EIAV and RSV budding in avian cells. (A) Triplicate plates of uninfected cells were transfected with CMV promoter-driven expression vectors containing the wild-type EIAV structural genes (pCMV.EIAVuk) or a protease mutant (pCMV.EIAVuk.pro-). In addition, triplicate plates of RSV-infected avian cells (positive control) and EIAV-infected equine cells (negative control) were seeded. The next day, plates from each set were pretreated for 90 min with DMSO (control), MG-132, or lactacystin, and the cells were metabolically labeled for 2.5 h with [35S]methionine, also in the presence of the drug. The cells and growth media were subsequently processed as described for Fig. 1. (B) The impact of lactacystin on budding was quantitated by phosphorimager, as described for Fig. 1. Averages from two experiments are shown, with error bars indicating the deviations of each measurement from the mean.
There are at least three ways to explain the ability of EIAV to resist the effects of proteasome inhibitors. First, this virus might utilize a completely different mechanism of budding. Because the same virus-encoded budding functions (i.e., M, L, and I) are present in all Gag proteins, including those of distantly related retroviruses (27), this explanation seems unlikely. Second, EIAV might utilize Ub-conjugating enzymes that have a greater efficiency than those used by RSV and HIV, allowing budding to continue even though free Ub is limiting. Until more is known about the specific host-encoded enzymes involved in EIAV budding, it is difficult to address this possibility. Third, EIAV might have acquired an additional function that supplants an absolute need for Ub modification. The availability of Gag proteins that are highly sensitive to proteasome inhibitors allowed us to search for novel budding functions encoded by EIAV.
Functions that enable EIAV to escape the effects of proteasome inhibitors might be encoded by any of the viral genes, and hence it was important to ascertain whether the protein solely responsible for budding (i.e., the Gag protein) contains the resistance activity. For this, avian cells were transfected with a plasmid that expresses EIAV Gag (designated EG; Fig. 4A) in the absence of all the other viral products. This protein was found to bud from avian cells with an efficiency lower than that of RSV Gag (down 70%), but its release was nevertheless dependent on the presence of its L domain (data not shown), as previously reported for mammalian cells (21, 22). Treatment of EG-expressing cells with MG-132 had no effect on budding beyond what was predicted for the release of particles in the absence of protease activity (compare Fig. 4C, bar EG, with Fig. 3B, bar EIAV, pro-, avian), even though the RSV control exhibited a striking reduction in budding (Fig. 4C, bar RG.GFP). Thus, if EIAV has acquired any additional budding functions, one must be contained within Gag.
