Abstract
We have previously employed an intercellular spreading strategy using herpes simplex virus type 1 (HSV-1) VP22 protein to enhance DNA vaccine potency because DNA vaccines lack the intrinsic ability to amplify in cells. Recently, studies have demonstrated that the protein encoded by UL49 of Marek's disease virus type 1 (MDV-1) exhibits some degree of homology to the HSV-1 VP22 protein and features the property of intercellular transport. We therefore generated a DNA vaccine encoding MDV-1 VP22 linked to a model antigen, human papillomavirus type 16 E7. We demonstrated that compared with mice vaccinated with DNA encoding wild-type E7, mice vaccinated with MDV-1 VP22/E7 DNA exhibited a significant increase in number of gamma-interferon-secreting, E7-specific CD8+-T-cell precursors as well as stronger tumor prevention and treatment effects. Furthermore, our data indicated that the antitumor effect was CD8 dependent. These results suggested that the development of vaccines encoding VP22 fused to a target antigen might be a promising strategy for improving DNA vaccine potency.
DNA vaccines have emerged as an attractive approach for generating antigen-specific immunotherapy (for reviews, see references 7, 26, 28, and 31). DNA is relatively stable and can be easily prepared and harvested in large quantities. In addition, naked plasmid DNA is relatively safe and can be administered repeatedly. Furthermore, because DNA can be maintained in cells to allow long-term expression of the encoded antigen, maintenance of immunologic memory is possible. Since naked DNA has no cell type specificity, it is important to find an efficient route for the administration of DNA vaccines into the appropriate target cells.
Intradermal administration of DNA vaccines via a gene gun is a convenient way of delivering such vaccines into professional antigen-presenting cells (APCs) in vivo. Professional APCs are the best candidate to mediate the presentation of antigens encoded by DNA vaccines to T cells. The gene gun strategy enables efficient delivery of DNA into epidermal bone marrow-derived APCs called Langerhans cells, which move into the lymphatic system via the draining lymph nodes (5). We have successfully used this system of DNA delivery to test various intracellular targeting strategies (2, 17).
One major limitation of DNA vaccines is their inability to amplify and spread in vivo as some replicating viral vaccine vectors are able to do. Therefore, a strategy that facilitates the spread of antigen may significantly enhance the potency of naked DNA vaccines. Recently, our group, as well as other researchers, has been able to enhance the potency of DNA vaccines by using human herpes simplex virus type 1 (HSV-1) (13, 25) or bovine herpesvirus 1 (24) VP22. VP22, a tegument protein that has demonstrated the property of intercellular transport, is capable of distributing protein to many surrounding cells (9). We showed that HSV-1 VP22 is capable of enhancing intercellular spreading of the linked protein, leading to augmentation of E7-specific CD8+-T-cell precursor numbers and enhancement of the antitumor effect in HSV-1 VP22/E7 DNA-vaccinated mice compared with wild-type E7 DNA-vaccinated mice (13). The success of this strategy warranted the consideration of other proteins with similar trafficking properties.
Recently, a tegument phosphoprotein of Marek's disease virus serotype 1 (MDV-1) has been shown to be capable of intercellular transport after exogenous application (8). MDV-1 UL49 codes for a protein of 249 amino acids (aa) with a molecular mass of 27.6 kDa (22). In addition, the protein encoded by MDV-1 UL49 exhibits homology to HSV-1 VP22 (20). We therefore investigated the novel use of MDV-1 VP22 linked to a model antigen (human papillomavirus type 16 [HPV-16] E7) in the context of a DNA vaccine (MDV-1 VP22/E7) and explored its ability to enhance E7-specific immune responses and antitumor effects. Our data indicated that the linkage of MDV-1 VP22 to E7 also led to an impressive increase in the number of E7-specific CD8+-T-cell precursors and generated significant potency against E7-expressing tumors in vaccinated mice. Mice vaccinated with MDV-1 VP22/E7 DNA generated E7-specific CD8+-T-cell precursors in numbers comparable to those produced by mice vaccinated with HSV-1 VP22/E7. Our results suggested that the fusion of MDV-1 VP22 DNA to a target antigen gene could significantly enhance DNA vaccine potency and that MDV-1 VP22 represents an alternative to HSV-1 VP22 for enhancing vaccine potency.
MATERIALS AND METHODS
Plasmid DNA construction.
We employed the pcDNA3 expression vector, which has been used effectively to investigate the correlation between the E7-specific T-cell-mediated immune response and the antitumor effect generated by various DNA vaccines (2). The generation of pcDNA3-E7 has been described previously (2). For the generation of pcDNA3-E7(E/B), which contains E7 with EcoRI and BamHI restriction sites on the flanking ends of the E7 DNA, we first used PCR to amplify the E7 fragment with pcDNA3-E7 and a set of primers, 5′-GGGGAATTCATGGAGATACACCTA-3′ and 5′-GGTGGATCCTTGAGAACAGATGG-3′. The amplified product was further cloned into the EcoRI and BamHI sites of pcDNA3. For the generation of pcDNA3-MDV-1 VP22, we first performed PCR to amplify the MDV-1 VP22 DNA fragment, using pUC18BE8.6, a DNA template containing MDV-1 UL49 (35) (kindly provided by Lucy F. Lee, United States Department of Agriculture, East Lansing, Mich.), and a set of primers, 5′-ATCTCTAGAATGGGGGATTCTGAAAGGCG-3′ and 5′-GATGAATTCTTCGCTATCACTGCTACGAT-3′. The amplified product was cloned into the unique EcoRI and XbaI cloning sites of the pcDNA3.1(−) expression vector (Invitrogen, Carlsbad, Calif.) downstream of the cytomegalovirus promoter. For the generation of pcDNA3-MDV-1 VP22/E7, the same PCR-amplified product (MDV-1 VP22) was cloned into the unique XbaI and EcoRI cloning sites of pcDNA3-E7(E/B). The MDV-1 VP22 gene was cloned in frame with the E7 gene. DNA sequencing confirmed that the vaccine encoded the MDV-1 VP22/E7 fusion protein.
