Abstract
The functional cooperation of equine herpesvirus 1 (EHV-1) glycoprotein M (gM) and the gene 10 (UL49.5) product was analyzed. Transient-transfection experiments using gM and UL49.5 expression plasmids as well as RK13 cell lines constitutively expressing UL49.5 (RK49.5) or gM (RKgM) demonstrated that the endo-β-N-acetylglucosaminidase H (endo H)-resistant mature form of gM was detectable only after coexpression of the two proteins. Deletion of the EHV-1 UL49.5-homologous gene 10 in strain KyA resulted in a small-plaque phenotype and up to 190-fold-reduced virus titers. The growth defects of the mutant KyAΔ49.5 virus, which were very similar to those of a gM-negative KyA virus, could be completely compensated for by growth of the mutant virus on RK49.5 cells or by repairing the deletion of gene 10 in the revertant virus KyAΔ49.5R. Analysis of cells infected with the UL49.5-negative EHV-1 demonstrated that gM was not transported to the trans-Golgi network in the absence of the UL49.5 product. In contrast, gM was efficiently transported and processed to the endo H-resistant mature form in KyAΔ49.5-infected RK49.5 cells. Furthermore, radioimmunoprecipitation experiments demonstrated that gM maturation was observed only if a 10,000-Mr protein was coprecipitated with gM in KyA- or KyAΔ49.5R-infected cells or virions. This protein was absent in cells infected with KyΔ49.5 or KyAΔgM, suggesting that it was the EHV-1 UL49.5 product. Taken together, our results demonstrate that the expression of the EHV-1 UL49.5 product is necessary and sufficient for gM processing and that it is required for efficient virus replication.
The Alphaherpesvirus Equine herpesvirus 1 (EHV-1) encodes a least 13 glycoproteins, most of which share high homology to those of herpes simplex virus type 1 or of its closest relatives suid herpesvirus 1 (pseudorabies virus [PRV]) and bovine herpesvirus 1 (BHV-1) (23, 29). PRV and BHV-1 together with EHV-1 and Varicella-zoster virus form the Varicellovirus genus within the Alphaherpesvirinae subfamily (30). Whereas the functions of several individual EHV-1 envelope proteins, such as glycoprotein B (gB), gC, gD, gE, and gM, have been analyzed in some detail (4, 18, 20-22), studies on the functions of the putative gE/gI, gH/gL, or gM/UL49.5 complexes have not yet been performed.
Complex formation between gM and the UL49.5 product, which is O glycosylated and therefore designated gN in PRV, Epstein-Barr virus (EBV), and human cytomegalovirus (HCMV), has been described for PRV (11), BHV-1 (31), EBV (14, 15), and HCMV (17). In the case of these viruses, gM and the UL49.5 product were shown to physically interact with each other by virtue of disulfide bonds that are probably formed between the first cysteine residue of the UL10 polypeptide, which is conserved throughout the UL10 homologs of all Herpesviridae sequenced to date, and one of the two equally highly conserved cysteine residues present within the UL49.5 homologs (31). Further, it could be shown for PRV that deletion of gM led to an absence of gN in the envelope of mature extracellular virions. In contrast, gM was efficiently incorporated into virions that lacked gN, and no growth defect after deletion of the UL49.5 (gN) gene from PRV was observed (11, 12). Similarly, deletion of the UL49.5 gene from HSV-1 or BHV-1 only marginally reduced virus titers (1, 16). In the case of EBV, gN processing was shown to be dependent on the expression of gM, whereas gM processing appeared to be unaffected in the absence of gN as assessed by experiments using CV-1 cells and T7 promoter-driven expression of gM and gN. Functional studies of the BLRF-1-negative EBV have demonstrated that gN is involved in an entry event following fusion and in virus egress from infected cells. The impaired egress was associated with an accumulation of nucleocapsids in condensed chromatin within the nuclei of infected cells (14). For HCMV, a member of the Betaherpesvirinae, transient-transfection experiments using gM and UL73 expression plasmids demonstrated that gM is necessary for processing and transport of the UL73 product, the UL49.5 (gN) homolog of HCMV (17).
Recent studies have provided more insight in the roles that gM or the gM/UL49.5 complex and the gE/gI complex play in the life cycle of EHV-1. The EHV-1 UL49.5-homologous protein is encoded by gene 10 of EHV-1. gM, a 50,000- to 55,000-Mr component of the virus envelope, is encoded by gene 52 (29). Concomitant deletion of gM together with gE and gI in EHV-1 led to a massive impairment in secondary virus envelopment at membranes of the trans-Golgi network (TGN) and subsequent virus egress which could be repaired by growth on a cell line expressing gM. These results indicated that gM or gM/UL49.5 and the gE/gI complex serve partially overlapping but distinct functions in virus egress (27). The results obtained with EHV-1 gE/I/M triple mutants closely mimicked those with PRV, in which the absence of gE and gM was responsible for massive growth defects affecting egress and cell-to-cell spread in cultured cells (2, 3). More recent work on the chicken Alphaherpesvirus Marek's disease virus (MDV) has suggested that one partner of gE/gI and/or gM/UL49.5 in cell-to-cell spread may be the product of the UL49 homolog (VP22), because MDV lacking gE, gI, or UL49 is nonviable in cultured cells (7, 26).
This study was performed to analyze the effect of a deletion of the UL49.5-homologous gene 10 in EHV-1 with respect to growth of a gE/gI-negative EHV-1 and the maturation of gM. Consistent with the absence of mature gM after infection of cells with the UL49.5-negative virus, a small-plaque phenotype and a 190-fold reduction of extracellular virus titers were observed in the absence of mature gM. The findings of this study demonstrate for the first time that a UL49.5-homologous protein is necessary for proper processing of its complex partner, the UL10 product, and for the function of gM and/or the gM/UL49.5 complex in virus egress and cell-to-cell spread.
