Abstract
Human immunodeficiency virus type 1 (HIV-1) heterogeneity contributes to the emergence of drug-resistant virus, escape from host defense systems, and/or conversion of the cellular tropism. To establish an in vitro system to address a heterogeneous virus population, we constructed a library of HIV-1 molecular clones containing a set of random combinations of zero to 11 amino acid substitutions associated with resistance to protease inhibitors by the HIV-1 protease. The complexity (2.1 × 105) of the HIV-1 library pNG-PRL was large enough to cover all of the possible combinations of zero to 11 amino acid substitutions (a total of 4,096 substitutions possible). The T-cell line MT-2 was infected with the HIV-1 library, and resistant viruses were selected after treatment by the protease inhibitor ritonavir (0.03 to 0.30 μM). The viruses that contained three to eight amino acid substitutions could be selected within 2 weeks. These results demonstrate that this HIV-1 library could serve as an alternative in vitro system to analyze the emergence of drug resistance and to evaluate the antiviral activity of novel compounds against multidrug-resistant viruses.
One of the most serious obstacles for efficient antiviral treatment is to prevent the emergence of mutant viruses that have a decreased susceptibility to drugs (11, 19, 32). Even several months of successful treatment with highly active antiretroviral therapy (HAART) have failed to completely suppress the development of drug-resistant viruses (22). One of the major targets for current antiviral drugs is the human immunodeficiency virus type 1 (HIV-1) protease. The HIV-1 protease contains 99 amino acids and functions in a dimeric form for processing of the Gag and Gag-Pol polyproteins during viral maturation at the late stage of the HIV-1 life cycle (8, 15, 24, 35). The most common resistance mechanism in HIV-1 viruses isolated from patients treated with protease inhibitors is through the emergence and accumulation of multiple amino acid substitutions within the viral protease (6, 10, 13, 19, 25, 34). On occasion, additional mutations in protease cleavage sites in the viral polyprotein are accompanied by these substitutions (7, 9). Various combinations of amino acid changes at specific positions throughout the protease sequence were obtained in vivo after selection with various protease inhibitors (6, 18, 31). However, the amino acid changes in the viral protease were not always consistent with those in vivo. Therefore, an in vitro system to address heterogeneous HIV-1 populations could be useful for further elucidation. Here, a novel in vitro method for the generation of a viral library with random mutations in a specific region of the HIV-1 genome has been developed in order to screen viruses with desirable properties, such as drug resistance.
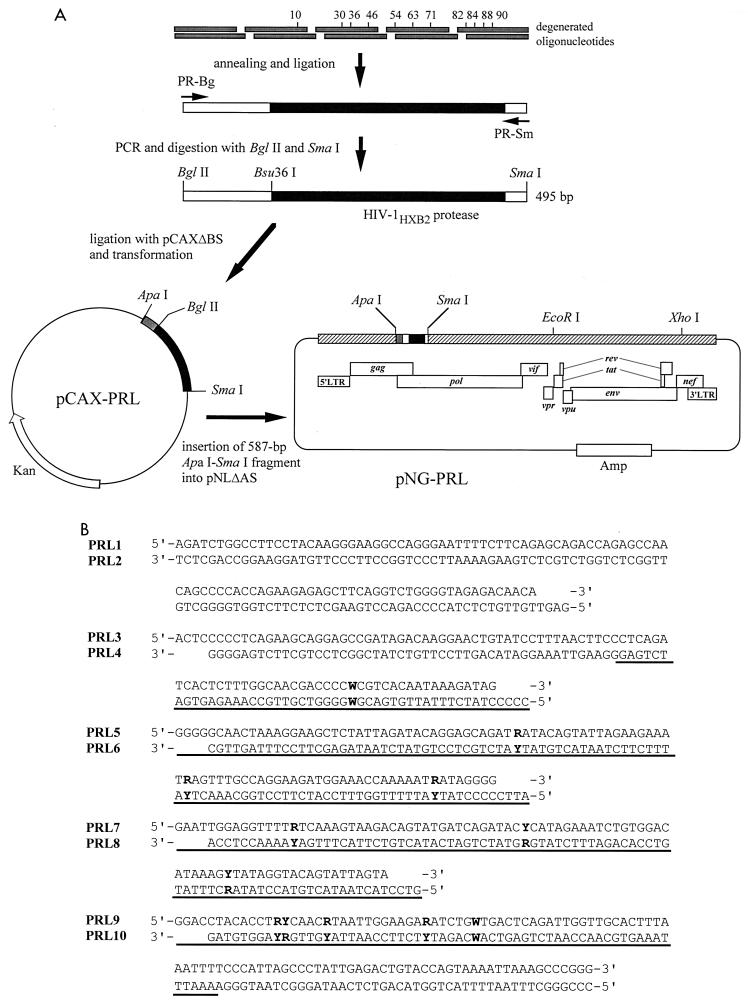