DNA vaccination.
Preparation of DNA-coated gold particles and gene gun particle-mediated DNA vaccination were performed according to previously described protocols (2). DNA-coated gold particles (1 μg of DNA/bullet) were delivered to the shaved abdominal regions of C57BL/6 mice with a helium-driven gene gun (Bio-Rad, Hercules, Calif.) with a discharge pressure of 400 lb/in2. Mice were vaccinated via gene gun with 2 μg of MDV-1 VP22, E7, MDV-1 VP22 mixed with E7 (MDV-1 VP22+E7 DNA), MDV-1 VP22/E7, or pcDNA3 (which has no insert). One week later, mice were boosted with the same regimen as the first vaccination.
Mice.
C57BL/6 mice (6- to 8-week-old females) from the National Cancer Institute (Frederick, Md.) were purchased and kept in the oncology animal facility of the Johns Hopkins Hospital (Baltimore, Md.). We have used these mice in previous DNA vaccine studies (2, 4, 13-15, 17). All animal procedures were performed according to approved protocols and in accordance with recommendations for the proper use and care of laboratory animals.
Intracellular cytokine staining.
Cell surface marker staining of CD8 or CD4, intracellular cytokine staining for gamma interferon (IFN-γ), and FACScan analysis were performed using conditions described previously (2). Prior to FACScan analysis, splenocytes from different vaccinated groups of mice were collected 8 days after the final vaccination and incubated for 20 h with either an E7 peptide (aa 49 to 57; RAHYNIVTF) (10) containing a major histocompatibility complex (MHC) class I epitope for detecting E7-specific CD8+-T-cell precursors (1 μg/ml) or an E7 peptide (aa 30 to 67; DSSEEEDEIDGPAGQAEPDRAHYNIVTFCCKCDSTLRL) (33) containing an MHC class II epitope for detecting E7-specific CD4+-T-cell precursors (10 μg/ml). The numbers of IFN-γ-secreting CD8+ and CD4+ T cells were determined by flow cytometry.
ELISA.
The levels of anti-HPV-16 E7 antibodies in sera were determined by a direct enzyme-linked immunosorbent assay (ELISA) as described previously (34). Briefly, wells of a 96-microwell plate were coated with 100 μl of a 10-μg/ml solution of bacterium-derived HPV-16 E7 protein and incubated at 4°C overnight. The wells were then blocked with phosphate-buffered saline (PBS) containing 20% fetal bovine serum. Sera were prepared from the mice on day 14 postimmunization, serially diluted in PBS, added to the ELISA wells, and incubated at 37°C for 2 h. After being washed with PBS containing 0.05% Tween 20, the plate was incubated with a 1/2,000 dilution of a peroxidase-conjugated rabbit anti-mouse immunoglobulin G antibody (Zymed, San Francisco, Calif.) at room temperature for 1 h. The plate was washed six times, and then 1-Step Turbo TMB-ELISA was used as a substrate for color development (Pierce, Rockford, Ill.); color development was stopped with 1 M H2SO4. The absorbance of the ELISA plate wells was then determined with a standard ELISA reader at 450 nm.
In vivo tumor protection experiment.
For the tumor protection experiment, mice (five per group) were vaccinated via gene gun with 2 μg of pcDNA3 (no insert), MDV-1 VP22, E7, MDV-1 VP22+E7, or MDV-1 VP22/E7 DNA. One week later, mice were boosted with the same regimen as was used in the first vaccination. One week after the last vaccination, mice were each challenged with 5 × 104 TC-1 tumor cells subcutaneously, in the right leg, and then monitored twice a week. The day of the first gene gun vaccination was considered the starting day for counting postoperative survival days.
In vivo tumor treatment experiment.
The day of intravenous tumor challenge was considered the starting day for this experiment. On the starting day, mice (five per group) were each intravenously challenged with 104 TC-1 tumor cells via the tail vein. Three days after the first challenge, mice were administered 2 μg of pcDNA3 (no insert), MDV-1 VP22, E7, MDV-1 VP22+E7 DNA, or MDV-1 VP22/E7 DNA via gene gun. One week after the first vaccination, these mice were boosted with the same regimen. Mice were sacrificed and lungs were explanted on day 24. The pulmonary nodules on the surfaces of the lungs in each mouse were counted and weighed by researchers blinded to sample identity. Compared with the percentage of tumor-free mice, used as a measurement with the subcutaneous tumor model, the mean number of tumor nodules in the pulmonary tumor model provides a more quantitative assessment of the antitumor effect generated by each vaccine. The pulmonary tumor model might allow the evaluation of subtle differences in antitumor effects that might not be appreciable when our subcutaneous tumor model is used.