MATERIALS AND METHODS
Virus and cells.
Strain KyA (kindly provided by D. J. O'Callaghan, Louisiana State University, Shreveport) was used in this study. KyA, which lacks the gE and gI genes (9), was grown on RK13 cells propagated in Dulbecco's minimal essential medium (DMEM) supplemented with 10% fetal calf serum. For isolation of a gene 10 (UL49.5)-negative KyA (KyAΔ49.5), 10 μg of recombinant plasmid pΔ49.5 was cotransfected with 1 to 10 μg of purified KyA DNA into RK13 cells. At 3 to 5 days after transfection, supernatants were harvested and plated on fresh cells. Recombinant fluorescent virus plaques were identified under a fluorescence microscope (Nikon), picked, and purified to homogeneity. Gene 10 revertant viruses were obtained after cotransfection of KyAΔ49.5 DNA and recombinant plasmid pE49.5 into RK13 cells. Revertant viruses were isolated by plaque purification of nonfluorescing virus plaques.
Plasmids.
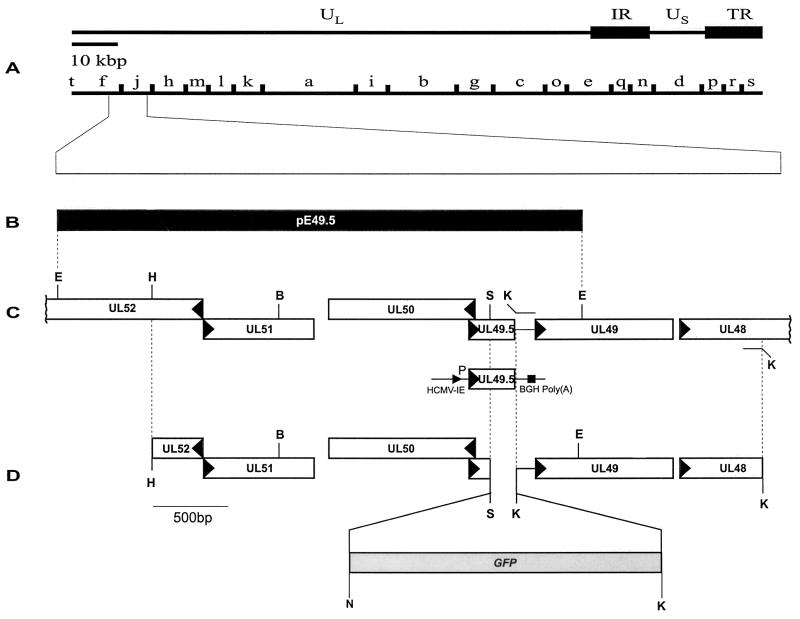
For construction of recombinant plasmid pc49.5, a PCR fragment (Table 1; Fig. 1) containing the EHV-1 gene 10 (UL49.5-homologous open reading frame [ORF] [29]) was cloned into the pcDNA3 vector (Invitrogen) under control of the HCMV immediate-early promoter-enhancer. To obtain recombinant plasmid pΔ49.5, a 2.2-kbp HindIII-SphI genomic fragment was cloned into vector pTZ18R (Amersham-Pharmacia) (Fig. 1). The SphI site is located at nucleotide position 146 of the 300-bp EHV-1 UL49.5-homologous gene 10. A 1.6-kbp fragment on the 3′ side of the UL49.5 ORF was amplified by PCR (Table 1) and cloned as a KpnI fragment in the vector containing the 5′ fragment. Finally, the green fluorescent protein (GFP) gene from plasmid pEGFP-N1 (Clontech) was released as an NspI-PstI fragment and ligated into the vector cut with SphI and PstI (Fig. 1). Plasmid pE49.5 contains a 3.5-kbp genomic fragment which encompasses gene 10 and was used for generation of the revertant virus (Fig. 1).
TABLE 1.
Primers used for generation of recombinant plasmids pc49.5 and pΔ49.5 and peptide sequences used for generation of gM-specific antisera
| Primer or peptide | Sequencea | Fragment size and/or positionb |
|---|---|---|
| Primers | ||
| 49.5up | 5′-ACATGGATCCATGCTGTCCACGAGATTCG-3′ | 300 bp (gene 10); 12084 |
| 49.5low | 5′-GACTGTCGACTCAATGCAGGTGTTGCAAC-3′ | 300 bp (gene 10); 12387 |
| pΔ49.5-for | 5′-ATCGGTACCTATGTATAACTCATCCC-3′ | 1.6 kbp; 12396 |
| pΔ49.5-rev | 5′-GACGGTACCATCACGTATATTTCTAGG-3′ | 1.6 kbp; 14059 |
| Peptides | ||
| 047 | H2N-TASLPQTGYPCFYGSLVDYTQKNHSVVD-COOH | 53-80 |
| 048 | H2N-LVRRYARGKECTAVAGCTRPTTTLI-COOH | 126-150 |
Boldface indicates restriction enzyme sites used for cloning into pcDNA3 or pTZ18R. Underlining indicates random 5′ overhangs to facilitate restriction enzyme digestion and cloning.
For the primers, nucleotide positions according to reference 29 are shown. For the peptides, the amino acid position in EHV-1 gM is shown.
FIG. 1.