FIG. 4.
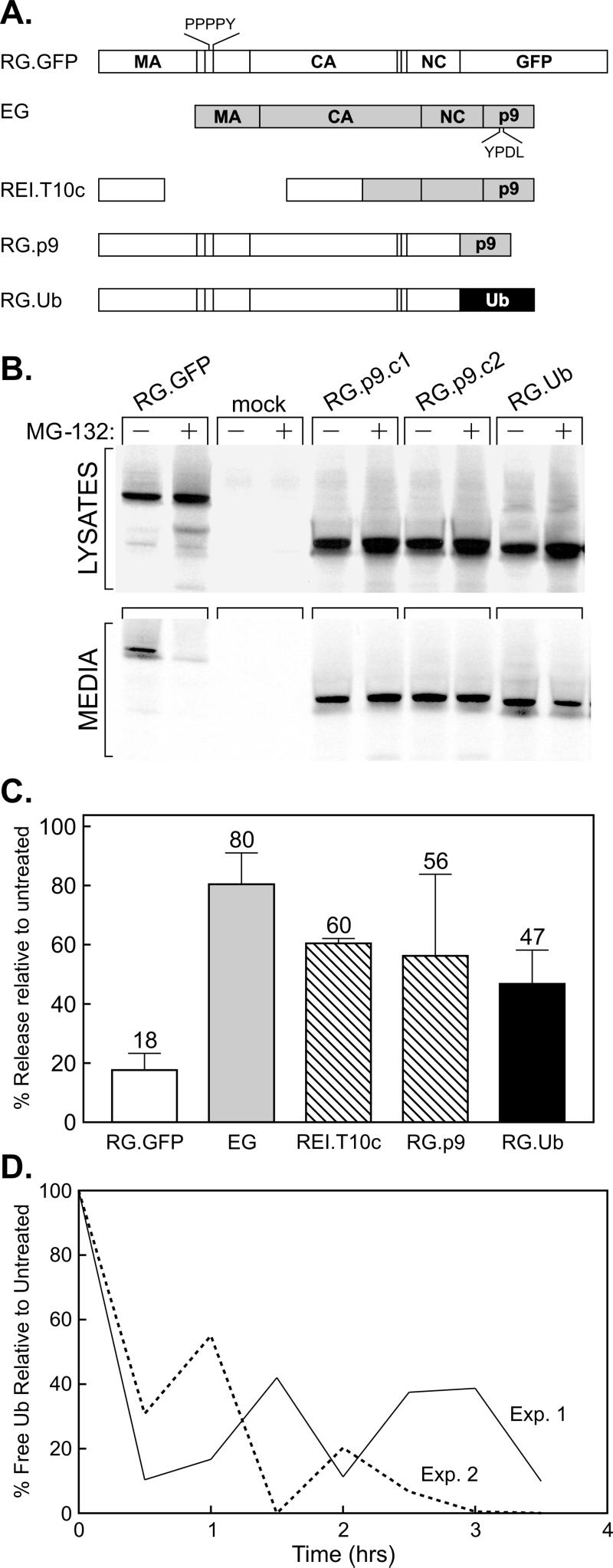
Drug resistance activity maps to a region within p9Gag. (A) Gag derivatives used for the mapping experiments. Open boxes, segments of the RSV Gag protein (along with GFP); gray boxes, segments of EIAV Gag. The relative positions of the RSV and EIAV late domains are indicated. (B) QT6 cells were transfected with the indicated DNAs. The next day, the cells were pretreated with either DMSO or 80 μM MG-132 for 90 min, before being metabolically labeled for 2.5 h with [35S]Met, also in the presence of the drug. The cells and growth media were subsequently processed as described for Fig. 1. (C) Phosphorimager analysis of budding efficiencies, calculated as described for Fig. 1. Averages from three experiments are shown for all constructs, with the exception of REI.T10c, which shows data from two experiments. (D) Reduction in free-Ub levels with time following exposure to 80 μM MG-132. Avian cell lysates were prepared at the indicated times and analyzed on an immunoblot using antibodies specific for Ub. Bands on the resulting autoradiograph were quantitated by means of densitometry.
If a simple resistance function is contained within the EIAV Gag protein, then it should be possible to move this activity to a Gag protein that is normally responsive to the inhibitors. We initially examined this issue by analyzing a previously described chimera named REI.T10c (18), which contains the M domain of RSV along with the I and L domains of EIAV (Fig. 4A). The budding of this chimera was much less sensitive to MG-132 than that of the RSV Gag control (RG.GFP), indicating that an additional function of EIAV is contained within its C-terminal half (Fig. 4C). We next linked only the p9 sequence of EIAV to the C terminus of the RSV control molecule in place of its GFP sequence (RG.p9; Fig. 4A), and this chimera was also found to be strikingly insensitive to proteasome inhibitors (Fig. 4B and C). Indeed, this chimera was even more resistant than a previously described derivative of RSV Gag (19) that has the Ub sequence linked to its C terminus (RG.Ub; Fig. 4). Immunoblotting cell extracts with antibodies specific for Ub (Fig. 4D) confirmed that MG-132 rapidly reduces the levels of free Ub in avian cells during the 4 h of total drug treatment (1.5 h of pretreatment plus 2.5 h of treatment during metabolic labeling). Taken together, these results suggest that the insensitivity of EIAV to proteasome inhibitors is Gag specific and maps to a region within p9Gag.