The strategy used to construct the HIV-1 library carrying a set of random amino acid substitutions in the viral protease is shown in Fig. 1A. We chose eight amino acid substitutions found in multidrug-resistant HIV-1 isolates from seven patients who had received various protease inhibitors, including indinavir, ritonavir, saquinavir, and amprenavir (L10I, M36I, M46I, I54V, L63P, A71V, V82A/T, and L90M) (Fig. 1B) (35). In addition to these mutations, we introduced nelfinavir resistance-associated mutations D30N and N88D (26), and I84V, associated with resistance to ritonavir, indinavir, saquinavir, or nelfinavir (18, 31).
FIG. 1.
Construction of HIV-1 library. (A) Schematic of the HIV-1 library construction carrying a set of random combinations of zero to 11 amino acids. (B) Oligonucleotide sequences used for insertion of 12 nucleotide substitutions in protease. (The residue at position 82 contains two sequential nucleotide substitutions.) LTR, long terminal repeat. The 5′ ends of the oligonucleotides, except for those of PRL1 and PRL10, were chemically phosphorylated. W = A or T, R = A or G, and Y = C or T. The protease-encoding region is underlined.
The 587-bp ApaI-SmaI fragment from HIV-1HXB2D (27) was subcloned into pCR7ΔZ digested with ApaI and SmaI, resulting in pCAX9. pCAX9ΔBS was created from pCAX9 by deleting a BglII-SmaI fragment and replacing it with a 25-bp linker. pCR7ΔZ was created from pCR-blunt (Invitrogen, Carlsbad, Calif.) by deleting a PmlI fragment containing a Zeocin cassette and replacing a 62-bp ApaI-HindIII fragment (multiple-cloning site) with an ApaI-HindIII fragment (multiple-cloning site, including an SmaI site) of pGEM7f(+) (Promega, Madison, Wis.). BglII-SmaI DNA fragments containing a set of random combination of mutations were prepared with a series of overlapping oligodeoxynucleotides comprising both DNA strands. Ten oligonucleotides (PRL1, PRL2, PRL3, PRL 4, PRL5, PRL6, PRL7, PRL8, PRL9, and PRL10) were commercially synthesized to generate five short DNA fragments with sticky ends (Fig. 1B). The respective pair of complementary oligonucleotides at 0.35 μg/μl was denatured for 2 min at 90°C in a heat block and then left in the heat block with the heat off for 30 min in 10 mM MgCl2 and 10 mM Tris-HCl (pH 8.0) at room temperature for annealing. The resultant short DNA fragments were designated as PRL1/2 (104 bp), PRL3/4 (95 bp), PRL5/6 (95 bp), PRL7/8 (83 bp), and PRL9/10 (108 bp), respectively. PRL3/4 contained one degenerated codon [CTC(Lys 10) or ATC(Ile 10)], PRL5/6 contained three degenerated codons [GAT(Asp 30) or AAT(Asn 30), ATG(Met 36) or ATA(Ile 36), or ATG(Met 46) or ATA(Ile 46)], PRL7/8 contained three degenerated codons [ATC(Ile 54) or GTC(Val 54), CTC(Leu 63) or CCC(Pro 63), or GCT(Ala 71) or GTT(Val 71)], and PRL9/10 contained four degenerated codons [GTC(Val 82), ACC(Thr 82), GCC(Ala 82), or ATC(Ile 82); ATA(Ile 84) or GTA(Val 84); AAT(Asn 88) or GAT(Asp 88); or TTG(Leu 90) or ATG(Met 90)]. Theoretically, the number of combinations of zero to 11 amino acid substitutions was 4,096 (amino acid residue 82 contained four possibilities, Val, Ala, Ile, or Thr and 10 other amino acid residues contained two possibilities each), indicating that