(A) Schematic illustration of the organization of the approximately 140-kbp KyA genome and the BamHI restriction map. (B) The 3.5-kbp EcoRI fragment (pE49.5) which contains the UL49.5-homologous gene 10 of EHV-1. (C) Plasmid pc49.5 was generated by insertion of the EHV-1 UL49.5-homologous gene 10 into vector pcDNA3 (Invitrogen). (D) Construction and structure of plasmid pΔ49.5 used for generation of the UL49.5-negative virus mutant. Restriction enzyme sites: B, BamHI; E, EcoRI; K, KpnI; N, NspI; S, SphI.
Generation of cell lines expressing EHV-1 UL49.5.
Ten micrograms of recombinant plasmid pc49.5 was transfected into RK13 cells, and cell clones developing under DMEM containing 500 μg of G-418 (Calbiochem) per ml were isolated and expanded for further analysis by cotransfection with recombinant plasmid pcgM, which encodes EHV-1 gM under control of the HCMV immediate-early promoter-enhancer (22).
Single-step growth kinetics and plaque diameter determinations.
Single-step growth kinetics were determined in triplicate by infecting 105 RK13, RK49.5, or RKgM cells (27) at a multiplicity of infection (MOI) of 3. Virus was allowed to attach for 2 h on ice, followed by a penetration period of 2 h at 37°C. At the indicated times after the temperature shift, supernatants of infected cells were harvested, and viral titers were determined by plating onto RK13 cells (27). Plaque diameters were measured after titration of virus mutants on different cell lines and 3 days of incubation at 37°C under a methylcellulose overlay. Cells were fixed with 5% formaldehyde and stained with crystal violet. For each combination of virus and cells, 150 plaques were measured and the average plaque diameters were determined. Values were calculated and compared to KyA plaque diameters, which were set to 100%. Average percentages of plaque diameters and standard deviations were determined from three independent experiments.
Electron microscopy.
For ultrastructural analyses, RK13 or RK49.5 cells were infected at an MOI of 1, fixed at different times postinfection (p.i.) (16 to 20 h) for 60 min with 2.5% glutaraldehyde buffered in 0.1 M Na-cacodylate (pH 7.2, 300 mosM) (Merck). Cells were then scraped off the flask, pelleted by low-speed centrifugation, and embedded in LMP agarose (Biozym). Small pieces were postfixed in 1.0% aqueous OsO4 (Polysciences Europe) and stained with uranyl acetate. After stepwise dehydration in ethanol, cells were cleared in propylene oxide, embedded in Glycid Ether 100 (Serva), and polymerized at 59°C for 4 days. Ultrathin sections were counterstained with uranyl acetate and lead salts and were examined by electron microscopy (Philips EM 400T).
Antibodies.
Monoclonal antibodies (MAbs) directed against EHV-1 gM were kindly provided by Lindsey Day and Richard Killington, University of Leeds, Leeds, United Kingdom (5, 27). As a TGN marker, a γ-adaptin MAb (Sigma) was used. Polyclonal rabbit anti-gM antisera 047 and 048 were produced after injection of the 047 or 048 peptide (Table 1), coupled to keyhole limpet hemocyanin (Sigma) and suspended in Freund's complete (first immunization) or incomplete (booster immunizations) adjuvant, for a total of six times into a New Zealand White rabbit (Charles River). An anti-gD 20-mer antibody (8) (kindly provided by Dennis J. O'Callaghan) and the anti-gB 3F8 MAb (20) were used as control antibodies. Secondary peroxidase-conjugated anti-mouse immunoglobulin G (IgG) antibodies (Sigma) as well as fluorescein isothiocyanate-conjugated anti-mouse IgG2b (Sigma) and Cy3-conjugated anti-mouse IgG1 antibodies (Amersham Biosciences) were used according to the manufacturers' instructions.
IIF, Western blotting, and RIPA.
For indirect immunofluorescence (IIF), RK13 cells were grown on six-well plates and subsequently infected at an MOI of 0.01 or transfected with gM or UL49.5 expression plasmids. Cells were fixed with acetone at 48 to 72 h p.i. or after transfection, and IIF and confocal laser scanning microscopy using a Zeiss LSM510 confocal microscope were done exactly as described previously (19, 23, 27). For Western blot analyses, cells were transfected or infected at an MOI of 2 with the various viruses. At 48 h after transfection or 16 h after infection, cell lysates were prepared. Extracellular virions were purified by sucrose gradient centrifugation (19, 22). Cell lysates were adjusted to equal protein concentrations of 5 mg per ml as determined with a bicinchoninic acid kit (Pierce) (24). Samples were heated at 56°C for 2 min in the presence of 2-mercaptoethanol unless otherwise stated, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to nitrocellulose membranes (Schleicher & Schuell) by the semidry method (13). Free binding sites on the sheets were blocked by addition of 10% skim milk in phosphate-buffered saline containing 0.05% Tween before the antibodies (suspended in phosphate-buffered saline containing 0.05% Tween) were added. Bound antibodies were detected with anti-rabbit (or anti-mouse) IgG-peroxidase conjugates (Sigma) and visualized by enhanced chemiluminescence (ECL; Pharmacia-Amersham). For radioimmunoprecipitation (RIPA), 106 infected or mock-infected cells (MOI = 1) were labeled at 16 h p.i. for 20 min with 200 μCi of Tran35S-label (ICN) after incubation for 20 min in Met- and Cys-free DMEM (Gibco-BRL). Radiolabeled extracellular virions were prepared by continuous labeling of infected RK13 cells with 250 μCi of Tran35S-label for 36 h and subsequent sucrose gradient purification. Cells or virions were lysed with detergent buffer (24), and precipitations after preclearing of the cell lysates were done using a mixture of 2 μl of the 047 peptide serum and 2 μl of the 048 peptide serum or with 50 μl of hybridoma supernatant of MAb 3F8. Antigen-antibody complexes were pelleted using Pansorbin cells (Calbiochem) according to the supplier's instructions. After three thorough washes in RIPA washing buffer (25), complexes were released from Pansorbin cells by addition of sample buffer and heating to 56°C for 2 min before SDS-PAGE.
