Abstract
The signal transducer and activator of transcription 3 (STAT-3), a member of the STAT family of proteins, binds to a large number of transcriptional control elements and regulates gene expression in response to cytokines. While it binds to its cognate nucleotide sequences, it has been recently shown to directly interact with other transcriptional factors in the absence of DNA. We report here one such novel interaction between STAT-3 and hepatocyte nuclear factor 3 (HNF-3) in the absence of DNA. We have identified a STAT-3 binding site within the core domain of hepatitis B virus (HBV) enhancer 1. The HBV enhancer 1 DNA-STAT-3 protein interaction is shown to be stimulated by interleukin-6 (IL-6) and epidermal growth factor, which leads to an overall stimulation of HBV enhancer 1 function and viral gene expression. Using mobility shift assays and transient transfection schemes, we demonstrate a cooperative interaction between HNF-3 and STAT-3 in mediating the cytokine-mediated HBV enhancer function. Cytokine stimulation of HBV gene expression represents an important regulatory scheme of direct relevance to liver disease pathogenesis associated with HBV infection.
Signal transducers and activators of transcription (STATs) represent a family of latent transcription factors that are activated upon tyrosine phosphorylation in response to extracellular signals such as cytokines and growth factors (1, 12, 54). Tyrosine-phosphorylated STATs form dimers or multimers through their SH2 domains and are transported into the nucleus, where they bind to the cognate DNA sequences (consensus sequence, TTCC(G/C)GGAA) and activate gene expression (9, 11, 24, 42, 51). A unique feature of STAT-mediated signaling is that the pathway from the cell membrane to the nucleus is traversed by a single molecule (22). The STAT proteins are subsequently inactivated by tyrosine dephosphorylation and are returned to the cytoplasm (18). The three-dimensional structure of STAT-3 has revealed four long helical coils, which include regions involved in protein-protein interaction. The DNA binding regions comprise 300 to 500 residues, a region of beta-sheet structures connected with unstructured loops, and the C-terminal region (75 residues). The latter is implicated in the transactivation function of the protein (4). STAT-3 has been shown to bind other transcriptional factors such as c-Jun, p300/CBP, and Sp1 (31, 37, 53). This mode of protein-protein interaction generally occurs to support synergism between the factors engaged in DNA-protein interactions at neighboring or overlapping sites. Since STAT-3 is activated by cytokine signaling, the protein-protein interactions are also regulated by cytokine signaling. Interleukin-6 (IL-6) and epidermal growth factors (EGF) have been shown to induce STAT-3 activation via tyrosine phosphorylation (27, 54). IL-6 is an important cytokine involved in the activation of acute-phase proteins in the liver (7). It has been shown that IL-6-mediated acute-phase protein (APP) regulation occurs through the JAK/STAT signaling pathway (32, 45). IL-6 also triggers the rapid activation and tyrosine phosphorylation of a latent transcription factor, acute-phase response factor (APRF), which plays an important role in the induction of multiple APPs (47). STAT-3 induces expression of a variety of genes following tissue injury or the acute phase (19, 20). Previously it was reported that the activation of liver-specific gene expression is mediated by STAT-3 and STAT-5 (6, 27). Binding sites for STAT-3 have been identified in a large number of promoters of acute-phase genes. Interestingly, STAT-3 was originally defined as an APRF (1, 48). STAT-3 has recently been shown to possess oncogenic potential (5). In a variety of tumors and transformed cell lines, STAT-3 is constitutively activated. Activation of STAT-3 is implicated in inducing genes, which are linked to both proliferation and apoptosis (5).
Hepatitis B virus (HBV) is a small DNA virus which replicates via a reverse transcriptase enzyme and predominantly infects human hepatocytes. HBV chronic hepatitis has the potential to proceed to the development of hepatocellular carcinoma (3). Expression of the HBV gene is regulated by the interaction between trans-acting cellular factors and enhancer element 1, located at a strategic site within the viral genome (Fig. 1). The enhancer element 1 spans nucleotides (nt) 970 to 1240 and has been shown to play an important role in regulating the activity of HBV promoter elements in a liver-specific manner (2, 23, 26, 44). The enhancer function in the hepatocytes results from the combinatorial action of both ubiquitous and liver-enriched transcriptional factors (14, 15, 17, 38). Chief among these are hepatocyte nuclear factor 3 (HNF-3), HNF-4, RXRα:PPAR heterodimers, and C/EBP. Mutational analysis supports the conclusion that a central region of enhancer 1 located at nt 1080 to 1165 (HBV subtype adw2), termed the enhancer core domain (29), plays a central role in enhancer 1 activity. This enhancer core domain harbors binding sites for HNF-3, HNF-4, the retinoic acid receptor RXRα, and nuclear factor 1 (NF-1) (8, 25). The HNF-3 isoforms (α, β, and γ) are liver-enriched members of the HNF-3/forkhead protein family (30, 49) and have been implicated in the regulation of tissue-specific gene expression and developmental processes (21, 30). We have previously demonstrated that HNF-3β is the predominant isoform of HNF-3 that interacts with the enhancer core domain (28). Using oligonucleotide chromatography with HNF-3 binding sequences, an additional cellular factor was copurified and interacted with the neighboring sequence (AAGGCCTT). This factor was tentatively designated the palindrome binding factor (PBF). The PBF nucleotide sequence bears striking homology to the consensus STAT-3 sequence. Using a myriad of approaches, STAT-3 interaction with the previously designated PBF site has been established and shown to respond to cytokine signaling. Our investigation further revealed a direct interaction between STAT-3 and HNF-3, thus expanding the repertoire of transcriptional factors STAT-3 interacts with. STAT-3 and HNF-3 binding cooperatively stimulates the enhancer function in response to IL-6 and EGF. Cytokine-mediated signaling of HBV gene expression has implications in the liver disease pathogenesis associated with HBV infection.
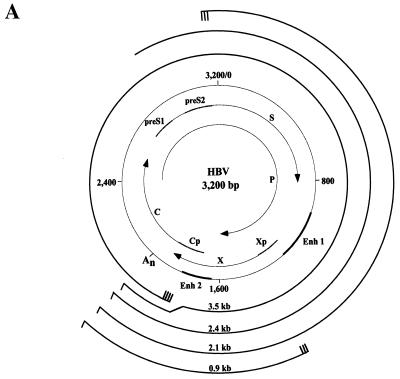
FIG. 1.