In vivo antibody depletion experiment.
In vivo antibody depletion has been described previously (21). Briefly, mice were each vaccinated with 2 μg of MDV-1 VP22/E7 DNA via gene gun, boosted 1 week later, and challenged with 5 × 104 TC-1 tumor cells subcutaneously. Depletions were started 1 week prior to tumor challenge. Monoclonal antibody (MAb) GK1.5 (6) was used for CD4 depletion, MAb 2.43 (29) was used for CD8 depletion, and MAb PK136 (19) was used for NK1.1 depletion. Depletion was terminated on day 70 after tumor challenge.
RESULTS
Comparison of amino acid sequence of MDV-1 UL49 (VP22 homologue) with that of HSV-1 VP22.
HSV-1 and MDV-1 are classified under the subfamily Alphaherpesviridae. The MDV-1 VP22 (UL49) and HSV-1 VP22 sequences were optimally aligned using the ClustalW method with a Blosum30 residue weight table as shown in Fig. 1. The results revealed approximately 25% similarity between the VP22s from HSV-1 and MDV-1, with the C-terminal halves of the sequences having a higher degree of similarity than the remaining portions of the sequences. Despite the limited degree of homology, the MDV-1 VP22 (UL49) protein has been shown to exhibit an intercellular spreading property similar to that demonstrated by HSV-1 VP22 (8).
FIG. 1.
Alignment of amino acid sequences of the VP22 tegument proteins from HSV-1 and MDV-1. Shaded regions represent matched amino acids found in at least three of the five aligned sequences. Horizontal lines represent gaps. Note that the MDV-1 VP22 and HSV-1 VP22 amino acid sequences were optimally aligned by the ClustalW(accurate) method with a Blosum30 residue weight table. The results revealed approximately 25% similarity between the MDV and HSV-1 VP22 sequences.
Vaccination with MDV-1 VP22/E7 fusion DNA significantly enhances E7-specific CD8+-T-cell-mediated immune responses.
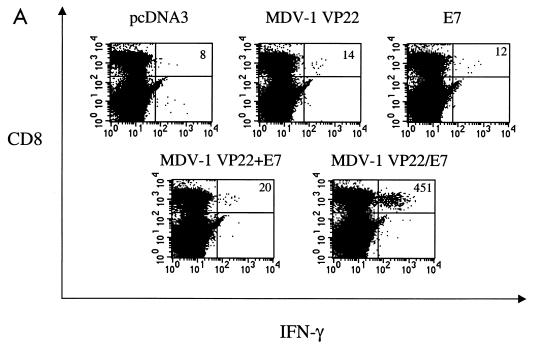
We have previously demonstrated that a DNA vaccine encoding HSV-1 VP22 linked to E7 generated significant enhancement of E7-specific CD8+-T-cell precursors in vaccinated mice (13). To determine whether vaccination of mice with the pcDNA3-MDV-1 VP22/E7 DNA vaccine could enhance the number of E7-specific CD8+-T-cell precursors, we performed intracellular cytokine staining on splenocytes from vaccinated mice. Splenocytes from naive or vaccinated groups of mice were incubated with the MHC class I (H-2Db)-restricted E7 peptide (aa 49 to 57) for detection of E7-specific CD8+ T cells. As shown in Fig. 2A, mice vaccinated with MDV-1 VP22/E7 DNA exhibited a significant increase in E7-specific IFN-γ+ CD8+-T-cell precursors (451 per 3 × 105 splenocytes) compared to mice vaccinated with wild-type E7 DNA (12 per 3 × 105 splenocytes). The physical linkage of MDV-1 VP22 to E7 was important for the observed enhancement of E7-specific CD8+-T-cell activity since MDV-1 VP22+E7 DNA did not generate a significant increase in the number of CD8+-T-cell precursors (20 per 3 × 105 splenocytes). Furthermore, the linkage of an irrelevant protein (such as green fluorescent protein or cytotoxic T-lymphocyte antigen 4) to E7 did not generate enhancement of E7-specific CD8+-T-cell activity (data not shown).
FIG. 2.