DISCUSSION
Proteasome inhibitors have the indirect effect of reducing the amount of free Ub in the cell, and because this host protein is needed in some way for efficient budding, at least for RSV and HIV, these drugs can inhibit virion release (19, 24). The results presented here show that the budding of EIAV is remarkably insensitive to proteasome inhibitors. Similar results showing at most a twofold reduction of budding have been obtained using pulse-chase labeling experiments (see the accompanying paper by Ott et al. [17]). We have mapped a resistance function to the p9 sequence of the EIAV Gag protein, and this activity can be transferred to the Gag protein of RSV, which is normally quite sensitive to the drugs. Rather than implying a different mechanism of budding, these results suggest that EIAV contains an additional function, lacking in RSV and HIV, that enables it to be released when Ub levels are limiting. To better characterize this activity, further mapping studies of the p9 sequence in the context of the RSV Gag-p9 chimera (RG.p9) will be needed. In addition, it will be important to ascertain whether other structural proteins of EIAV (e.g., Pol or Env) or other portions of its Gag protein contain functions that are also capable of providing resistance to these drugs.
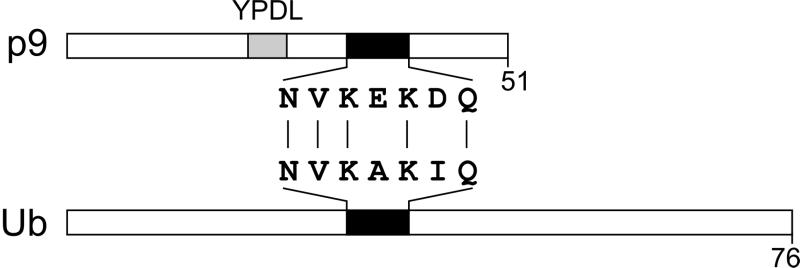
Although much more information is needed, it is intriguing that the p9 sequence contains a short sequence that bears striking (but imperfect) similarity to the surface-exposed helix in Ub (Fig. 5). It is imaginable that EIAV is resistant to the effects of proteasome inhibitors because its Gag protein already contains the function of Ub that is essential for budding. However, since EIAV contains Ub, some of which is conjugated to p9 (see the accompanying paper by Ott et al. [17]), in an amount that is similar to those for other retroviruses, it seems likely that its mechanism of budding is fundamentally the same. Selective pressure for capturing an extra function required for release could have arisen if this virus normally encounters cells with low levels of Ub. Consistent with this idea of redundancy, p9 mutants that are competent for budding and infectivity in cell cultures even though they lack C-terminal residues that encompass the Ub-like helix have been described (2).
FIG. 5.
A ubiquitin-like motif within EIAV p9Gag. Alignment of EIAV p9Gag and ubiquitin showing the region of similarity (black rectangles) between the two proteins. The position of the L domain is also indicated (gray rectangle).
Role of Ub in retrovirus budding.
What is the function of Ub in retrovirus budding? Two general models have been proposed. According to one, proteasome inhibition results in the accumulation of misfolded or aberrant Gag proteins (also called defective ribosomal products or DRiPs), which interfere with assembly and budding (24). According to this model, Ub is solely needed to tag the misfolded proteins for degradation by the proteasome. This hypothesis is difficult to reconcile with the finding that EIAV is resistant to the drugs since there is no reason to expect that the Gag protein of this virus would fold with a vastly superior efficiency. Indeed, the efficiency of budding from avian cells for EIAV Gag is reduced compared to that for RSV Gag (down 70%), yet it is far more resistant to the drugs. Moreover, the p9 sequence of EIAV can bestow on RSV Gag the ability to resist drug treatments. Although we cannot completely rule out the hypothesis that some Gag proteins are misfolded and need to be targeted for destruction by the proteasome, it seems unlikely that this is the primary role for Ub in budding.
The other hypothesis is that Ub plays an active role, perhaps serving to recruit additional host proteins to the site of budding. In support of this idea, both overexpression of Ub in trans and fusion of Ub to the C terminus of Gag stimulate budding when the proteasome is shut down and is thus unable to degrade misfolded proteins (19). The only thing that is known about the specific role for Ub in this general model is that it is needed very late in budding, after the Gag protein arrives on the plasma membrane. Perhaps the function provided by Ub in budding is in some way related to the role it plays in the endocytosis of receptor-ligand complexes (10, 15, 25). In both endocytosis and budding, monoubiquitination is hypothesized to provide the means for recruitment of additional host factors that are needed for subsequent steps in the pathway.