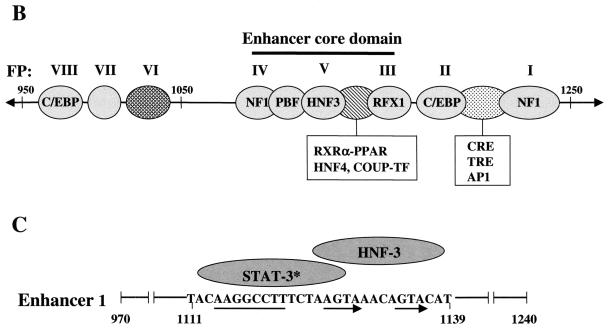
Schematic representations of the HBV genome and enhancer 1 element. (A) The viral genome is numbered (0 to 3,200 bp) according to the adw2 subtype of HBV. S, C, P, and X represent the viral genes encoding the surface antigen, core or e antigen, DNA polymerase, and X proteins, respectively. The HBV promoters (preS1, preS2, Cp, and Xp) and enhancers (Enh 1 and Enh 2) are indicated. An represents the single polyadenylation site used by all of the HBV RNAs. Each viral transcript is labeled with its respective length in kilobases (kb) and a poly(A) tail at the 3′ end. Multiple transcription initiation sites are indicated at the 5′ end of the 3.5-, 2.1-, and 0.9-kb RNAs. (B) Protein binding sites on Enh 1 are shown and footprint designations previously defined (38) are indicated above the corresponding sites. (C) The core domain spans nt 1080 to 1165 and is composed of the STAT-3 binding site, which is located at nt 1111 to 1127. The HNF-3 binding site spans nt 1124 to 1139. The DNA sequence of the STAT-3/HNF-3 region is shown with the corresponding cellular factors as indicated above. The thin line and arrows below the STAT-3-HNF-3 region indicate the positions of STAT-3 and FPV direct repeats, respectively.
MATERIALS AND METHODS
Plasmids and oligonucleotides.
Construction of plasmids containing the wild-type footprint V (FPV) of the HBV enhancer 1 and a point mutant FPV has been described previously (28). The FPV spans nt 1111-1139 within enhancer 1. The point mutation in FPV, designated as FPV′, is indicated by lowercase letters (5′-TACAAGGCCTTTCTAAcgAAACAtcACAT-3′). The wild-type STAT-3 (GATCCTTCTGGGAATTCCTAGATC), mutant STAT-3 (GATCCTTCTGGGCCGTCCTAGATC), wild-type PBF (CACTTACAAGGCCTTTCTAAGTAA), and mutant PBF (CACTTACAAGGCCgggCTAAGTAA) oligonucleotides were synthesized from Santa Cruz Biotechnology. The plasmid pHnLuc contains the HBV full-length enhancer 1-X-promoter sequences, and the plasmid p′SLuc contains mutated PBF-STAT-3 binding sites within the FPV (TACAAGGCCgggCTAAGTAAACAGTACAT); both are linked to luciferase reporter gene sequences.
Bacterial expression of phosphorylated STAT-3 proteins.
Tyrosine-phosphorylated STAT-3 was purified from bacterial culture (Epicurian Coli) that coexpresses STAT-3 and JAK kinase (4). Bacterial cultures were grown in the presence of ampicillin and tetracycline and induced in the presence of 1 mM isopropyl-β-d-thiogalactopyranoside for the expression of STAT-3. The harvested cells were suspended in M9 medium (1 mM MgSO4, 0.2% glucose, 0.1% Casamino Acids, 0.5 mg of 1% thiamine-HCl, 0.01 mg of 3-β-indoleacrylic acid/ml, 50 μg of ampicillin/ml, 12.5 μg of tetracycline/ml). The pellet was resuspended in ice-cold extraction buffer (20 mM HEPES [pH 7.6], 0.1 M KCl, 10% glycerol, 1 mM EDTA, 10 mM MnCl2, 20 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride [PMSF]). The lysis was performed by sonication, followed by centrifugation. Ammonium sulfate was added at 35% saturation. The precipitated protein was collected by centrifugation, suspended in a buffer (20 mM HEPES [pH 7.0], 200 mM NaCl, 10 mM MgCl2, 5 mM DTT, 0.5 mM PMSF), and dialyzed against the same buffer.
Preparation of nuclear extracts.
Nuclear lysates were prepared from human hepatoma cell line 7 (Huh-7) cells, and cells were transfected with v-Src expression vector, which constitutively phosphorylates STAT-3 (50, 55). Cells were lysed in hypotonic buffer (20 mM HEPES [pH 7.9], 10 mM KCl, 0.1 mM Na3VO4, 1 mM EDTA, 10% glycerol, 1 mM PMSF, 3 mg of aprotinin/ml, 1 mg of pepstatin/ml, 20 mM NaF, 1 mM DTT with 0.2% NP-40) on ice for 10 min. After centrifugation at 4°C (13,000 rpm in a 5415D microcentrifuge) for 1 min, the nuclear pellet was resuspended in high-salt buffer (hypotonic buffer with 20% glycerol and 420 mM NaCl) at 4°C by rocking for 30 min following centrifugation. The supernatant was collected and stored at −80°C in aliquots.
EMSA.
The FPV, PBF, and STAT-3 consensus sequences were radiolabeled at the 5′ end with [γ-32P]ATP by T4 polynucleotide kinase. About 20,000 cpm of gel-purified probe was incubated with bacterially expressed phosphorylated STAT-3- or v-Src-transfected lysates in electrophoretic mobility shift assay (EMSA) buffer (20 mM Tris-HCl [pH 7.9], 10 mM MgCl2, 50 mM KCl, 16.7 μg of poly(dI-dC)/ml, 1 mM EDTA, 1 mM DTT, 1 μM leupeptin) for 20 min on ice. Competition analyses carried out in the presence of 100-fold molar excesses of unlabeled competitor oligonucleotide were preincubated for 20 min on ice prior to the addition of radiolabeled probe. The DNA-protein complexes were resolved by 5% polyacrylamide gel electrophoresis (PAGE) in 0.5× Tris-borate-EDTA buffer. The gels were dried and subjected to autoradiography.
DNase I footprinting.
The DNase I footprinting assay was carried out essentially as described by Kosovsky et al. (28) in the presence of an enhancer 1 DNA fragment spanning nt 1074 to 1308 of the HBV genome. The enhancer 1 was 5′ end labeled by T4 polynucleotide kinase in the presence of [γ-32P]ATP (7,000 Ci/mmol; ICN Pharmaceuticals, Inc.). Approximately 25,000 cpm of the probe was incubated with bacterially synthesized STAT-3 protein and subjected to DNase I digestion followed by 8%-urea PAGE. Gels were dried and exposed to X-ray film for 24 to 48 h at −80°C.
Purification of GST fusion proteins.
The glutathione transferase (GST) and GST-HNF-3 fusion proteins were purified by the procedure recommended by Pharmacia. Briefly, the bacterial cultures expressing these fusion proteins were centrifuged and the pellets were resuspended in lysis buffer (20 mM HEPES [pH 7.9], 1 mM EDTA, 1% Triton X-100, 1% Tween 20, 1% NP-40, 1 mM PMSF, 1 mM ATP, 1 mM DTT) with 1 M NaCl. After sonication, the lysates were centrifuged and the supernatant was incubated with glutathione-Sepharose beads (Pharmacia) for 1 h at 4°C. The beads were washed several times with lysis buffer containing decreasing concentrations of NaCl (1.0 to 0.2 M). The washed beads were equilibrated with binding buffer (150 mM KCl, 6 mM MgCl2, 25 mM HEPES [pH 7.9], 10% glycerol, 0.1% NP-40, 1 mM ATP, 1 mM DTT, 1 mM PMSF, 10 μg of leupeptin/ml, 9 μg of aprotinin/ml) for 30 min at 4°C.