Deglycosylation of cell lysates.
Deglycosylation experiments were performed by suspension of infected-cell lysates (from 106 cells) in 100 μl of de-N-deglycosylation buffer (50 mM K3PO4 [pH 7.2], 50 mM EDTA, 0.6% [vol/vol] CHAPS, 0.1% SDS) and subsequent addition of peptide-N-glycosidase (PNGase) F (0.4 U) or endo H (2 mU) (Roche Biochemicals). Control lysates were left untreated. After 16 h of incubation at 37°C, lysates were separated by SDS-10% PAGE, transferred to nitrocellulose, and examined by Western blotting. De-O-glycosylations were done with neuraminidase and O-glycosidase exactly as previously described (12, 19), except that immunoprecipitates with the 047 and 048 rabbit antibodies or the anti-gD antibody as a control were used.
RESULTS
Coexpression of EHV-1 UL49.5 and gM results in gM maturation.
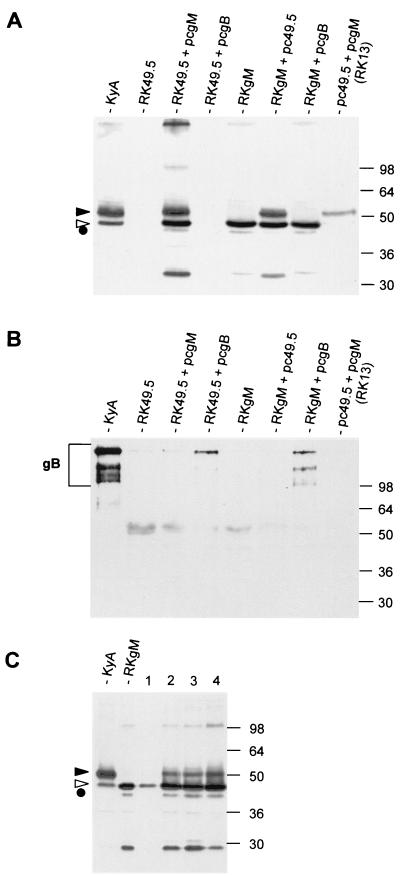
The results of preliminary transfection experiments using a gM expression plasmid had suggested that the coexpression of the EHV-1 UL49.5 gene can result in gM maturation. To analyze the putative UL49.5-gM interaction in more detail, RK13 cells were cotransfected with plasmids expressing either EHV-1 UL49.5 (pc49.5) or gM (pcgM) (22) or were cotransfected with both expression plasmids. Whereas no reactivity with the gM-specific MAb P18/A8 (5, 27) was observed in cells transfected with pc49.5, the 46,000- to 48,000-Mr endo H-sensitive form of gM was observed in cells transfected with pcgM only (27) (data not shown). In contrast, in RK13 cells cotransfected with both pc49.5 and pcgM, both high-mannose-containing gM and, predominantly, the mature 50,000- to 55,000-Mr gM moiety were detectable (Fig. 2A). The same observation, i.e., maturation of gM to the 50,000- to 55,000-Mr protein, was made when cell line RKgM (27) was transfected with pc49.5 or when a cell line constitutively expressing EHV-1 UL49.5 (RK49.5) was transfected with pcgM (Fig. 2A). In contrast, the endo H-resistant 50,000- to 55,000-Mr gM protein was absent in RKgM cells cotransfected with a gB expression plasmid (pcgB) (20). The expression of gB in pcgB-transfected RK13 cells but not in those transfected with pcgM or pc49.5 was verified by Western blot analysis using MAb 3F8 (20). Especially the di- and oligomeric forms of EHV-1 gB were detectable in the respective cell lysates (Fig. 2B), because the cell lysates had been heated at 56°C, which leaves most of the gB complexes intact (20). From a number of transfection experiments it became apparent that gM processing was much more efficient after cotransfection of UL49.5 and gM expression plasmids than after transfection of gM- or UL49.5-expressing cells with the complementary expression plasmid. An explanation for this observation may be that efficient processing of gM requires cotranslational complex formation, which can be impaired if one of the complex partners is preexistent. From the results of the transfection experiments, we concluded that processing of EHV-1 gM into the mature endo H-resistant form requires the UL49.5 product.
FIG. 2.
Western blot analyses of RK13, RK49.5, and RKgM cells transfected with pc49.5, pcgM, or pcgB. Forty-eight hours after transfection, cells were harvested and lysed, and protein extracts were separated by SDS-10% PAGE followed by Western blotting using anti-gM antibody P18/A8 (A and C) or anti-gB antibody 3F8 (B). Cells lines used for the transfections are given. The gM precursor (circle), the high-mannose gM (open arrowhead), and the fully glycosylated gM (solid arrowhead) are indicated. Individual cell clones which constitutively expressed the UL49.5 protein were identified by the presence of processed gM after transfection of pcgM (C). Cell line 4 was chosen for further studies and termed RK49.5. Mature and immature gM are indicated by solid and open arrowheads, respectively (A and C); gB-specific bands are marked by a bracket (B). The sizes of a prestained molecular weight marker (SeaBlue; Novex) are given in thousands.
Generation of an RK13 cell line constitutively expressing UL49.5.