each recombinant comprised 0.024% of the library. These five DNA fragments (0.25 μg each) were ligated by T4 DNA ligase (New England Biolabs, Inc., Beverly, Mass.), and the resultant 493-bp DNA fragment was purified via 1.5% agarose electrophoresis. One hundred nanograms of the purified DNA fragment was used for PCR as the DNA template with Pfx DNA polymerase (Invitrogen, Carlsbad, Calif.). The upstream primer was PRL-Bg (5′-GGA AGA TCT GGC CTT CCT ACA AGG GAA GGC CAG GG-3′), and the downstream primer was PRL-Sm (5′-CAT CCC GGG CTT TAA TTT TAC TGG TAC AGT CTC AAT AG-3′). PCR was performed with the Gene Amp PCR system 9700 (Applied Biosystems, Inc., Foster City, Calif.). The PCR product (5 μg) was purified by Concert Rapid PCR Purification System (Invitrogen) and digested with BglII and SmaI and ligated to the BglII and SmaI sites of pCAX9ΔBS (10 μg). pCAX9ΔBS was constructed from pCAX9 by replacing a BglII-SmaI fragment with a 25-bp linker. The ligation mixture was purified by Microcon-30 (Millipore Corp., Bedford, Mass.) and used to transform Escherichia coli strain DH5α by electroporation with BTX ECM 600 (BTX, San Diego, Calif.) at 2.5 kV as described previously (17), to generate pCAX-PRL containing 3.4 × 106 independent clones with multiple mutations in the BglII-SmaI region. After purification of pCAX-PRL DNA, the ApaI-SmaI fragment from 15 μg of the plasmid was inserted into the ApaI and SmaI sites of pNLΔAS (20 μg). pNLΔAS was created from pNL4-3Sma (35) by replacing the ApaI-SmaI fragment with a 28-bp linker. pNL4-3Sma, in which the SmaI site has been introduced in the pol gene of pNL4-3 (1), was a gift from Takamasa Ueno (Kumamoto University). The ligation products (0.2 μg) were transformed into E. coli strain JM109 by electroporation as described above. Finally the HIV-1 library pNG-PRL contained 2.1 × 105 independent clones with a 496-bp pol DNA fragment (ligation efficiency, 65%) (Table 1 and Fig. 2). The pNG-WT clone was constructed by replacement of the ApaI-SmaI fragment with that of pCAX9 (containing the wild-type HIV-1HXB2D protease gene). Incorporation of a random combination of the mutations in the protease-encoding sequence was confirmed, and an average of 0.08% of unintended nucleotide changes were also detected (Fig. 2). The size of the library was able to cover the complexity of all recombinant molecular clones.
TABLE 1.
HIV-1 pNG-PRL library carrying combinations of zero to 11 amino acid substitutions
| Plasmid | No. of combinations of amino acid substitutions in protease | No. of independent clonesa | No. of clones containing a mutated protease fragment | % of clones with ligation efficiencyb |
|---|---|---|---|---|
| 4,096 | ||||
| pCAX-PRL | 4.5 × 106 | 3.4 × 106 | 76 | |
| pNG-PRL | 3.1 × 105 | 2.1 × 105 | 65 |
After transformation, a 1/1,000 volume of the cell suspension was inoculated on a Luria-Bertani agar plate containing 100 μg of ampicillin per ml incubated overnight. Numbers of independent clones were counted.
Ligation efficiency was obtained by >40 independent clones.
FIG. 2.