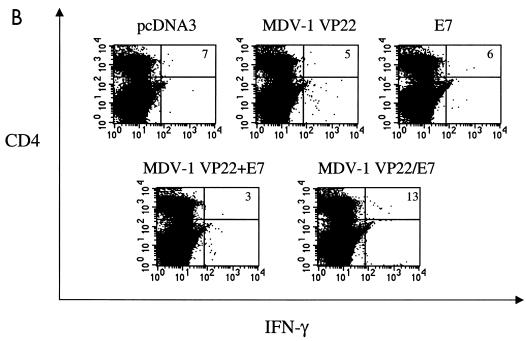
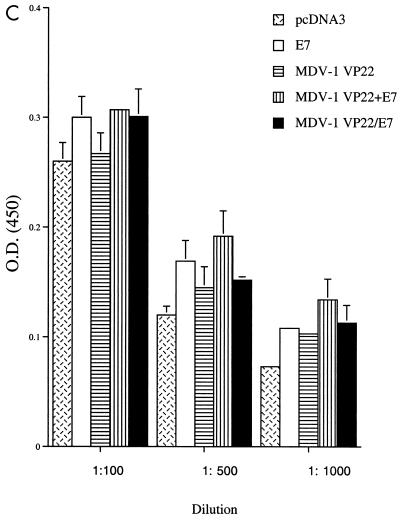
Immunologic responses generated by various recombinant DNA vaccines. (A) Results of flow cytometry analysis of IFN-γ-secreting, E7-specific CD8+-T-cell precursors. Mice were immunized and splenocytes were collected and cultured as described in Materials and Methods. Note that vaccination of mice with MDV-1 VP22/E7 DNA generated a larger number of IFN-γ+ CD8+-T-cell precursors than did vaccination with any of the other constructs. The results shown are from one representative experiment of three that were performed. (B) Results of flow cytometry analysis of IFN-γ-secreting, E7-specific CD4+-T-cell precursors. (C) Results of an ELISA to determine E7-specific antibodies in the sera of the various vaccinated mice. Mice were immunized and sera were collected and cultured as described in Materials and Methods. O.D. (450), optical density at 450 nm.
While addition of MDV-1 VP22 to E7 led to enhanced E7-specific CD8+-T-cell activity, we did not detect a significant difference in the number of E7-specific, IFN-γ-secreting CD4+ T cells (Fig. 2B) among the various vaccination groups. Using a direct ELISA, we found that there was a slight increase in the E7-specific antibody response in mice vaccinated with E7-containing DNA vaccines compared to mice vaccinated with the empty vector (pcDNA3). We did not detect a significant difference in E7-specific antibody responses in the sera of mice vaccinated with E7, MDV-1 VP22+E7, or MDV-1/E7 DNA (Fig. 2C).
Vaccination with a chimeric MDV-1 VP22/E7 DNA vaccine enhances protection of mice against growth of TC-1 tumors.
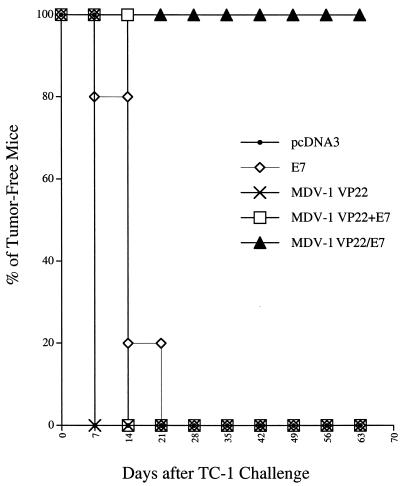
To determine whether the observed enhancement of E7-specific CD8+-T-cell-mediated immunity translated to a significant E7-specific antitumor effect, we performed an in vivo tumor protection experiment using a previously characterized E7-expressing tumor model, TC-1 (21). As shown in Fig. 3, 100% of mice receiving the MDV-1 VP22/E7 DNA vaccine remained tumor free 65 days after TC-1 challenge. In contrast, all of the mice receiving pcDNA3 (no insert), wild-type E7, or MDV-1 VP22+E7 DNA developed tumors within 21 days after tumor challenge. We also observed that fusion of E7 to MDV-1 VP22 was required for antitumor immunity, since MDV-1 VP22+E7 DNA did not generate enhancement of tumor protection.
FIG. 3.
Tumor protection experiments and the effect of MDV-1 VP22/E7 on subcutaneous tumor cell growth. In the in vivo tumor protection experiments, mice were immunized and challenged subcutaneously with TC-1 tumor cells as described in Materials and Methods. The data shown here are from one representative experiment of two that were performed.
Treatment with MDV-1 VP22/E7 fusion DNA eradicates established E7-expressing tumors in the lungs.
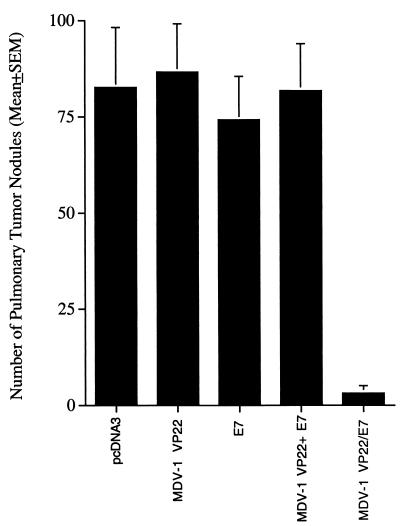
We then investigated the therapeutic potential of the chimeric MDV-1 VP22/E7 DNA construct in treating TC-1 tumors, using a lung hematogenous spread model (16). As shown in Fig. 4A, mice vaccinated with MDV-1 VP22/E7 DNA exhibited fewer pulmonary nodules (mean ± standard error of the mean, 3.2 ± 1.9) than mice vaccinated with wild-type E7 DNA (74.2 ± 11.27) or MDV-1 VP22 DNA (86.6 ± 12.5). Also, mice vaccinated with MDV-1 VP22/E7 DNA exhibited a lower average lung weight than mice vaccinated with wild-type E7 DNA or MDV-1 VP22 DNA (data not shown). Taken together, the results from our tumor protection and treatment experiments indicated that linkage of MDV-1 VP22 to E7 significantly enhanced antitumor effects on TC-1 tumors.