Effect of MG-132 on EIAV maturation.
Previous studies have shown that the EIAV protease is resistant to a wide spectrum of inhibitors of HIV type 1 (HIV-1) protease. Only one protease inhibitor (HBY-793) showed similar inhibitory potential against EIAV protease (20). In this study, we observed that MG-132 has a potent effect on EIAV (but not RSV) protease activity. Two lines of evidence suggest a direct effect of MG-132 on EIAV protease. First, when virions released from MG-132-treated cells were centrifuged to remove the drug prior to incubation at 37°C for 3 h, the viral protease activity was partially restored relative to that of unwashed controls (data not shown). A caveat to this experiment is that the virus may incorporate a cellular protease whose activity is inhibited by MG-132. Second, others have shown that in vitro incubation of purified EIAV protease with MG-132 inhibits the enzyme's ability to cleave substrates (D. Ott, unpublished observations). These data suggest that MG-132 directly and reversibly interferes with the EIAV protease. In contrast, previous studies have shown that MG-132 interferes with Gag polyprotein processing of HIV-1, but not via a direct effect on HIV-1 protease activity (24).
In summary, we have identified a retrovirus with a mechanism of particle release that is highly resistant to proteasome inhibitors. This phenotype can be explained by the presence of an additional function within the p9 sequence of the Gag protein, and this activity is reminiscent of the function provided by Ub for other retroviruses such as RSV and HIV. These observations may provide further insight into the mechanism by which Ub triggers budding and in turn may shed light on additional host factors that are involved in late virus-budding events.
Acknowledgments
We thank Rebecca Craven for her thoughtful comments regarding the manuscript, David Ott and Ulrich Schubert for graciously sharing their findings on EIAV prior to publication, and Volker Vogt and Marc Johnson for their many stimulating conversations during the course of this investigation.
This work was supported National Institutes of Health grants to J.W.W. (CA47482) and R.C.M. (CA49296).
REFERENCES
- 1.Bowzard, J. B., R. J. Visalli, C. B. Wilson, E. M. Callahan, R. J. Courtney, and J. W. Wills. 2000. Membrane targeting properties of a herpesvirus tegument protein-retrovirus Gag chimera. J. Virol. 74:8692-8699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, C., F. Li, and R. C. Montelaro. 2001. Functional roles of equine infectious anemia virus Gag p9 in viral budding and infection. J. Virol. 75:9762-9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Craven, R. C., R. N. Harty, J. Paragas, P. Palese, and J. W. Wills. 1999. Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras. J. Virol. 73:3359-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cunningham, T. P., R. C. Montelaro, and K. E. Rushlow. 1993. Lentivirus envelope sequences and proviral genomes are stabilized in Escherichia coli when cloned in low-copy-number plasmid vectors. Gene 124:93-98. [DOI] [PubMed] [Google Scholar]
- 5.Donello, J. E., A. A. Beeche, G. J. Smith, G. R. Lucero, and T. J. Hope. 1996. The hepatitis B virus posttranscriptional regulatory element is composed of two subelements. J. Virol. 70:4345-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garnier, L., J. W. Wills, M. F. Verderame, and M. Sudol. 1996. WW domains and retrovirus budding. Nature 381:744-745. [DOI] [PubMed] [Google Scholar]
- 7.Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harty, R. N., J. Paragas, M. Sudol, and P. Palese. 1999. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J. Virol. 73:2921-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hicke, L. 2001. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2:195-201. [DOI] [PubMed]
- 11.Jayakar, H. R., K. G. Murti, and M. A. Whitt. 2000. Mutations in the PPPY motif of vesicular stomatitis virus matrix protein reduce virus budding by inhibiting a late step in virion release. J. Virol. 74:9818-9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for Gag budding from cells. Proc. Natl. Acad. Sci. USA 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein, R. L., E. M. Meyer, A. L. Peel, A. Zolotukhin, C. Meyers, N. Muzyczka, and M. A. King. 1998. Neuron-specific transduction in the rat septohippocampal or nigrostriatal pathway by recombinant adeno-associated virus vectors. Exp. Neurol. 150:183-194. [DOI] [PubMed] [Google Scholar]