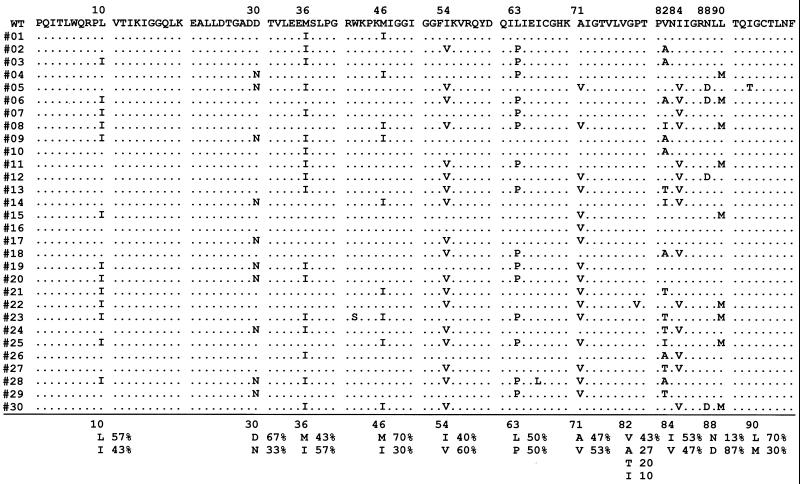
DNA sequences of 30 protease recombinants of the HIV-1 pNG-PRL library. WT, wild type. Thirty randomly selected clones were isolated and sequenced. Conserved nucleotides are indicated by a dot. For DNA sequencing, InstaGene (Bio-Rad Laboratories) was used for preparation of DNA from HIV-1-infected MT-2 cells. A 706-bp fragment containing a viral protease-encoding sequence was amplified by PCR with primers PR5 (5′-GTT AAG TGT TTC AAT TGT GGC AAA GAA GGG C-3′) and PR12 (5′-CTC TGT ACA AAT TTC TAC TAA TGC-3′). The PCR products were purified via 1% agarose electrophoresis and cloned into StuI-digested pCRΔZ vector created from pCR-blunt (Invitrogen) by deletion of a PmlI fragment containing a Zeocin cassette. Cloned sequences were sequenced with ABI Prism 377 (Applied Biosystems, Inc., Foster City, Calif.).
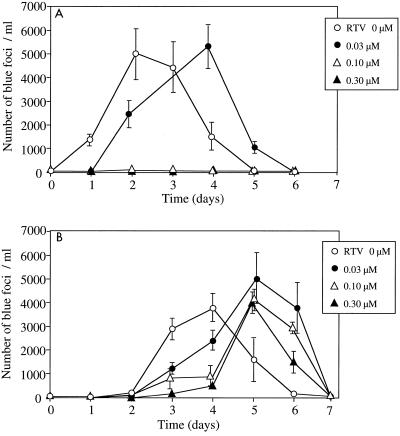
To prepare HIV-1 library, 293T cells were transfected with the HIV-1 library DNA by calcium phosphate precipitation method (5). The library virus was propagated with T-cell line MT-2 and stored at −80°C until use. Replication kinetics of the virus mixture was determined and compared to that of parental virus (Fig. 3). MT-2 cells were used for growth kinetics and selection of protease inhibitor-resistant virus because of their high susceptibility to HIV-1 infection. Infectious virions in the supernatant were evaluated by a single-round viral infectivity assay. HeLa-CD4-LTR-β-gal cells (2 × 104) were inoculated in 24-well plates and incubated for 12 h at 37°C in 5% CO2. The cells were infected with 500 μl of the supernatant for 2 h and incubated for 48 h. Then the cells were treated with 0.5% glutaraldehyde for 5 min and stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid), as described previously (16). The production of infectious virions with wild-type protease in MT-2 cells peaked on day 2 in the absence of ritonavir (Fig. 3A), while the virus carrying mutations in protease was delayed 2 days in reaching the maximum level of infectious virions (Fig. 3B). For sequencing, the generated library virus was transferred to fresh MT-2 cells on days 4 and 8 postinfection. The proviral DNA was subjected to PCR on day 12 to amplify the protease-encoding region (Fig. 4A). HIV-1 clones isolated 2 weeks postinfection contained two to seven amino acid substitutions in protease. L10I, M36I, I54V, L63P, A71V, V82A, and I84V could be maintained in the library population, indicating that these amino acid substitutions did not impair viral replication with primary mutations V82A, I84V, and V82A/I84V. These results were consistent with the previous observation that secondary mutations such as L10I, M36I, I54V, L63P, and A71V restore virus infectivity to the levels comparable to wild type (3, 16, 30). The levels of D30N, N88D, V82T, and L90M could be observed at less than 10% in the clones, which showed that insertion of these mutations in the protease leads to a disadvantage in viral fitness in the absence of specific protease inhibitors.