Immunoprecipitation and Western blot analysis.
Exponentially growing Huh-7 cells transfected with v-Src expression vector, which activates STAT-3 (55), were harvested, and cell extracts were prepared by incubation in radioimmune precipitation buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1 mM sodium orthovanadate, 1 mM sodium formate, 1 mM PMSF, 10 μg of aprotinin/ml, 10 μg of leupeptin/ml) for 30 min on ice. Immunoprecipitation was performed with anti-STAT-3 serum for 4 h. The immune complexes were incubated with protein A-Sepharose, washed three times with radioimmune precipitation buffer, and boiled for 5 min in SDS-PAGE sample buffer. The samples were subjected to SDS-PAGE. Gels were electroblotted onto polyvinylidene difluoride (PVDF) membrane (Amersham) in 25 mM Tris-192 mM glycine-20% methanol by electrophoresis. Membranes were treated for 1 h in blocking buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.3% polyvinyl pyrrolidone, 0.5% [wt/vol] Tween 20), probed with anti-HNF-3 serum overnight, and washed twice for 10 min with blocking buffer followed by incubation with secondary antibody for 45 min. After an additional washing step with blocking buffer, immunoblots were visualized using an ECL detection system (Amersham).
Northern blot analysis.
Total RNA was isolated using RNA STAT-60 (Tel-Test, Inc., Friendswood, Tex.) from HepG2 cell line 2.2.15, which stably expresses all HBV genes (43), and treated with EGF and IL-6 at various time intervals. Northern blot analysis was performed according to standard methods. Equal amounts (15 μg) of electroblotted RNA were hybridized with γ-32P-labeled HBV DNA (48 bp of X cDNA sequence) overnight and washed twice in 2× SSC (1× SSC is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) and 0.1% SDS, followed by 1× SSC, 0.1% SDS for 15 min, and finally 0.1× SSC and 0.1% SDS for 2 × 10 min at 65°C before being subjected to autoradiography.
Luciferase assay.
For transient transfections, Huh-7 cells were plated at a density of ∼5 × 105 cells/60-mm dish and maintained in Dulbecco's modified Eagle's medium containing 10% fetal calf serum, penicillin (75 units/ml), and streptomycin (50 units/ml) at 37°C. Cells (∼50% confluent) were transfected with 1 μg of luciferase reporter plasmid by using Lipofectin reagent (Life Technologies). Thirty-six hours after transfection, cells were serum starved overnight before the addition of IL-6 (30 ng/ml) and EGF (100 ng/ml) for 15 min. The cells were harvested, and cellular lysates were analyzed for luciferase expression using a luminometer (13).
RESULTS
STAT-3 binds to the core domain of HBV enhancer 1.
Kosovsky et al. (28) have previously described the interaction of a factor, tentatively designated PBF, which copurified with HNF-3 and bound to the nucleotide sequence nt 1111 to 1121 (5′-AAGGCCTT-3′) within HBV enhancer 1. This PBF-binding sequence bore striking homology to the consensus STAT sequence, in particular to STAT-3. To investigate possible interaction between the STAT-3 protein and the PBF sequence that resides within the FPV region of enhancer 1, EMSAs were carried out. Tyrosine-phosphorylated STAT-3 was purified from bacteria that were doubly transformed with STAT-3 and JAK kinase vectors. Using the purified STAT-3 proteins, an EMSA was carried out to demonstrate its interaction with the PBF site of HBV enhancer 1 (36). The results shown in Fig. 2A clearly demonstrate the formation of a DNA-protein complex between FPV and STAT-3 proteins. The specificity of this interaction was confirmed by using unlabeled competitor oligonucleotides representing the FPV sequence, consensus STAT-3, and PBF. In each case, a mutant version (see Materials and Methods) of the competitor nucleotides was also included. FPV encompasses a 29 nt sequence (nt 1111 to 1139). The PBF site is located within the footprint that includes the nucleotide sequence between 1111 and 1121. It is clear from the results (Fig. 2A) that mutant oligonucleotides fail to compete with STAT-3-FPV DNA complex formation. To further confirm the presence of STAT-3 in the DNA-protein complexes observed in Fig. 2A, an antibody against STAT-3 was included during the EMSA. A supershift of DNA-protein complex in the presence of STAT-3 is shown in Fig. 2B, whereas an unrelated serum failed to produce a similar supershift of the complex. The direct interaction between STAT-3 proteins and PBF oligonucleotide probe during the EMSA is shown in Fig. 2C. These results clearly establish direct binding of STAT-3 to the PBF nucleotide sequence within the FPV of viral enhancer 1.
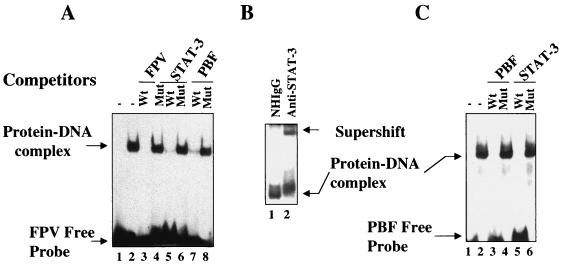
FIG. 2.
STAT-3 binds within HBV enhancer 1. (A) The EMSA was carried out in the presence of 32P-labeled FPV probe and STAT-3 protein synthesized in bacteria. Lane 1, probe alone; lane 2, 1 μg of STAT-3 protein; lanes 3, 5 , and 7 , 100-fold excesses of wild-type unlabeled FPV, STAT-3, and PBF oligonucleotides, respectively; lanes 4, 6, and 8, 100-fold excesses of mutant FPV, STAT-3, and PBF oligonucleotides, respectively. (B) The EMSA was carried out as described above but with the inclusion of normal human IgG (lane 1) and anti-STAT-3 serum (lane 2). (C) The EMSA was carried out in the presence of 32P-labeled PBF probe. Lane 1, probe alone; lane 2, 1 μg of STAT-3 protein; lanes 3 and 5, 100-fold excesses of unlabeled wild-type PBF and STAT-3 oligonucleotides, respectively; lanes 4 and 6, 100-fold excesses of mutant PBF and STAT-3 oligonucleotides, respectively.
Next, we used lysates from Huh-7 cells that were transfected with the v-Src expression vector during the EMSA. The STAT-3 in the hepatoma cell lysates was able to form DNA-protein complexes that were completely inhibited in the presence of a 100 M excess of wild-type FPV, STAT-3, and PBF unlabeled competitor oligonucleotides. The results shown in Fig. 3 further support the STAT-3 binding at PBF site shown in Fig. 2. To confirm the presence of STAT-3 and HNF-3 in the DNA-protein complex, an antibody against STAT-3 and HNF-3 was included during the EMSA (Fig. 3, lanes 11 and 12).