The full maturation of gM in the presence of the UL49.5 product was exploited to identify and isolate cell clones constitutively expressing UL49.5, because an antibody against the UL49.5 protein of EHV-1 was not available. After transfection of pc49.5 into RK13 cells, G418-resistant cell clones were isolated and subsequently transfected with pcgM. UL49.5-expressing cell clones were identified by the detection of the mature 50,000- to 55,000-Mr gM moiety (Fig. 2A). One G418-resistant cell clone (clone 4) in which high quantities of mature gM were observed after transfection of pcgM was termed RK49.5 and used for all further transcomplementation and deglycosylation experiments (Fig. 2A and C).
Generation and analysis of a UL49.5-negative KyA.
Cotransfection of RK13 cells with recombinant plasmid pΔ49.5, in which gene 10, the UL49.5-homologous gene, was replaced with the GFP gene, and KyA DNA resulted in virus progeny that induced the formation of fluorescent plaques. The transfection progeny viruses were plaque purified on RK49.5 cells three times until all plaques stained homogenously green under the fluorescence microscope. Southern blot analysis confirmed the correct insertion of the GFP ORF instead of the UL49.5-homologous gene 10 in the UL49.5-negative KyA, which was termed KyAΔ49.5 (Fig. 1). A gene 10 revertant virus, KyAΔ49.5R, was generated by cotransfection of KyAΔ49.5 DNA with recombinant plasmid pE49.5 (Fig. 1) into RK13 cells and selection for nonfluorescing plaques. The correct reinsertion of the UL49.5 gene in the revertant virus was also confirmed by Southern blot analysis (data not shown).
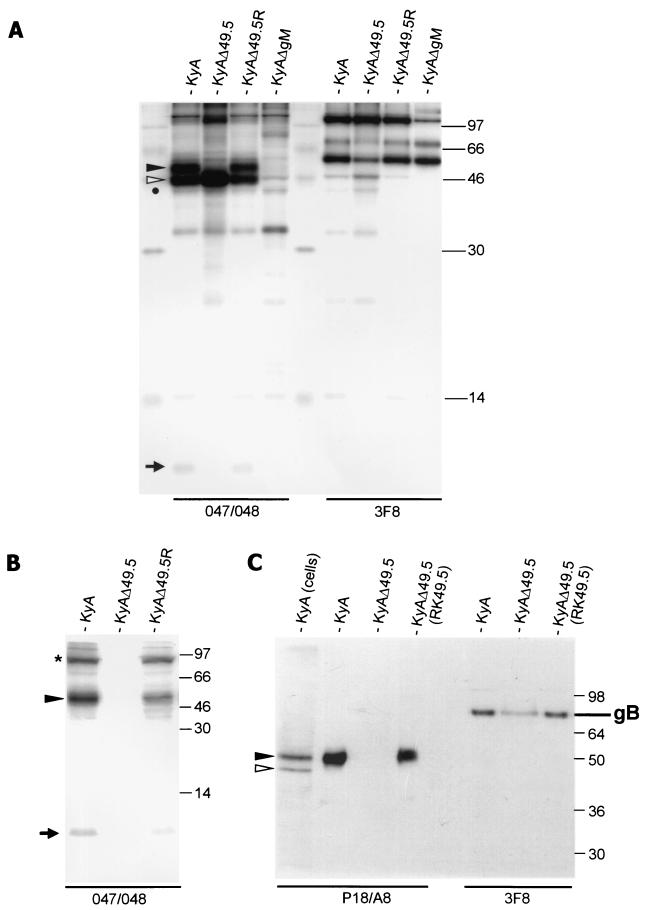
KyAΔ49.5, KyAΔ49.5R, KyAΔgM (27), or parental KyA virus was used to infect RK13 cells and to perform RIPA with the 047 and 048 anti-gM peptide antisera (Table 1) (27). In RK13 cells, a 10,000-Mr protein was coprecipitated with the two gM-specific proteins after infection with the parental KyA or UL49.5 revertant virus, but no such protein was observed in precipitates of RK13 cells infected with KyAΔ49.5 or KyAΔgM (Fig. 3A). In addition, only the immature form of gM was detectable in RK13 cells infected with the UL49.5-negative mutant virus (Fig. 3A), whereas both the immature and mature forms of gM were precipitated with the gM-specific antibody in cells infected with KyA or revertant KyAΔ49.5R virus. The 10,000-Mr protein could also be coprecipitated with the gM-specific 047 or 048 antiserum from extracellular KyA or KyAΔ49.5R virions purified over sucrose gradients, but not from KyAΔ49.5 virions (Fig. 3B). Because the 10,000-Mr protein was present in immunoprecipitates of wild-type KyA or UL49.5 revertant viruses but not in that of KyAΔ49.5, and because the apparent Mr of the putative UL49.5 product corresponded well to the calculated Mr of 10,801, we concluded that gM and the UL49.5 protein product form a heterodimer in infected cells and virions. The fact that the apparent Mr of the UL49.5 product remained unaltered after treatment with neuraminidase and O-glycosidase and after treatment with PNGase F (data not shown) suggested that the EHV-1 UL49.5 product is not glycosylated.
FIG. 3.