- 14.Mimnaugh, E. G., H. Y. Chen, J. R. Davie, J. E. Celis, and L. Neckers. 1997. Rapid deubiquitination of nucleosomal histones in human tumor cells caused by proteosome inhibitors and stress response inducers: effects on replication, transcription, translation, and the cellular stress response. Biochemistry 36:14418-14429. [DOI] [PubMed] [Google Scholar]
- 15.Nakatsu, F., M. Sakuma, Y. Matsuo, H. Arase, S. Yamasaki, N. Nakamura, T. Saito, and H. Ohno. 2000. A di-leucine signal in the ubiquitin moiety: possible involvement in ubiquitin-mediated endocytosis. J. Biol. Chem. 275:26213-26219. [DOI] [PubMed] [Google Scholar]
- 16.Ott, D. E., L. V. Coren, T. D. Copeland, B. P. Kane, D. G. Johnson, R. C. Sowder II, Y. Yoshinaka, S. Oroszlan, L. O. Arthur, and L. E. Henderson. 1998. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J. Virol. 72:2962-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ott, D. E., L. V. Coren, R. C. Sowder II, J. Adams, K. Nagashima, and U. Schubert. 2002. Equine infectious anemia virus and the ubiquitin-proteasome system. J. Virol. 76:3038-3044. [DOI] [PMC free article] [PubMed]
- 18.Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 69:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Powell, D. J., D. Bur, A. Wlodawer, A. Gustchina, S. L. Payne, B. M. Dunn, and J. Kay. 1996. Expression, characterization and mutagenesis of the aspartic proteinase from equine infectious anemia virus. Eur. J. Biochem. 241:664-674. [DOI] [PubMed] [Google Scholar]
- 21.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puffer, B. A., S. C. Watkins, and R. C. Montelaro. 1998. EIAV Gag polyprotein late domain recruits the cellular AP-2 adapter protein complex to facilitate viral budding. J. Virol. 72:10218-10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Putterman, D., R. B. Pepinsky, and V. M. Vogt. 1990. Ubiquitin in avian leukosis virus particles. Virology 176:633-637. [DOI] [PubMed] [Google Scholar]
- 24.Schubert, U., D. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. Princiotta, J. Bennink, H.-G. Krausslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with Gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97:13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shih, S. C., K. E. Sloper-Mould, and L. Hicke. 2000. Monoubiquitination carries a novel internalization signal that is appended to activated receptors. EMBO J. 19:187-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strack, B., A. Calistri, M. A. Accola, G. Palú, and H. G. Göttlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97:13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swanstrom, R., and J. W. Wills. 1997. Retroviral gene expression. II. Synthesis, processing, and assembly of viral proteins, p. 263-334. In R. Weiss, N. Teich, H. Varmus, and J. M. Coffin (ed.), Retroviruses. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y. [PubMed]
- 28.VerPlank, L., F. L. T. J. Bouamr, B. Agresta, A. Kikonyogo, J. Leis, and C. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type I Pr55Gag. Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vogt, V., and M. N. Simon. 1999. Mass determination of Rous sarcoma virus virions by scanning transmission electron microscopy. J. Virol. 73:7050-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weldon, R. A., Jr., C. R. Erdie, M. G. Oliver, and J. W. Wills. 1990. Incorporation of chimeric Gag protein into retroviral particles. J. Virol. 64:4169-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiang, Y., C. E. Cameron, J. W. Wills, and J. Leis. 1996. Fine mapping and characterization of Rous sarcoma virus pr76gag late assembly domain. J. Virol. 70:5695-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72:4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan, B., S. Campbell, E. Bacharach, A. Rein, and S. P. Goff. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 74:7250-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan, B., X. Li, and S. Goff. 1999. Mutations altering the Moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18:4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zolotukhin, S., M. Potter, W. W. Hauswirth, J. Guy, and N. Muzyczka. 1996. A “humanized” green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J. Virol. 70:4646-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]