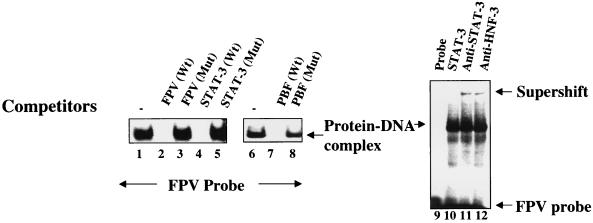
FIG. 3.
Interaction between STAT-3 and FPV of HBV enhancer 1. The EMSA was carried out in the presence of 32P-labeled FPV probe and lysates from Huh-7 cells transfected with v-Src. Lanes 1 and 6, 5 μg of lysates as a source of STAT-3; lanes 2, 4, and 7, 100-fold excesses of wild-type FPV, STAT-3, and PBF oligonucleotides, respectively; lanes 3, 5, and 8, 100-fold excesses of mutant FPV, STAT-3, and PBF oligonucleotides, respectively; lanes 11 and 12, protein-DNA complex incubated with anti-STAT-3 and anti-HNF-3 serum, respectively.
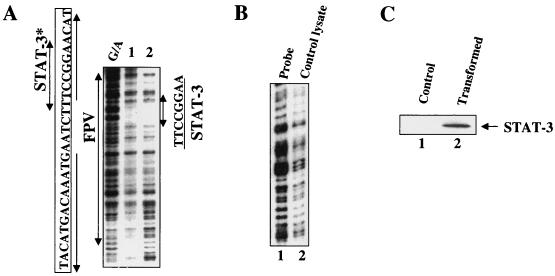
Next, DNase I protection analysis was carried out to identify the nucleotide sequences occupied by STAT-3. A footprint was formed at nt 5′-AAGGCCTT-3′ by using the bacterially synthesized STAT-3 protein preparation (Fig. 4A, lanes 1 and 2). Shown in Fig. 4C is the Western blot of the bacterial lysates expressing STAT-3 protein. Lysates from bacteria lacking STAT-3 did not produce a footprint at that sequence within the FPV (Fig. 4B, lane 2). Together, these results establish STAT-3 interaction at the previously described PBF site, which resides within the FPV.
FIG. 4.
DNase I protection analysis of STAT-3 binding. (A) Samples were analyzed in the presence of the enhancer 1 probe (25,000 cpm; labeled at nt 1308). Lanes 1 and 2 contained 50 and 100 μg of STAT-3 protein, respectively. G/A, sequencing ladder. The STAT-3 binding site is shown (nt 1111 to 1121) with a double-headed arrow indicating an 8-bp palindrome. ∗, previously designated as the PBF binding site (28). (B) DNase I protection assay. Lane 1, probe; lane 2, probe incubated with bacterial lysates lacking STAT-3 expression vector. (C) Western blot analysis. Bacterial lysates lacking STAT-3 vector (lane 1), doubly transformed with STAT-3 and JAK vectors (lane 2) were subjected to SDS-7.5% PAGE followed by immunoblot analysis using anti-STAT-3 polyclonal antibodies.
EGF and IL-6 stimulate HBV enhancer function through STAT-3.
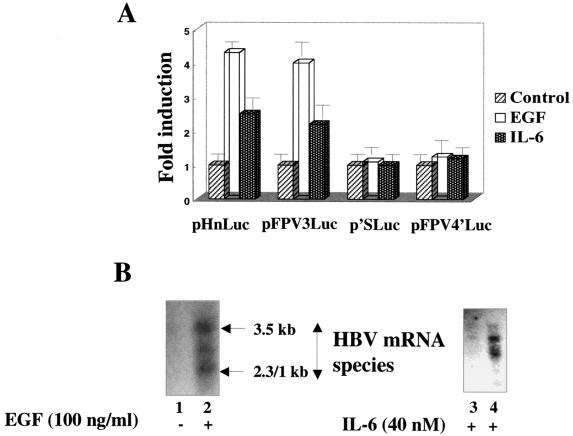
Since STAT-3 activation is triggered by cytokine signaling (54), we addressed this issue using transient schemes of transfection. Four target luciferase reporter vectors were employed in this study. These include a full-length HBV enhancer 1-X promoter (pHnLuc), a STAT-3 containing a multimeric FPV sequence (pFPV3Luc), a mutant version of the FPV sequence (pFPV4′Luc) and a mutation in the STAT-3/PBF binding site (p′SLuc), all of which were cloned upstream of the luciferase reporter gene. Huh-7 cells were transfected with these vector DNAs. Thirty-six hours after transfection, cells were serum starved overnight and then treated with EGF (100 ng/ml) and IL-6 (30 ng/ml) for 15 min. Lysates were prepared, and luciferase expression was measured. The results presented in Fig. 5A demonstrate an approximately fourfold stimulation of FPV3Luc activity in response to EGF. IL-6 signaling produced a relatively lower level (2.5- to 2.2-fold) of stimulation of the STAT-3 site associated with the HBV enhancer. The FPV point mutation and STAT-3 and PBF site mutations did not respond to either cytokine (Fig. 5A).
FIG. 5.
(A) Cytokines upregulate the STAT-3-dependent stimulation of HBV enhancer 1 in vivo . Huh-7 cells were transiently transfected with pHnLuc, pFPV3Luc, pFPV4′Luc, and p′SLuc reporter plasmids, followed by stimulation with EGF and IL-6 as described in Materials and Methods. The experiments were performed in duplicate. Luciferase activity is expressed as fold induction over the baseline level of activity observed during transfection of these vectors without EGF and IL-6 treatment. Each reporter plasmid is indicated below the graph. (B) Cytokine-mediated stimulation of HBV mRNA biosynthesis. Equal amounts of RNA from 2.2.1.5 cells either untreated or treated with EGF and IL-6 were analyzed by Northern blot analysis. RNA was transferred onto a nitrocellulose filter and probed with 32P-labeled 48-bp HBV DNA representing the X gene. Lane 1, RNA before stimulation was used as control; lane 2, incubation with EGF (6 h); lanes 3 and 4, stimulation with IL-6 for 6 and 12 h, respectively. Arrows indicate the 3.5- and 2.3- and 2.1-kb HBV mRNA species.
Since HBV enhancer 1 is generally believed to regulate overall HBV gene expression, we set out to determine the effect of EGF on the viral mRNA biosynthesis. In this analysis, we performed Northern blot analysis of a HepG2 2.2.15 cell line that stably expresses the whole HBV genome. The 2.2.15 cell line contains a dimer molecule of the HBV genome, thus achieving the continuity of the circular HBV DNA genome (43). HepG2 2.2.15 cells were incubated with EGF and IL-6. A typical pattern of the HBV transcripts shows a significant increase in the presence of EGF and IL-6 (Fig. 5B, lanes 2 and 4). This analysis indicates the abilities of EGF and IL-6 to positively influence overall HBV gene expression. Additional STAT-3 cognate sequences were not readily apparent elsewhere in the HBV genome.