(A) RIPA analysis of RK13 cells infected with wild-type KyA, the UL49.5- (gene 10)-negative KyAΔ49.5 mutant, the UL49.5 revertant virus, or gM-negative KyAΔgM (27). (B) RIPA of purified radiolabeled KyA, KyAΔ49.5, or KyAΔ49.5R virions The antibodies used for precipitations were anti-gB antibody 3F8 and the anti-gM polyclonal antibodies 047 and 048. Mature and immature gM are indicated by full and open arrowheads, respectively; 2-mercaptoethanol-resistant gM dimers (24, 27) are indicated by an asterisk. The putative UL49.5 (gene 10) product is indicated by an arrow. The sizes of a molecular weight marker (14C marker; Gibco-BRL) are given in thousands. (C) Western blot analysis of purified KyA and KyAΔ49.5 virions prepared from RK13 or RK49.5 cells by using anti-gM MAb P18/A8 or anti-gB MAb 3F8. Whereas the 72,000- to 75,000-Mr large subunit of gB could be detected with MAb 3F8 in all virion preparations, gM was detected in KyA virions and KyAΔ49.5 virions prepared from RK49.5 cells. gM was absent from UL49.5-negative virions prepared on RK13 cells. Samples were heated for 2 min at 56°C for detection of gM and for 3 min at 95°C for detection of gB. In the first lane, KyA-infected RK13 cells were loaded. The sizes of a prestained molecular weight marker (SeaBlue; Novex) are given in thousands.
The incomplete processing of gM in the absence of the UL49.5 product was also examined by performing deglycosylation experiments with RK13 and RK49.5 cells infected with either KyA or KyAΔ49.5. Whereas the endo H-resistant 50,000- to 55,000-Mr gM moiety was not observed in RK13 cells infected with KyAΔ49.5, the mature glycoprotein was readily detected in KyAΔ49.5-infected complementing RK49.5 cells (data not shown). Because EHV-1 gM was not fully processed in the absence of the UL49.5 product, it was absent in KyAΔ49.5 virions purified from RK13 cells, whereas the 72,000- to 75,000-Mr subunit of gB was readily detected in purified KyAΔ49.5 virions by using MAb 3F8 after heating of the samples to 95°C (Fig. 3C). In contrast, gM was efficiently incorporated into UL49.5-negative virions by growth on complementing RK49.5 cells (Fig. 3C), demonstrating that incorporation of gM into extracellular mature virions requires the UL49.5 product.
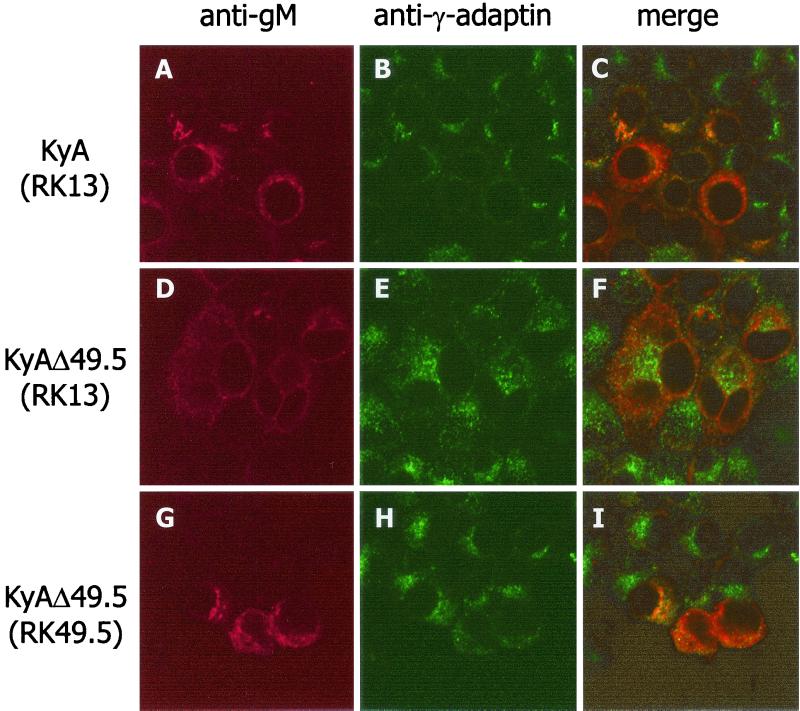
The intracellular distribution of gM in the absence of the UL49.5 product was also examined by colocalization studies with IIF and a TGN-specific anti-γ-adaptin MAb and anti-gM or gB MAbs, followed by confocal laser scanning microscopy (19, 23). Whereas gB localization to the TGN was readily detectable in both KyA- and KyAΔ49.5-infected cells (data not shown), gM was detectable in this compartment in KyA-infected cells (Fig. 4A to C) but not in RK13 cells infected with KyAΔ49.5, where gM exhibited a strong signal around the nuclear rim and was dispersed throughout the cytoplasm, and no colocalization of γ-adaptin and gM signals was observed (Fig. 4D to F). The observed gM distribution was highly reminiscent of that previously demonstrated for mutant EHV-1 gM molecules, which were retained in the endoplasmic reticulum (27). Colocalization of gM with γ-adaptin signals was restored in RK49.5 cells infected with KyAΔ49.5 (Fig. 4G to I). From these results we concluded that incomplete processing of EHV-1 gM in the absence of the UL49.5 product was caused by an inhibition of gM transport to the TGN.
FIG. 4.
Confocal laser scanning microscopy of RK13 (A to F) or RK49.5 (G to I) cells infected with KyA (A to C) or KyAΔ49.5 (D to I). gM was detected with MAb P18/A8 and visualized with anti-mouse IgG1-Cy3 conjugate (A, D, and G), and TGN vesicles were stained with γ-adaptin MAb followed by anti-mouse IgG2b-fluorescein isothiocyanate antibody (B, E, and H). Merges of the signals recorded separately for each channel (green, 505 to 530 nm; red, >585 nm) are shown in panels C, F, and I. The yellow color in C and G indicates colocalization of gM and γ-adaptin signals. Panels represent views of 60 by 60 μm.