FIG. 4.
Tumor treatment experiments and the effect of MDV-1 VP22/E7 on pulmonary tumor cell growth. The mean number of lung tumor nodules generated in each vaccinated group was determined. For the in vivo tumor treatment experiments, mice (five per group) were challenged intravenously in the tail vein and vaccinated as described in Materials and Methods.
CD8+ T cells, but not CD4+ T cells or NK cells, are important for antitumor effect generated by the MDV-1 VP22/E7 DNA vaccine.
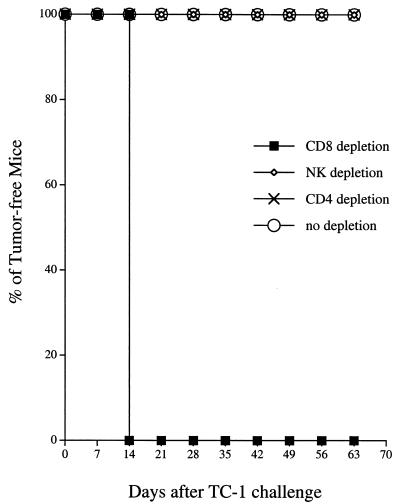
To determine the subset of lymphocytes that is important for the antitumor effect generated by the MDV-1 VP22/E7 DNA vaccine, we performed in vivo antibody depletion experiments. As shown in Fig. 5, all MDV-1 VP22/E7 DNA-vaccinated mice depleted of CD8+ T cells and all unvaccinated naive mice grew tumors within 14 days after tumor challenge. In contrast, all MDV-1 VP22/E7 DNA-vaccinated mice depleted of CD4+ T cells and NK cells remained tumor free 70 days after tumor challenge. These results suggested that CD8+ T cells, but not CD4+ T cells or NK cells, were essential for the antitumor immunity generated by the VP22/E7 DNA vaccine.
FIG. 5.
In vivo antibody depletion experiment. An in vivo antibody depletion experiment was performed to determine the effect of lymphocyte subsets on the potency of the MDV-1 VP22/E7 DNA vaccine. Mice were vaccinated, challenged with TC-1, and depleted of the relevant subset of lymphocytes as described in Materials and Methods. Note that depletion of CD8+ T cells, but not of CD4+ T cells or NK cells, led to significant tumor growth in MDV-1 VP22/E7 DNA-vaccinated mice.
DISCUSSION
In this study, we observed that MDV-1 VP22 enhanced DNA vaccine potency when linked to a model antigen, E7. Despite the limited similarity between the amino acid sequences of MDV-1 UL49 (VP22) and HSV-1 VP22 (approximately 25%), the MDV-1 VP22/E7 and HSV-1 VP22/E7 (13) DNA vaccines generated comparable degrees of E7-specific CD8+-T-cell-mediated immune response. The property of intercellular spreading shared by these VP22 homologues may differ from that of other molecules with trafficking potential. We recently observed that DNA vaccines encoding E7 linked to molecules derived from other proteins with trafficking properties (such as human immunodeficiency virus Tat protein, membrane-translocating sequence, and the third helix of the Antennapedia homeodomain in Drosophila) did not generate CD8+-T-cell-mediated immune responses as potent as those generated by HSV-1 VP22/E7 (13) or MDV-1 VP22/E7.
Alignment of the amino acid sequences of MDV-1 VP22 and HSV-1 VP22 revealed that the carboxyl-terminal halves of the sequences exhibit a considerably higher degree of similarity than the amino-terminal halves, suggesting that the C-terminus region may be important for VP22 function. Studies have shown that the C-terminus region is important for intercellular spreading (9) and nuclear localization (27). Deletion mutants linked to fluorescent proteins might be used to identify the precise region of MDV-1 VP22 that is responsible for intercellular spreading. Recently, HSV-1 VP22's cytoskeleton binding, nuclear localization, chromatin binding, nuclear membrane binding, and intercellular transport domains were mapped using such deletion experiments (1). Mapping the functional domains that are important for the ability of MDV-1 VP22 and other homologues to enhance vaccine potency would be an important endeavor for the future.
We found that vaccination with MDV-1 VP22/E7 generated an immune response and antitumor effect comparable to those provided by HSV-1 VP22/E7 DNA, although a more definitive conclusion requires a head-to-head comparison. Meanwhile, a notable difference between the MDV-1 VP22/E7 and HSV-1 VP22/E7 DNA vaccines in terms of the effector cells responsible for antitumor immunity was observed. While CD8+ T cells and NK cells were found to be important for the antitumor effect generated by HSV-1 VP22/E7 DNA (13), our data revealed that the antitumor effect generated by MDV-1 VP22/E7 DNA required only CD8+ T cells. These data imply that the two vaccines may activate different subsets of effector cells in the vaccinated host and may have different immunologic or antitumor mechanisms. It would be interesting to perform additional experiments to evaluate qualitative and/or quantitative differences in NK cells in mice vaccinated with MDV-1 VP22 or its homologues.