Direct interaction between STAT-3 and HNF-3.
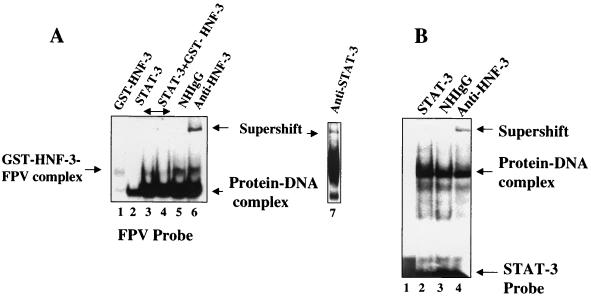
Since the putative STAT-3 site resides close to the previously defined HNF-3 binding site within FPV (28), we investigated possible cooperative interactions between these proteins. Bacterially synthesized STAT-3 and GST affinity-purified HNF-3 proteins were used in the EMSA. GST-HNF-3 at a certain low concentration barely formed the DNA-protein complex (Fig. 6A, lane 1). In the presence of increasing concentrations of HNF-3, there was a significant increase in the intensity of the STAT-3-FPV DNA complex (compare Fig. 6, lane 2 with lanes 3 and 4). To determine if HNF-3 was also present in the complex, an anti-HNF-3 serum was included during the EMSA, which resulted in retardation of DNA-protein complexes (Fig. 6, lane 6). Similarly, anti-STAT-3 serum further shifted the complex (Fig. 6, lane 7). A normal human immunoglobulin G (IgG) failed to produce a similar supershift (Fig. 6, lane 5). Such cooperation was not observed with GST protein, and anti-GST serum did not further shift the migration of the complex (data not shown). Since the probe used in these assays contained FPV sequences, which also include the HNF-3 binding site, it is not surprising that the DNA-protein complex contained HNF-3 protein. We next used a STAT-3 consensus oligonucleotide probe devoid of an HNF-3 binding site during the EMSA. Huh-7 cell lysates transfected with v-Src vector, which constitutively activates STAT-3 (50, 55), were used in this analysis. The results presented in Fig. 6B show that HNF-3 was also present in the DNA-protein complex, which includes the STAT-3 protein and a STAT-3 consensus oligonucleotide as evidenced by a supershift of the DNA-protein complex (Fig. 6, lane 4). GST-HNF-3 itself did not bind the STAT-3 probe in the EMSA (data not shown). These results suggest that HNF-3 and STAT-3 probably form a protein-protein complex without binding to the DNA.
FIG. 6.
Cooperation of interaction between STAT-3 and HNF-3. (A) The EMSA was carried out in the presence of 32P-labeled FPV probe. Binding of FPV probe with recombinant GST-HNF-3 (lane 1); with STAT-3 synthesized in bacteria (lane 2); both STAT-3 and increasing concentration of HNF-3 (lanes 3 and 4); same as shown in lane 4 but incubated with normal human IgG (lane 5); or with anti-HNF-3 serum (lane 6) and with anti-STAT-3 serum (lane 7). (B) STAT-3-HNF-3 complex interacts with STAT-3 oligonucleotide probe. The EMSA was carried out in the presence of 32P-labeled STAT-3 oligonucleotide probe and nuclear lysates from HepG2 cells transfected with v-Src vector. Lane 1, probe alone; lane 2, 5 μg of lysates (source of STAT-3-HNF-3 complex); DNA-protein complex treated with normal human IgG (lane 3) or with anti-HNF-3 serum (lane 4).
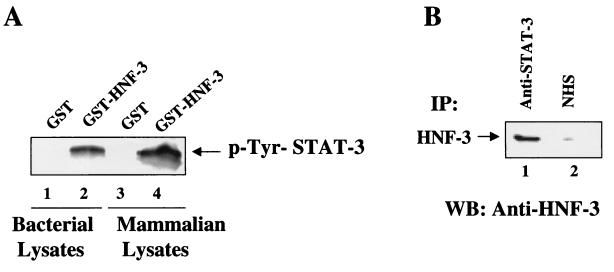
To examine the direct physical interaction between STAT-3 and HNF-3 in the absence of DNA, v-Src-transfected Huh-7 lysates were incubated with GST-HNF-3 protein. GST-HNF-3 bound proteins were immobilized on GST beads, washed extensively, and eluted. The eluate was then subjected to SDS-PAGE and immunoblotted with anti-STAT-3 antibody. The results are shown in Fig. 7A, in which STAT-3 is shown to interact with GST-HNF-3 (lane 4) but not with GST (lane 3). When bacterially synthesized STAT-3 was examined for direct binding to GST-HNF-3, similar results were obtained (Fig. 7, lanes 1 and 2). To determine if such complexes are formed in vivo, v-Src-transfected Huh-7 lysates were immunoprecipitated with anti-STAT-3 serum. The immunoprecipitates were fractionated by SDS-PAGE and electroblotted onto a PVDF membrane. The membrane was then incubated with anti-HNF-3 serum and visualized by ECL, a commercial detection kit. The results shown in Fig. 7B demonstrate that HNF-3 and STAT-3 form a heteromeric complex in vivo . Together, these results clearly support the view that there is a direct physical interaction between HNF-3 and STAT-3 proteins in Huh-7 cells in the absence of STAT-3 cognate DNA sequences.
FIG. 7.
(A) Direct interaction between STAT-3 and HNF-3. Nuclear extract from Huh-7 cells transfected with v-Src expression vector or STAT-3 synthesized in bacteria was incubated with GST (lanes 1 and 3) and GST-HNF-3 (lanes 2 and 4) bound to glutathione-Sepharose beads. STAT-3 bound to HNF-3 was extensively washed and subjected to SDS-7.5% PAGE followed by immunoblot analysis using anti-STAT-3 polyclonal antibodies. (B) In vivo interaction between STAT-3 and HNF-3. Huh-7 v-Src-transfected lysates were immunoprecipitated (IP) with anti-STAT-3 serum (lane 1) and normal human serum (NHS) (lane 2) fractionated by SDS-PAGE and electroblotted onto a PVDF membrane. The membrane was incubated with anti-HNF-3 serum followed by incubation with a secondary antibody and analyzed by ECL, a commercial detection kit.The arrow indicates the coimmunoprecipated HNF-3 in the STAT-3-HNF-3 in vivo complex.