KyAΔ49.5 is impaired in cell-to-cell spread virus and egress.
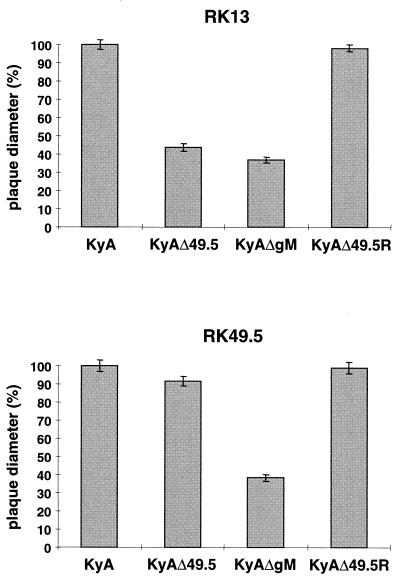
The next series of experiments addressed the growth properties of KyAΔ49.5 in comparison to parental KyA or revertant virus. Plating of the different viruses on RK13 and RK49.5 cells demonstrated that cell-to-cell spread of the UL49.5-negative mutant was severely impaired in the absence of the protein, and only 37% of the plaque diameters of parental KyA or revertant virus were reached (Fig. 5). This defect in cell-to-cell spread of infectivity was completely restored by plating of KyAΔ49.5 on RK49.5 cells constitutively expressing the UL49.5 protein (Fig. 5) but not on RKgM cells, which constitutively express gM and which were able to rescue plaque diameters of the gM-negative KyA mutant (27).
FIG. 5.
Plaque diameters of KyA, KyAΔ49.5, KyAΔ49.5R, and KyAΔgM on RK13 and RK49.5 cells. Shown are means and standard deviations of diameters of 150 plaques measured for each virus-cell line combination. Plaque diameters of KyA were set to 100%.
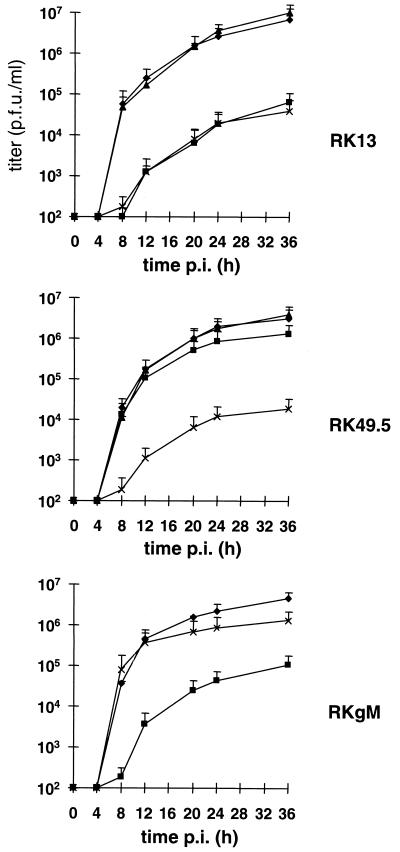
Single-step growth kinetics of mutant KyAΔ49.5 virus and wild-type KyA or the UL49.5 revertant virus were determined. The individual viruses were used to infect RK13, RK49.5, or RKgM cells at an MOI of 3, and the results of the experiments are summarized in Fig. 6. KyAΔ49.5 grown on RK13 cells exhibited a reduction of virus titers of up to 190-fold compared to titers of parental KyA or rescuant KyAΔ49.5R virus (Fig. 6). This defect in virus maturation and egress was virtually completely reversed when KyAΔ49.5 was grown on complementing RK49.5 cells but not when it was grown on RKgM cells (Fig. 6). On RKgM cells, the impaired growth of the gM-negative KyAΔgM virus which was observed on RK13 cells, and was virtually indistinguishable from that of the UL49.5-negative KyAΔ49.5 virus, could be completely rescued (Fig. 6). From these results we concluded that efficient growth of the gE/gI-negative EHV-1 strain KyA in cultured cells requires the UL49.5 product and that deletion of the gene 10 (UL49.5-homologous) ORF resulted in a massive impairment of cell-to-cell spread and virus egress that was identical to that of a gM-negative KyA virus.
FIG. 6.
Single-step growth kinetics of KyA (⧫), KyAΔ49.5 (▪), KyAΔ49.5R (▴), and KyAΔgM (×) on RK13, RK49.5, or RKgM cells. At the indicated times p.i., virus titers were determined on RK13 cells as described in Materials and Methods. Shown are means and standard deviations from three independent experiments.
KyAΔ49.5 virus maturation is blocked at the stage of secondary envelopment.