FIG. 3.
Viral growth of HIV-1 library in the presence or absence of protease inhibitor. After infection of 5 × 105 MT-2 cells with HIV-1NG-WT (A) or HIV-1NG-PRL (B) (200 ng of p24gag antigen) for 3 h, MT-2 cells were incubated in the absence or in the presence of 0.03, 0.10, or 0.30 μM ritonavir (RTV). The supernatant was stored at −80°C until assay. Infectious virions in the supernatant were evaluated by using a single-round viral infectivity assay. Error bars indicate the standard deviation of three experiments.
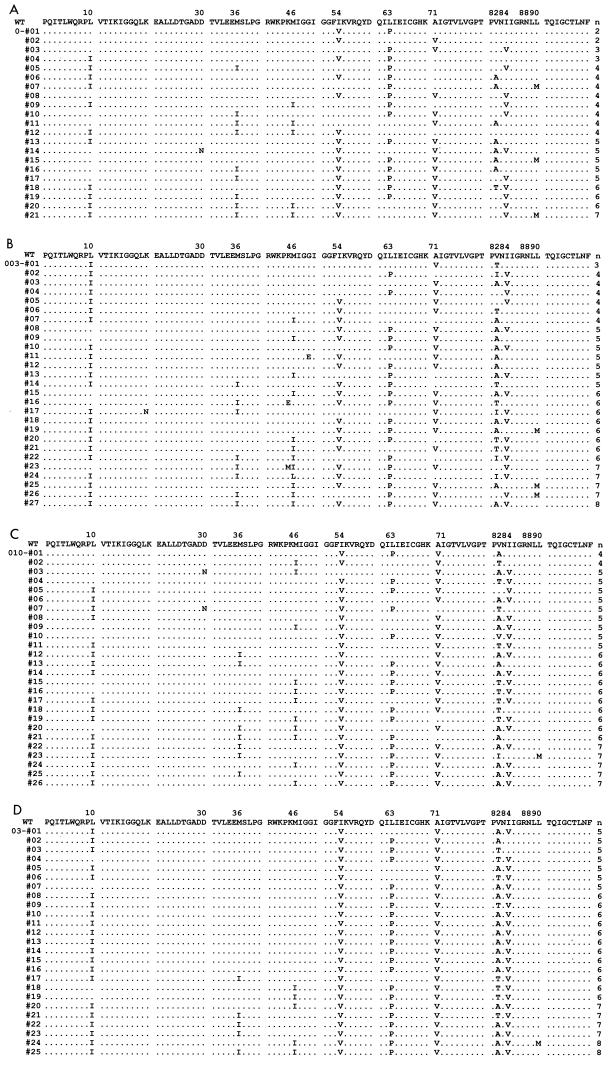
FIG.4.
Combination of amino acid substitutions in viral protease selected with ritonavir on day 12 after infection. Protease sequences of selected HIV-1NG-PRL isolates in the absence (A) or presence (B) of 0.03, 0.10 (C), and 0.30 (D) μM ritonavir are shown. WT, wild type. For DNA sequencing, InstaGene (Bio-Rad Laboratories, Hercules, Calif.) was used for preparation of DNA from HIV-1-infected MT-2 cells. A 706-bp fragment containing a viral protease-encoding sequence was amplified by PCR with primers PR5 (5′-GTT AAG TGT TTC AAT TGT GGC AAA GAA GGG C-3′) and PR12 (5′-CTC TGT ACA AAT TTC TAC TAA TGC-3′). The PCR products, including protease-encoding sequences and the cleavage sites NC/p1, p1/p6gag, TFP/p6pol, and PR/RT (protease/reverse transcriptase), were purified and subcloned in pCRΔZ vectors as described in Fig. 1A.