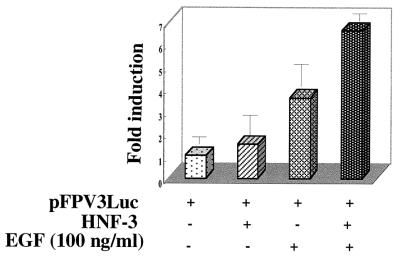
Next, we examined cooperative interaction between HNF-3 and STAT-3 in a cell-based assay. Huh-7 cells incubated with EGF, which activates STAT-3, stimulated luciferase reporter activity under the control of the HBV FPV (STAT-3) sequence. When cells were cotransfected with the HNF-3 expression vector and FPV-containing luciferase reporter plasmid and incubated in the presence of EGF, a 6.5-fold stimulation was observed (Fig. 8). The FPV mutant reporter construct did not show any activation (data not shown). Similar results were obtained with IL-6, which also stimulates STAT-3 (data not shown).
FIG. 8.
Cooperative activation of HBV Enh 1 by STAT-3 and HNF-3. Huh-7 cells were transiently transfected with 1 μg each of pFPV3Luc reporter plasmid and HNF-3 vectors. At 36 h after transfection, the cells were serum starved overnight and then stimulated with EGF (100 ng/ml) for 15 min. The cells were harvested and analyzed for luciferase activity. Each transfection was carried out in duplicate and repeated at least three times.
In summary, these studies show that STAT-3 binds to its cognate sequence within enhancer 1 at FPV. HNF-3 and STAT-3 heteromeric complexes formed in vivo further contribute to stimulation of the HBV enhancer function.
DISCUSSION
This study was initially undertaken to demonstrate the binding of STAT-3 to a suspect sequence, previously identified as the PBF site, that displayed striking homology to the STAT-3 site. Evidence for this interaction was obtained using a myriad of standard approaches which unambiguously establish the binding of STAT-3 to the PBF site (PBF site, 5′AAGGCCTT3′; consensus STAT-3 site, 5′TTCC(G/C)GGAA3′) in a critical domain within the HBV enhancer. Enhancer 1 is believed to control overall HBV gene expression in a liver-specific manner and is the sequence to which a large number of liver-specific and ubiquitous transcription factors have been shown to bind (29) (Fig. 1B).
A cooperative interaction between STAT-3 and HNF-3 during both the EMSA and transient transfection was observed. These results demonstrate synergism between STAT-3 and HNF-3 in bringing about a stimulatory role in these interactions. A further augmentation of STAT-3-mediated transcription was observed in the presence of IL-6 and EGF. Therefore, we investigated the mechanism responsible for STAT-3-mediated enhancer 1 activity, since HBV infection initially results in acute hepatitis. Our transient transfection data revealed the stimulation of HBV enhancer 1 via STAT-3 in response to EGF and IL-6 (Fig. 5A). Previously, a similar level of induction of the reporter gene expression under the control of HBV enhancer by IL-6 has been reported (35).
We further demonstrate a direct protein-protein interaction between HNF-3 and STAT-3 in vivo. The ability of STAT-3 to bind other transcription factors such as c-Jun, CBP, Sp1, SRF, AP1, and CREB has been reported previously (31, 37, 40, 53). This novel property of STAT-3 to enter into heteromeric complexes with other transcription factors is intriguing and may suggest its participation in the formation of preenhanceosome. In that scenario, STAT-3 is implicated in fulfilling a key role in active enhanceosome assembly (33, 39). Our future work will examine this issue in close detail by mapping the interactive sites of each protein and identifying the exact amino acid residues in the STAT-3 molecule involved in this phenomenon in light of the X-ray structure of STAT-3 (4).
In transient transfection, the reporter plasmids containing point mutations in the STAT-3 (p′SLuc) or the HNF-3 (pFPV4′Luc) binding site did not show any stimulation. This result was expected for the STAT-3-mutated site, but the basal level of stimulation with the HNF-3-mutated site suggests that the transcription factor binding site studied here functions as a component of an interdependent transcription control system in the HBV enhancer. Analysis of the competitive binding of STAT-3 protein to FPV probe during the EMSA demonstrates that both the unlabeled STAT-3 and PBF oligonucleotides are competing for the STAT-3 protein. DNase I footprint analysis shows the palindrome region of the HBV enhancer 1 (-AAGGTTCC-) that is occupied by STAT-3.
Since many cytokines can activate STAT molecules, the role of STATs in growth control is crucial. Several studies suggest that STAT-3 may be involved in promoting cell growth, since IL-6-mediated activation of STAT-3 is required in liver regeneration (10). The biological mechanism by which STAT-3 contributes to oncogenesis remains to be determined. Two possible mechanisms have been postulated. (i) Constitutive activation of STAT-3 signaling may directly stimulate cell proliferation, because constitutively activated STAT-3 has been reported in cells and tissues from a number of human tumors (5, 52). This possibility is consistent with the finding that numerous growth factors (e.g., EGF and platelet-derived growth factor) activate signaling by STATs, including STAT-3 (46, 54). (ii) STAT-3 may contribute to cell transformation by preventing apoptosis. This possibility is supported by the finding that activation of STAT-3 leads to an antiapoptotic response upon IL-6 stimulation in a murine myeloid cell line. Dominant negative STAT-3 lacks the antiapoptotic property (16). Since STAT-3 signaling is implicated in the control of cell differentiation, proliferation, and apoptosis (10, 34), the biological consequences of constitutive STAT-3 activation are likely to be complex and specific to the cell type being studied. This activity may also confer resistance to conventional therapies that rely on apoptotic mechanisms to kill tumor cells, thereby indirectly increasing cell numbers. Thus, identification and characterization of the nuclear genes regulated by STAT-3 should provide new insight into the specific events leading to the loss of normal growth control and the process of malignant transformation. In a recent study, it was observed that the X protein encoded by the HBV genome constitutively activated STAT-3 (46a). This finding implicates a potential role of HBV gene expression in regulating STAT-3 in HBV-associated liver disease pathogenesis.
The present study identifies STAT-3 as an important factor in overall HBV gene expression. The role of cytokine-mediated STAT-3 action in acute and chronic hepatitis is of significance in hepatitis B infection, as these activities determine the onset of liver disease. The interaction between HNF-3 and STAT-3 indicates the complex nature of the ways in which STAT-3 functions as an activator of the HBV gene expression.
Acknowledgments
We thank R. Costa for anti-HNF-3 serum and Brent H. Cochran for anti-STAT-3 serum. Bacterial STAT-3 expression vector was a gift of C. M. Muller.
This research was supported by NIH grants CA 78769 and CA 64415 to A.S.
REFERENCES
- 1.Akira, S., Y. Nishio, M. Inoue, M. Wang, X. J. Wei, S. Matsusaka, T. Yoshida, T. Sudo, M. Naruto, and T. Kishimoto. 1994. Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell 77:6371.. [DOI] [PubMed] [Google Scholar]
- 2.Antonucci, T. K., and W. J. Rutter. 1989. Hepatitis B virus (HBV) promoters are regulated by the HBV enhancer in a tissue-specific manner. J. Virol. 63:579-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beasely, R. P., and L. Y. Hwang. 1984. Epidemiology of hepatocellular carcinoma. In G. N. Vyas et al. (ed.), Viral hepatitis and liver diseases. Grune & Stratton, New York, N.Y.