Several factors may account for the observed enhancement of E7-specific CD8+-T-cell activity in mice administered DNA vaccines encoding MDV-1 VP22/E7 or other VP22 homologues linked to E7. First, since ballistic DNA delivery can introduce DNA directly into dermal professional APCs, the intercellular spreading of the MDV-1 VP22 and HSV-1 VP22 protein within the epidermis raises the possibility of generating an increased number of APCs that present the linked protein. This notion is supported by our previous study using an HSV-1 VP22 deletion mutant, HSV-1 VP22 (aa1-267), which lacks the property of intercellular spreading (13). Linkage of HSV-1 VP22 (aa1-267) to antigen fails to enhance DNA vaccine potency. Second, we have previously shown that cells transfected with DNA encoding HSV-1 VP22/E7 directly enhance the presentation of E7 through the MHC class I pathway to an E7-specific CD8+-T-cell line (13), suggesting that direct priming may be an important mechanism for enhancing E7-specific CD8+-T-cell activity. Third, we have demonstrated that dendritic cells pulsed with lysates of cells transfected with naked HSV-1 VP22/E7 RNA replicons exhibit a significantly higher percentage of specific lysis by an E7-specific CD8+-T-cell line than dendritic cells pulsed with lysates of cells transfected with wild-type E7 RNA replicons (3), indicating that cross-priming may be another potential mechanism for the enhancement of E7-specific CD8+-T-cell responses mediated by VP22/E7. Finally, DNA vaccines may influence the biology of professional APCs at vaccination sites through the effects of CpG islands (18, 30, 32) or induced cytokines (12, 18), which in turn may affect vaccine-mediated, antigen-specific CD8+-T-cell activity.
We have previously developed several intracellular targeting strategies that are capable of enhancing MHC class I and class II presentation of antigens encoded by DNA vaccines. For example, an endosomal-lysosomal targeting strategy using E7 chimerically linked to a signal peptide (Sig) and the endosomal-lysosomal sorting signal (derived from lysosome-associated membrane protein 1) was found to significantly enhance MHC class II presentation of antigen to CD4+ T cells (17). Furthermore, we have used several different molecules, including Mycobacterium tuberculosis heat shock protein 70 (2), domain II of Pseudomonas exotoxin A (14), and calreticulin (4), linked to antigen in the context of DNA vaccines to significantly enhance MHC class I presentation of the E7 antigen to CD8+ T cells. The vaccine potency of these intracellular approaches may be further enhanced if the intercellular spreading strategy is also incorporated into the vaccine design.
Safety is an important issue to address before considering broader applications of this vaccine. Although DNA may integrate into the host genome, it is estimated that the frequency of integration is much lower than that of spontaneous mutation and should not pose any real risk (23). Potential risks associated with the presence of the HPV-16 oncogenic E7 protein in host cells may be alleviated by introducing mutations into E7 DNA so that the resulting E7 protein cannot bind with pRB (11) but still maintains most of its antigenicity. Another strategy is to use a shuffled E7 gene, which can alleviate concerns of oncogenicity associated with E7 (25). Another concern about DNA vaccination is the generation of autoimmune disease against host DNA. However, we examined vital organs in MDV-1 VP22/E7-vaccinated mice and did not observe any significant pathological changes. These results indicated that MDV-1 VP22/E7 could be an effective DNA vaccine with minimal detrimental side effects.
In summary, our results indicated that the fusion of MDV-1 VP22 to the HPV-16 E7 gene generated powerful E7-specific CD8+-T-cell-mediated immune responses and antitumor effects against HPV-16 E7-expressing murine tumors. The VP22 strategy can potentially be applied to other cancer systems with known tumor-specific antigens. It would be valuable to compare VP22 homologues and determine whether the extent of intercellular spreading of linked antigen encoded by DNA vaccine can be correlated with the degree of antigen-specific immunity and the ensuing antitumor effect.
Acknowledgments
This work was supported by NIH grants 5 POI 34582-01, U19 CA 72108-02, RO1 CA72631-01, and RO1 CA83706-01; the Cancer Research Institute; and the American Cancer Society.
We thank Robert J. Kurman, Drew M. Pardol, and Keerti V. Shah for helpful discussions. We also thank Richard B. S. Roden for critical review of the manuscript and Willson Kwok for superb technical support.