The 50% inhibitory concentration (IC50) of wild-type HIV-1NG-WT for ritonavir was 0.030 μM in MT-2 cells (data not shown). Therefore, HIV-1NG-WT could replicate with partial replicative suppression in the presence of ritonavir at 0.03 μM (Fig. 3A). However, wild-type virus could not be detected in the 27 HIV-1NG-PRL clones on day 12 after infection. The clones contained three to eight amino acid substitutions, including L10I, M36I, M46I, I54V, L63P, A71V, V82A/T/I, I84V, and L90 M (Fig. 4B). All clones contained at least one primary mutation—V82A, V82T, or I84V—which could confer ritonavir resistance to HIV-1. There were no clones with identical sets of mutations, indicating that various combinations of these substitutions plus the primary mutation(s) could lead to equal or higher fitness compared to the wild type in the presence of 0.03 μM ritonavir. Ritonavir at 0.10 μM could completely block replication of wild-type virus (Fig. 3A), but could not suppress replication of HIV-1NG-PRL (Fig. 3B). Infectious virions were first detected in the culture media at day 3 and peaked on day 5, suggesting that the restricted virus population of HIV-1NG-PRL could survive in this condition. The HIV-1 clones selected with 0.10 μM ritonavir contained a set of four to eight amino acid substitutions including M36I(8/26), M46I(11/26), I54V(25/26), L63P(9/26), A71V(23/26), V82A/T/I(13/26, 6/26, 4/26), I84V(19/26), and L90M(3/26), which had been reported as a ritonavir-associated resistance mutation in patients (Fig. 4B). The combination of mutations treated with 0.30 μM ritonavir resulted in a similar tendency, although all clones contained I54V, A71V, and V82A/T, and the variety of the mutations decreased (Fig. 4C). Previously, in order to estimate the emergence of protease inhibitor-resistant virus in vivo, induction of drug-resistant viruses was performed in vitro. However, the accumulated amino acid substitutions in the viral protease were not always consistent with those found in vivo. Earlier studies found that K20R, L33F, M36I, I54V/L, V82A/T/S, and L90M mutations were not induced by ritonavir in vitro (22, 24, 31). Mutations M36I, M46I, I54V, A71V, V82A/T, I84V, and L90 M in the HIV-1 library were associated with ritonavir resistance found in vivo. With the HIV-1 library, the virus population selected with ritonavir contained combinations of multiple mutations, including M36I, I54V, V82A/T, and L90 M.
It is known that the low fidelity of HIV-1 reverse transcriptase results in a heterogeneous population of HIV-1 in vivo. Based on the estimated mutation rate of HIV-1 (3.4 × 105/bp per replication cycle) and the genome size (104 bp), mutations occur at every position of the genome several times per day (20), although some of them allow impairment of viral replication or a decrease in infectivity. The heterogeneous virus population led to the emergence of drug resistance (29), escape from the host defense system (4, 13), and a variety of cellular tropisms (2, 36). Up until this point, there were few in vitro methods to mimic the heterogeneous HIV-1 population found in vivo. We sought to develop a useful alternative system for screening and analysis of biological properties of HIV-1. Previously, extremely low transformation efficiencies in ligation mixtures containing a large plasmid including HIV-1 proviral DNA had been an obstacle for constructing a useful HIV-1 library. We were able to develop a large HIV-1 library (2.1 × 105 clones) containing a set of random combination of zero to 11 amino acid substitutions in viral protease by using a highly efficient transformation method (15).
It is reported that additional mutations in protease cleavage sites within the viral polyprotein are occasionally accompanied by accumulated mutations in proteases both in vitro and in vivo (7, 9). The viruses selected by ritonavir did not contain any mutations in cleavage sites NC/p1, p1/p6gag, TFP/p6pol, or PR/RT (protease/reverse transcriptase) (data not shown). In order to improve an HIV-1 library to mimic the acquisition of resistant viruses in vivo, it may be necessary to incorporate mutations into the protease cleavage sites, in addition to the protease amino acid substitutions. These HIV-1 libraries may serve as an alternative assay system to elucidate how protease inhibitor resistance mechanisms are acquired and for the development of novel compounds that are designed to avoid cross-resistance to the current generation of the protease inhibitors.
Acknowledgments
We thank Fonda Newcomb for critical review of the manuscript, Yosuke Maeda for valuable discussion, and Mitsuko Honda for technical assistance.