- 4.Becker, S., B. Groner, and C. W. Muller. 1998. Three-dimensional structure of the Stat3β homodimer bound to DNA. Nature 349:145-151. [DOI] [PubMed] [Google Scholar]
- 5.Bromberg, J. F., M. H. Wrzeszczynska, G. Devgan, Y. Zhao, R. G. Pestell, C. Albanese, and J. E. Darnell, Jr. 1999. Stat3 as an oncogene. Cell 98:295-303. [DOI] [PubMed] [Google Scholar]
- 6.Cantwell, C. A., E. Sterneck, and P. F. Johnson. 1998. Interleukin-6-specific activation of the C/EBPδ gene in hepatocyte is mediated by Stat3 and Sp1. Mol. Cell. Biol. 18:2108-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castell, J. V., M. J. Gomez-Lechon, M. David, R. Fabra, R. Trullenque, and P. C. Heinrich. 1990. Acute-phase response of human hepatocytes: recognition of acute-phase protein synthesis by interleukin-6. Hepatology 12:1179-1186. [DOI] [PubMed] [Google Scholar]
- 8.Chen, M., S. Hieng, X. Qian, R. Costa, and J.-H. Qu. 1994. Regulation of hepatitis B virus EN1 enhancer activity by hepatocyte-enriched transcription factor HNF3. Virology 205:127-132. [DOI] [PubMed] [Google Scholar]
- 9.Chen, X., U. Vinkemeier, Y. Zhao, D. Jeruzalmi, J. E. Darnell, Jr., and J. Kuriyan. 1998. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell 93:827-839. [DOI] [PubMed] [Google Scholar]
- 10.Cressman, D. E., L. E. Greenbaum, R. A. DeAngelis, G. Ciliberto, E. E. Furth, V. Poli, and R. Taub. 1996. Liver failure and defective hepatocyte regeneration in interleukin-6-deficient mice. Science 274:1379-1383. [DOI] [PubMed] [Google Scholar]
- 11.Darnell, J. E., Jr., I. M. Kerr, and G. R. Stark. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415-1421. [DOI] [PubMed] [Google Scholar]
- 12.Darnell, J. E., Jr. 1997. STATs and gene regulation. Science 277:1630-1635. [DOI] [PubMed] [Google Scholar]
- 13.de Wet, J. R., K. V. Wood, M. Deluka, D. R. Helinski, and S. Subramaniam. 1987. Firefly luciferase gene: structure and expression in mammalian cells. Mol. Cell. Biol. 7:723-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dikstein, R., O. Faktor, and Y. Shaul. 1990. Hierarchic and cooperative binding of the rat liver nuclear protein C/EBP at the hepatitis B virus. Mol. Cell. Biol. 10:4427-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dikstein, R., O. Faktor, R. Ben-Levy, and Y. Shaul. 1990. Functional organization of the hepatitis B virus enhancer. Mol. Cell. Biol. 10:3683-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukada, T., M. Hibi, Y. Yamanaka, M. Takahashi-Tezuka, Y. Fujitani, T. Yamaguchi, K. Nakajima, and T. Hirano. 1996. Two signals are necessary for cell proliferation induced by a cytokine receptor gp130: involvement of STAT3 in anti-apoptosis. Immunity 5:449-460. [DOI] [PubMed] [Google Scholar]
- 17.Garcia, A. D., P. Ostapchuk, and P. Hearing. 1993. Functional interaction of nuclear factors EF-C, HNF-4, and RXRα with hepatitis B virus enhancer 1. J. Virol. 67:3940-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haspel, R. L., G. M. Salditt, and J. E. Darnell, Jr. 1996. The rapid inactivation of nuclear tyrosine phosphorylated Stat1 depends on a protein tyrosine phosphatase. EMBO J. 15:6262-6268. [PMC free article] [PubMed] [Google Scholar]
- 19.Heinrich, P. C., G. Dufhues, S. Flohe, F. Horn, E. Krause, A. Kruttgen, L. Legres, D. Lenz, C. Lutticken, H. Schooltink, T. Stoyan, H. Conradt, and S. Rose-John. 1991. Interleukin-6, its hepatic receptor and the acute phase response in liver, p. 129-145. In H. Sies, L. Flohe, and G. Zimmer (ed.), Molecular aspects of inflammation. Springer-Verlag, Berlin, Germany.
- 20.Heinrich, P. C., F. Horn, L. Graeve, E. Dittrich, I. Kerr, G. Muller-Newen, J. Grotzinger, and A. Wollmer. 1998. Interleukin-6 and related cytokines: effect on the acute phase reaction. Z. Ernaehrwiss. 37:43-49. [PubMed] [Google Scholar]
- 21.Hellqvist, M., M. Mahlapuu, L. S. Samuelsson, S. Enerback, and P. Carlsson. 1996. Differential activation of lung-specific genes by two forkhead proteins, FREAC-1 and FREAC-2. J. Biol. Chem. 271:4482-4490. [DOI] [PubMed] [Google Scholar]
- 22.Hoey, T., and U. Schindler. 1998. STAT structure and function in signaling. Curr. Opin. Genet. Dev. 8:582-587. [DOI] [PubMed] [Google Scholar]
- 23.Honigwachs, J., O. Faktor, R. Dikstein, Y. Shaul, and O. Laub. 1989. Liver-specific expression of hepatitis B virus is determined by the combined action of the core gene promoter and the enhancer. J. Virol. 63:919-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horvath, C. M., Z. Wen, and J. E. Darnell, Jr. 1995. A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev. 9:984-994. [DOI] [PubMed] [Google Scholar]
- 25.Huan, B., and A. Siddiqui. 1992. Retinoid X receptor RXRα binds to and trans-activates the hepatitis B virus enhancer. Proc. Natl. Acad. Sci. USA 89:9059-9063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jameel, S., and A. Siddiqui. 1986. The human hepatitis B virus enhancer requires trans-acting cellular factor(s) for activity. Mol. Cell. Biol. 6:710-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jamison, S. R., K. Chen, and S. Cohen. 1995. Epidermal growth factor induces the tyrosine phosphorylation and nuclear translocation of Stat 5 in mouse liver. Proc. Natl. Acad. Sci. USA 92:4215-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kosovsky, M. J., B. Huan, and A. Siddiqui. 1996. Purification and properties of rat liver nuclear proteins that interact with the hepatitis B virus enhancer 1. J. Biol. Chem. 271:21859-21869. [DOI] [PubMed] [Google Scholar]
- 29.Kosovsky, M. J., I. Qadri, and A. Siddiqui. 1998. The regulation of hepatitis B virus gene expression: an overview of the cis- and trans-acting components, p. 21-50. In R. Koshy and W. H. Caselmann (ed.), Hepatitis B virus. Imperial College Press, London, United Kingdom.