REFERENCES
- 1.Aints, A., H. Guven, G. Gahrton, C. I. Smith, and M. S. Dilber. 2001. Mapping of herpes simplex virus-1 VP22 functional domains for inter- and subcellular protein targeting. Gene Ther. 8:1051-1056. [DOI] [PubMed]
- 2.Chen, C. H., T. L. Wang, C. F. Hung, Y. Yang, R. A. Young, D. M. Pardoll, and T. C. Wu. 2000. Enhancement of DNA vaccine potency by linkage of antigen gene to an HSP70 gene. Cancer Res. 60:1035-1042. [PubMed] [Google Scholar]
- 3.Cheng, W.-F., C.-F. Hung, C.-Y. Chai, K.-F. Hsu, L. He, M. Ling, and T.-C. Wu. 2001. Enhancement of Sindbis virus self-replicating RNA vaccine potency by linkage of herpes simplex virus type 1 VP22 protein to antigen. J. Virol. 75:2368-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng, W. F., C. F. Hung, C. Y. Chai, K. F. Hsu, L. He, M. Ling, and T. C. Wu. 2001. Tumor-specific immunity and antiangiogenesis generated by a DNA vaccine encoding calreticulin linked to a tumor antigen. J. Clin. Investig. 108:669-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Condon, C., S. C. Watkins, C. M. Celluzzi, K. Thompson, and L. D. Falo, Jr. 1996. DNA-based immunization by in vivo transfection of dendritic cells. Nat. Med. 2:1122-1128. [DOI] [PubMed] [Google Scholar]
- 6.Dialynas, D. P., Z. S. Quan, K. A. Wall, A. Pierres, J. Quintans, M. R. Loken, M. Pierres, and F. W. Fitch. 1983. Characterization of the murine T cell surface molecule, designated L3T4, identified by monoclonal antibody GK1.5: similarity of L3T4 to the human Leu-3/T4 molecule. J. Immunol. 131:2445-2451. [PubMed] [Google Scholar]
- 7.Donnelly, J. J., J. B. Ulmer, J. W. Shiver, and M. A. Liu. 1997. DNA vaccines. Annu. Rev. Immunol. 15:617-648. [DOI] [PubMed] [Google Scholar]
- 8.Dorange, F., S. El Mehdaoui, C. Pichon, P. Coursaget, and J. F. Vautherot. 2000. Marek's disease virus (MDV) homologues of herpes simplex virus type 1 UL49 (VP22) and UL48 (VP16) genes: high-level expression and characterization of MDV-1 VP22 and VP16. J. Gen. Virol. 81:2219-2230. [DOI] [PubMed] [Google Scholar]
- 9.Elliott, G., and P. O'Hare. 1997. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell 88:223-233. [DOI] [PubMed] [Google Scholar]
- 10.Feltkamp, M. C., H. L. Smits, M. P. Vierboom, R. P. Minnaar, B. M. de Jongh, J. W. Drijfhout, J. ter Schegget, C. J. Melief, and W. M. Kast. 1993. Vaccination with cytotoxic T lymphocyte epitope-containing peptide protects against a tumor induced by human papillomavirus type 16-transformed cells. Eur. J. Immunol. 23:2242-2249. [DOI] [PubMed] [Google Scholar]
- 11.Heck, D. V., C. L. Yee, P. M. Howley, and K. Munger. 1992. Efficiency of binding the retinoblastoma protein correlates with the transforming capacity of the E7 oncoproteins of the human papillomaviruses. Proc. Natl. Acad. Sci. USA 89:4442-4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hemmi, H., O. Takeuchi, T. Kawai, T. Kaisho, S. Sato, H. Sanjo, M. Matsumoto, K. Hoshino, H. Wagner, K. Takeda, and S. Akira. 2000. A Toll-like receptor recognizes bacterial DNA. Nature 408:740-745. [DOI] [PubMed] [Google Scholar]
- 13.Hung, C.-F., W.-F. Cheng, C.-Y. Chai, K.-F. Hsu, L. He, M. Ling, and T.-C. Wu. 2001. Improving vaccine potency through intercellular spreading and enhanced MHC class I presentation of antigen. J. Immunol. 166:5733-5740. [DOI] [PubMed] [Google Scholar]
- 14.Hung, C.-F., W.-F. Cheng, K.-F. Hsu, C.-Y. Chai, L. He, M. Ling, and T.-C. Wu. 2001. Cancer immunotherapy using a DNA vaccine encoding the translocation domain of a bacterial toxin linked to a tumor antigen. Cancer Res. 61:3698-3703. [PubMed] [Google Scholar]
- 15.Hung, C. F., K. F. Hsu, W. F. Cheng, C. Y. Chai, L. He, M. Ling, and T. C. Wu. 2001. Enhancement of DNA vaccine potency by linkage of antigen gene to a gene encoding the extracellular domain of Fms-like tyrosine kinase 3-ligand. Cancer Res. 61:1080-1088. [PubMed] [Google Scholar]
- 16.Ji, H., E. Y. Chang, K. Y. Lin, R. J. Kurman, D. M. Pardoll, and T. C. Wu. 1998. Antigen-specific immunotherapy for murine lung metastatic tumors expressing human papillomavirus type 16 E7 oncoprotein. Int. J. Cancer 78:41-45. [DOI] [PubMed] [Google Scholar]
- 17.Ji, H., T.-L. Wang, C.-H. Chen, C.-F. Hung, S. Pai, K.-Y. Lin, R. J. Kurman, D. M. Pardoll, and T.-C. Wu. 1999. Targeting HPV-16 E7 to the endosomal/lysosomal compartment enhances the antitumor immunity of DNA vaccines against murine HPV-16 E7-expressing tumors. Hum. Gene Ther. 10:2727-2740. [DOI] [PubMed] [Google Scholar]