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology and the Ministry of Health, Labor and Welfare, Japan.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. [DOI] [PubMed] [Google Scholar]
- 3.Borman, A. M., S. Paulous, and F. Clavel. 1996. Resistance of human immunodeficiency virus type 1 protease inhibitors: selection of resistance mutations in the presence and absence of the drug. J. Gen. Virol. 77:419-426. [DOI] [PubMed] [Google Scholar]
- 4.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205-211. [DOI] [PubMed] [Google Scholar]
- 5.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Condra, J. H., W. A. Schleif, O. M. Blahy, L. J. Gabryelski, D. J. Graham, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash et al. 1995. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 374:569-571. [DOI] [PubMed] [Google Scholar]
- 7.Côté, H. C. F., Z. L. Brumme, and P. R. Harrigan. 2001. Human immunodeficiency virus type 1 protease cleavage site mutations associated with protease inhibitor cross-resistance selected by indinavir, ritonavir, and/or saquinavir. J. Virol. 75:589-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Debouck, C. 1992. The HIV-1 protease as a therapeutic target for AIDS. AIDS Res. Hum. Retroviruses 8:153-164. [DOI] [PubMed] [Google Scholar]
- 9.Doyon, L., G. Croteau, D. Thibeault, F. Poulin, L. Pilote, and D. Lamarre. 1996. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J. Virol. 70:3763-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El-Farrash, M. A., M. J. Kuroda, T. Kitazaki, T. Masuda, K. Kato, M. Hatanaka, and S. Harada. 1994. Generation and characterization of a human immunodeficiency virus type 1 (HIV-1) mutant resistant to an HIV-1 protease inhibitor. J. Virol. 68:233-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erickson, J. W., and S. K. Burt. 1996. Structural mechanisms of HIV drug resistance. Annu. Rev. Pharmacol. Toxicol. 36:545-571. [DOI] [PubMed] [Google Scholar]
- 12.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 13.Ho, D. D., T. Toyoshima, H. Mo, D. J. Kempf, D. Norbeck, C.-M. Chen, N. E. Wideburg, S. K. Burt, J. W. Erickson, and M. K. Singh. 1994. Characterization of human immunodeficiency virus type 1 variants with increased resistance to a C2-symmetric protease inhibitor. J. Virol. 68:2016-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inoue, H., H. Nojima, and H. Okayama. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23-28. [DOI] [PubMed] [Google Scholar]
- 15.Kaplan, A. H., J. A. Zack, M. Knigge, D. A. Paul, D. J. Kempf, D. W. Norbeck, and R. Swanstrom. 1993. Partial inhibition of the human immunodeficiency virus type 1 protease results in aberrant virus assembly and the formation of noninfectious particles. J. Virol. 67:4050-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobori, M., and H. Nojima. 1993. A simple treatment of DNA in a ligation mixture prior to electroporation improves transformation frequency. Nucleic Acids Res. 21:2782.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuiken, C. L., B. Foley, B. Hahn, B. Korber, F. McCutchan, P. A. Marx, J. W. Mellors, J. I. Mullins, J. Sodroski, and S. Wolinsky (ed.). 2000. Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N. Mex.
- 19.Louis, J. M., I. T. Weber, J. Tozser, G. M. Clore, and A. M. Gronenborn. 2000. HIV-1 protease: maturation, enzyme specificity, and drug resistance. Adv. Pharmacol. 49:111-146. [DOI] [PubMed] [Google Scholar]
- 20.Mansky, L. M., and H. M. Temin. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 69:5087-5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Markowitz, M., H. Mo, D. J. Kempf, D. W. Norbeck, T. N. Bhat, J. W. Erickson, and D. D. Ho. 1995. Selection and analysis of human immunodeficiency virus type 1 variants with increased resistance to ABT-538, a novel protease inhibitor. J. Virol. 69:701-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez-Picado, J., M. P. DePasquale, N. Kartsonis, G. J. Hanna, J. Wong, D. Finzi, E. Rosenberg, H. F. Gunthard, L. Sutton, A. Savara, C. J. Petropoulos, N. Hellmann, B. D. Walker, D. D. Richman, R. Siliciano, and R. T. D'Aquila. 2000. Antiretroviral resistance during successful therapy of HIV type 1 infection. Proc. Natl. Acad. Sci. USA 97:10948-10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Molla, A., M. Korneyeva, Q. Gao, S. Vasavanonda, P. J. Schipper, H. M. Mo, M. Markowitz, T. Chernyavskiy, P. Niu, N. Lyons, A. Hsu, G. R. Granneman, D. D. Ho, C. A. Boucher, J. M. Leonard, D. W. Norbeck, and D. J. Kempf. 1996. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat. Med. 2:760-766. [DOI] [PubMed] [Google Scholar]