- 30.Lai, E., K. L. Clark, S. K. Burley, and J. E. Darnell, Jr. 1993. Hepatocyte nuclear factor 3/fork head or “winged helix” proteins: a family of transcription factors of diverse biologic function. Proc. Natl. Acad. Sci. USA 90:10421-10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Look, D. C., M. R. Pelletier, R. M. Tidwell, W. T. Roswit, and M. J. Holtzman. 1995. Stat1 depends on transcriptional synergy with Sp1. J. Biol. Chem. 270:30264-30267. [DOI] [PubMed] [Google Scholar]
- 32.Lutticken, C., U. M. Wegenka, J. Yuan, J. Buschmann, C. Schindler, A. Ziemiecki, A. G. Harpur, and A. F. Wilks. 1994. Association of transcription factor APRF and protein kinase Jak1 with the interleukin-6 signal transducer gp130. Science 263:89-92. [DOI] [PubMed] [Google Scholar]
- 33.Mirkovich, J. T., T. Decker, and J. E. Darnell, Jr. 1992. Interferon induction of gene transcription analyzed by in vivo footprinting. Mol. Cell. Biol. 12:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakajima, K., Y. Yamanaka, K. Nakae, H. Kojima, M. Ichiba, N. Kiuchi, T. Kitaoka, T. Fukada, M. Hibi, and T. Hirano. A central role for Stat3 in IL-6-induced regulation of growth and differentiation in M1 leukemia cells. EMBO J. 15:3651-3658. [PMC free article] [PubMed]
- 35.Ohno, H. S., S. Kaneko, K. Kobayashi, and S. Murakani. 1997. Human hepatitis B virus enhancer 1 is responsive to human interleukin-6. J. Med. Virol. 52:413-418. [DOI] [PubMed] [Google Scholar]
- 36.Patel, N., S. Jameel, H. Isom, and A. Siddiqui. 1989. Interactions between nuclear factors and the hepatitis B virus enhancer. J. Virol. 63:5293-5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paulson, M., S. Pisharody, L. P. S. Guadagno, A. L. Mui, and D. E. Levy. 1999. Stat protein transactivation domains recruit p300/CBP through widely divergent sequences. J. Biol. Chem. 274:25343-25349. [DOI] [PubMed] [Google Scholar]
- 38.Reinhold, W., L. Emens, A. Itkes, M. Blake, I. Ichinose, and M. Zajac-Kaye. 1995. The Myc intron-binding polypeptide associates with RFX1 in vivo and binds to the major histocompatibility complex class II promoter region, to the hepatitis B virus enhancer, and to regulatory regions of several distinct viral genes. Mol. Cell. Biol. 15:3041-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robertson, L. M., T. K. Kerppola, M. Vendrell, D. Luk, R. J. Smeyne, C. Bocchiaro, J. I. Morgan, and T. Curran. 1995. Regulation of c-fos expression in transgenic mice requires multiple interdependent transcriptional control elements. Neuron 14:241-252. [DOI] [PubMed] [Google Scholar]
- 40.Sadowski, H. B., K. Shuai, J. E. Darnell, Jr., and M. J. Gilman. 1993. A common nuclear signal transduction pathway activated by growth factor and cytokine receptors. Science 261:1739-1744. [DOI] [PubMed] [Google Scholar]
- 41.Schaefer, T. S., L. K. Sanders, and D. Nathans. 1995. Cooperative transcriptional activity of Jun and Stat3 beta, a short form of Stat3. Proc. Natl. Acad. Sci. USA 92:9097-9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schindler, C., and J. E. Darnell, Jr. 1995. Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu. Rev. Biochem. 64:621-651. [DOI] [PubMed] [Google Scholar]
- 43.Sells, M. A., M.-L. Chen, and G. Acs. 1987. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc. Natl. Acad. Sci. USA 84:1005-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shaul, Y., W. J. Rutter, and O. Laub. 1985. A human hepatitis B enhancer element. EMBO J. 4:427-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stahl, N., T. G. Boulton, T. Farruggella, N. Y. Ip, S. Davis, B. A. Witthuhn, F. W. Quelle, O. Silvennoinen, G. Barbieri, and S. Pellegrini. 1994. Association and activation of Jak-Tyk kinase by CNTF-LIF-OSM-IL-6 beta receptor components. Science 263:92-95. [DOI] [PubMed] [Google Scholar]
- 46.Vignais, M.-L., H. B. Sadowski, D. Watling, N. C. Rogers, and M. Gilman. 1996. Platelet-derived growth factor induces phosphorylation of multiple JAK family kinases and STAT proteins. Mol. Cell. Biol. 16:1759-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46a.Waris, G., K. W. Huh, and A. Siddiqui. 2001. Mitochondrially associated hepatitis B virus X protein constitutively activates transcription factors STAT-3 and NF-κB via oxidative stress. Mol. Cell. Biol. 21:7721-7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wegenka, U. M., J. Buschmann, C. Lutticken, P. C. Heinrich, and F. Horn. 1993. Acute-phase response factor, a nuclear factor binding to acute-phase response elements, is rapidly activated by interleukin-6 at the posttranslational level. Mol. Cell. Biol. 13:276-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wegenka, U. M., C. Lutticken, J. Buschmann, J. Yuan, F. Lottspeich, W. Muller-Esterl, C. Schindler, E. Roeb, P. C. Heinrich, and F. Horn. 1994. The interleukin-6-activated acute-phase response factor is antigenically and functionally related to members of the signal transducer and activator of transcription (STAT) family. Mol. Cell. Biol. 14:3186-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weigel, D., and H. Jackle. 1990. The fork head domain: a novel DNA binding motif of eukaryotic transcription factors? Cell 63:655-656. [DOI] [PubMed] [Google Scholar]
- 50.Wen, Z., Z. Zhong, and J. E. Darnell, Jr. 1995. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 82:241-250. [DOI] [PubMed] [Google Scholar]
- 51.Xu, X., Y. L. Sun, and T. Hoey. 1996. Cooperative DNA binding and sequence-selection recognition conferred by the STAT amino-terminal domain. Science 273:794-797. [DOI] [PubMed] [Google Scholar]
- 52.Yu, C. L., D. J. Meyer, G. S. Campbell, A. C. Larner, C. Carter-Su, J. Schwartz, and R. Jove. 1995. Enhanced DNA-binding activity of a Stat3-related protein in cells transformed by the Src oncoprotein. Science 269:81-83. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, X., M. H. Wrzeszczynska, C. M. Horvath, and J. E. Darnell, Jr. 1999. Interacting regions in Stat3 and c-Jun that participate in cooperative transcriptional activation. Mol. Cell. Biol. 19:7138-7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhong, Z., Z. Wen, and J. E. Darnell, Jr. 1994. Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science 264:95-98. [DOI] [PubMed] [Google Scholar]
- 55.Zong, C., R. Yan, A. August, J. E. Darnell, Jr., and H. Hanafusa. 1996. Unique signal transduction of Eyk: constitutive stimulation of the JAK-STAT pathway by an oncogenic receptor-type tyrosine kinase. EMBO J. 15:4515-4525. [PMC free article] [PubMed] [Google Scholar]