- 18.Klinman, D. M., G. Yamshchikov, and Y. Ishigatsubo. 1997. Contribution of CpG motifs to the immunogenicity of DNA vaccines. J. Immunol. 158:3635-3639. [PubMed] [Google Scholar]
- 19.Koo, G. C., F. J. Dumont, M. Tutt, J. Hackett, Jr., and V. Kumar. 1986. The NK-1.1− mouse: a model to study differentiation of murine NK cells. J. Immunol. 137:3742-3747. [PubMed] [Google Scholar]
- 20.Koptidesova, D., J. Kopacek, V. Zelnik, N. L. Ross, S. Pastorekova, and J. Pastorek. 1995. Identification and characterization of a cDNA clone derived from the Marek's disease tumour cell line RPL1 encoding a homologue of alpha-transinducing factor (VP16) of HSV-1. Arch. Virol. 140:355-362. [DOI] [PubMed] [Google Scholar]
- 21.Lin, K.-Y., F. G. Guarnieri, K. F. Staveley-O'Carroll, H. I. Levitsky, T. August, D. M. Pardoll, and T.-C. Wu. 1996. Treatment of established tumors with a novel vaccine that enhances major histocompatibility class II presentation of tumor antigen. Cancer Res. 56:21-26. [PubMed] [Google Scholar]
- 22.Lupiani, B., L. F. Lee, and S. M. Reddy. 2001. Protein-coding content of the sequence of Marek's disease virus serotype 1. Curr. Top. Microbiol. Immunol. 255:159-190. [DOI] [PubMed] [Google Scholar]
- 23.Nichols, W. W., B. J. Ledwith, S. V. Manam, and P. J. Troilo. 1995. Potential DNA vaccine integration into host cell genome. Ann. N. Y. Acad. Sci. 772:30-39. [DOI] [PubMed] [Google Scholar]
- 24.Oliveira, S. C., J. S. Harms, R. R. Afonso, and G. A. Splitter. 2001. A genetic immunization adjuvant system based on bvp22-antigen fusion. Hum. Gene Ther. 12:1353-1359. [DOI] [PubMed] [Google Scholar]
- 25.Osen, W., T. Peiler, P. Ohlschlager, S. Caldeira, S. Faath, N. Michel, M. Muller, M. Tommasino, I. Jochmus, and L. Gissmann. 2001. A DNA vaccine based on a shuffled E7 oncogene of the human papillomavirus type 16 (HPV 16) induces E7-specific cytotoxic T cells but lacks transforming activity. Vaccine 19:4276-4286. [DOI] [PubMed] [Google Scholar]
- 26.Pardoll, D. M., and A. M. Beckerleg. 1995. Exposing the immunology of naked DNA vaccines. Immunity 3:165-169. [DOI] [PubMed] [Google Scholar]
- 27.Ren, X., J. S. Harms, and G. A. Splitter. 2001. Bovine herpesvirus 1 tegument protein VP22 interacts with histones, and the carboxyl terminus of VP22 is required for nuclear localization. J. Virol. 75:8251-8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson, H. L., and C. A. Torres. 1997. DNA vaccines. Semin. Immunol. 9:271-283. [DOI] [PubMed] [Google Scholar]
- 29.Sarmiento, M., A. L. Glasebrook, and F. W. Fitch. 1980. IgG or IgM monoclonal antibodies reactive with different determinants on the molecular complex bearing Lyt2 antigen block T cell-mediated cytolysis in the absence of complement. J. Immunol. 125:2665-2672. [PubMed] [Google Scholar]
- 30.Sato, Y., M. Roman, H. Tighe, D. Lee, M. Corr, M. D. Nguyen, G. J. Silverman, M. Lotz, D. A. Carson, and E. Raz. 1996. Immunostimulatory DNA sequences necessary for effective intradermal gene immunization. Science 273:352-354. [DOI] [PubMed] [Google Scholar]
- 31.Shedlock, D. J., and D. B. Weiner. 2000. DNA vaccination: antigen presentation and the induction of immunity. J. Leukoc. Biol. 68:793-806. [PubMed] [Google Scholar]
- 32.Sparwasser, T., E. S. Koch, R. M. Vabulas, K. Heeg, G. B. Lipford, J. W. Ellwart, and H. Wagner. 1998. Bacterial DNA and immunostimulatory CpG oligonucleotides trigger maturation and activation of murine dendritic cells. Eur. J. Immunol. 28:2045-2054. [DOI] [PubMed] [Google Scholar]
- 33.Tindle, R. W., G. J. Fernando, J. C. Sterling, and I. H. Frazer. 1991. A “public” T-helper epitope of the E7 transforming protein of human papillomavirus 16 provides cognate help for several E7 B-cell epitopes from cervical cancer-associated human papillomavirus genotypes. Proc. Natl. Acad. Sci. USA 88:5887-5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu, T.-C., F. G. Guarnieri, K. F. Staveley-O'Carroll, R. P. Viscidi, H. I. Levitsky, L. Hedrick, K. R. Cho, T. August, and D. M. Pardoll. 1995. Engineering an intracellular pathway for MHC class II presentation of HPV-16 E7. Proc. Natl. Acad. Sci. USA 92:11671-11675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yanagida, N., S. Yoshida, K. Nazerian, and L. F. Lee. 1993. Nucleotide and predicted amino acid sequences of Marek's disease virus homologues of herpes simplex virus major tegument proteins. J. Gen. Virol. 74:1837-1845. [DOI] [PubMed] [Google Scholar]