- 24.Oroszlan, S., and R. B. Luftig. 1990. Retroviral proteinases. Curr. Top. Microbiol. Immunol. 157:153-185. [DOI] [PubMed] [Google Scholar]
- 25.Palmer, S., R. W. Shafer, and T. C. Merigan. 1999. Highly drug-resistant HIV-1 clinical isolates are cross-resistant to many antiretroviral compounds in current clinical development. AIDS 13:661-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patick, A. K., H. Mo, M. Markowitz, K. Appelt, B. Wu, L. Musick, V. Kalish, S. Kaldor, S. Reich, D. Ho, and S. Webber. 1996. Antiviral and resistance studies of AG1343, an orally bioavailable inhibitor of human immunodeficiency virus protease. Antimicrob. Agents Chemother. 40:292-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ratner, L., W. Haseltine, R. Patarca, K. J. Livak, B. Starcich, S. F. Josephs, E. R. Doran, J. A. Rafalski, E. A. Whitehorn, K. Baumeister, L. Ivanoff, S. R. J. Petteway, M. L. Pearson, J. A. Lautenberger, T. S. Papas, J. Ghrayeb, N. T. Chang, R. C. Gallo, and F. Wong-Staal. 1985. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature 313:277-284. [DOI] [PubMed] [Google Scholar]
- 28.Richman, D. D. 1993. HIV drug resistance. Annu. Rev. Pharmacol. Toxicol. 33:149-164. [DOI] [PubMed] [Google Scholar]
- 29.Rose, R., Y. Gong, J. Greytok, C. Bechtold, B. Terry, B. Robinson, M. Alam, R. Colonno, and P. Lin. 1996. Human immunodeficiency virus type 1 viral background plays a major role in development of resistance to protease inhibitors. Proc. Natl. Acad. Sci. USA 93:1648-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmit, J. C., L. Ruiz, B. Clotet, A. Raventos, J. Tor, J. Leonard, J. Desmyter, E. De Clercq, and A. M. Vandamme. 1996. Resistance-related mutations in the HIV-1 protease gene of patients treated for 1 year with the protease inhibitor ritonavir (ABT-538). AIDS 10:995-999. [DOI] [PubMed] [Google Scholar]
- 31.Shafer, R. W., D. Stevenson, and B. Chan. 1999. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 27:348-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.St Clair, M. H., J. L. Martin, G. Tudor-Williams, M. C. Bach, C. L. Vavro, D. M. King, P. Kellam, S. D. Kemp, and B. A. Larder. 1991. Resistance to ddI and sensitivity to AZT induced by a mutation in HIV-1 reverse transcriptase. Science 253:1557-1559. [DOI] [PubMed] [Google Scholar]
- 33.Tessmer, U., and H.-G. Kräusslich. 1998. Cleavage of human immunodeficiency virus type 1 proteinase from the N-terminally adjacent p6* protein is essential for efficient Gag polyprotein processing and viral infectivity. J. Virol. 72:3459-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tisdale, M., R. E. Myers, B. Maschera, N. R. Parry, N. M. Oliver, and E. D. Blair. 1995. Cross-resistance analysis of human immunodeficiency virus type 1 variants individually selected for resistance to five different protease inhibitors. Antimicrob. Agents Chemother. 39:1704-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshimura, K., R. Kato, K. Yusa, M. F. Kavlick, V. Maroun, A. Nguyen, T. Mimoto, T. Ueno, M. Shintani, J. Falloon, H. Masur, H. Hayashi, J. Erickson, and H. Mitsuya. 1999. JE-2147: a dipeptide protease inhibitor (PI) that potently inhibits multi-PI-resistant HIV-1. Proc. Natl. Acad. Sci. USA 96:8675-8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang, J., A. Gupta, R. Dave, N. M. Yime, B. Zerhouni, and K. Saha. 2001. Isolation of primary HIV-1 that target CD8+ T lymphocytes using CD8 as a receptor. Nat. Med. 7:65-72. [DOI] [PubMed] [Google Scholar]