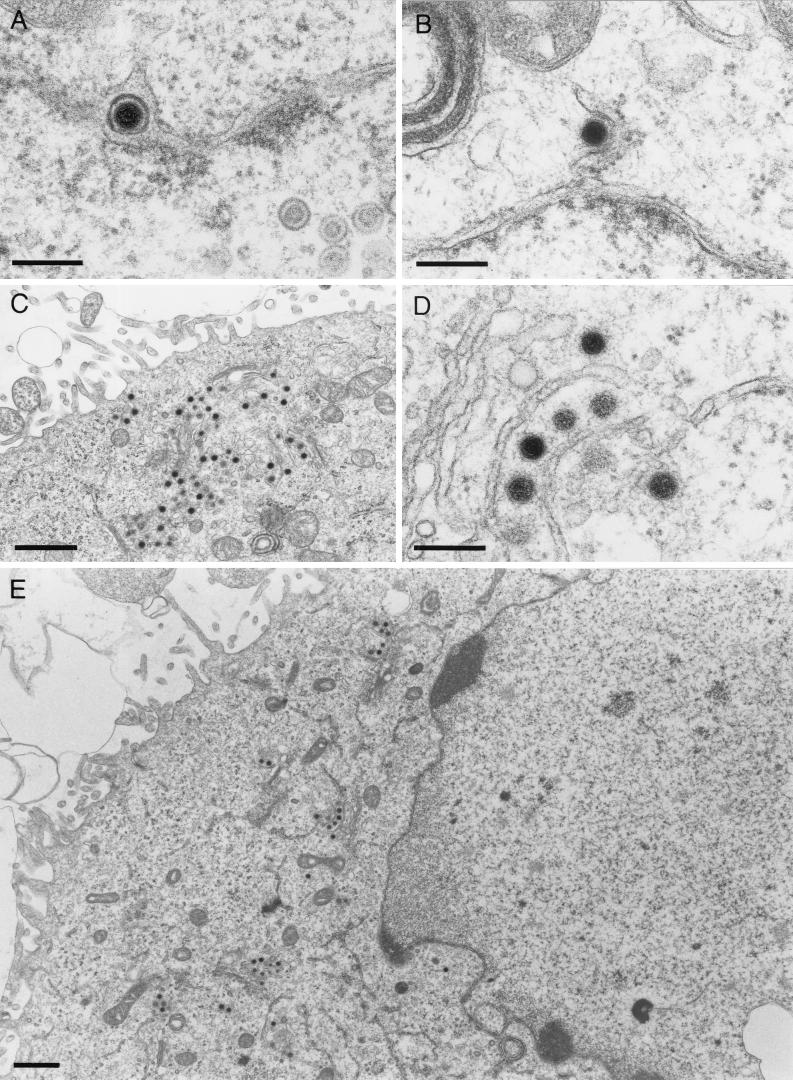
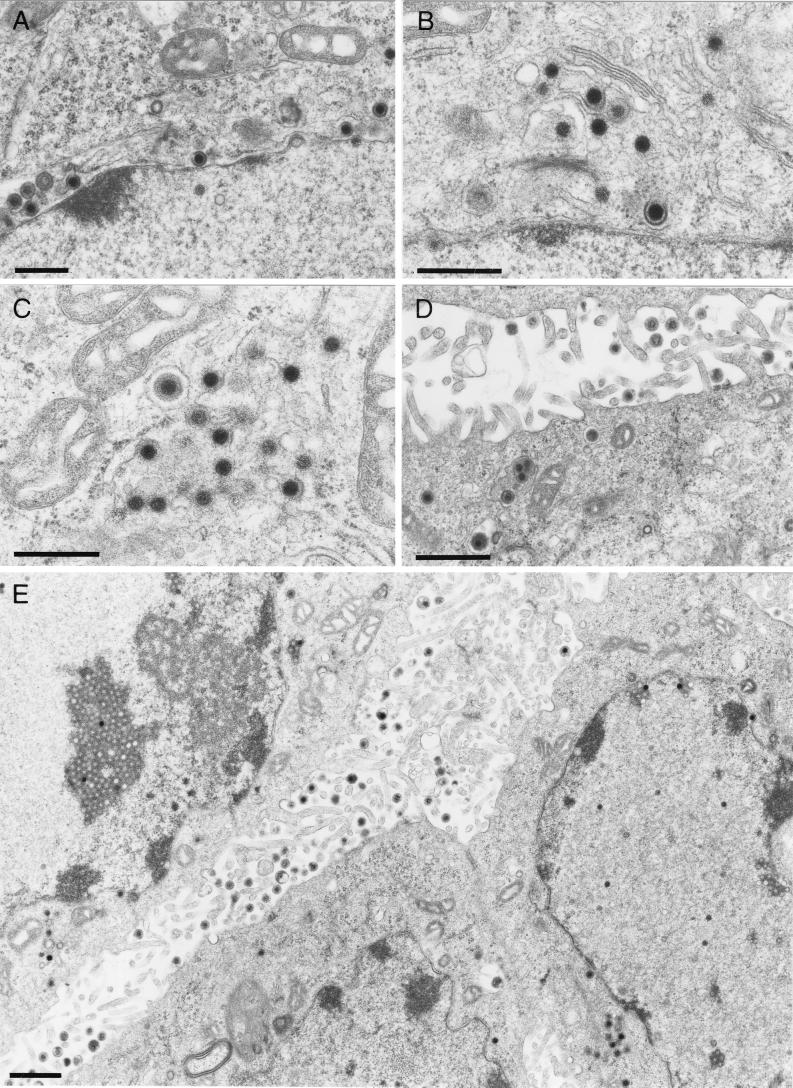
To further examine the reason for the massive impairment of virus egress and cell-to-cell spread of UL49.5-negative KyA, ultrastructural studies were performed. These studies revealed that UL49.5-negative KyAΔ49.5 exhibited a clearly visible defect in secondary envelopment at presumably TGN membranes. Nucleocapsid formation in infected-cell nuclei, primary budding at the inner lamella of the nuclear membrane, and de-envelopment at the outer lamella of the nuclear membrane, however, did not appear to be different from those observed in KyA-infected cells (Fig. 7A and B). Budding of nucleocapsids at membranes of the TGN was observed only very rarely, and de-enveloped nucleocapsids accumulated in the Golgi region (Fig. 7C). Higher magnification of the accumulated nucleocapsids revealed a clearly detectable rim of fuzzy material, presumably tegument proteins, around cytoplasmic nucleocapsids which was similar to that observed after deletion of gM (Fig. 7D) (27). Very rarely, virions released into the extracellular space were observed (Fig. 7E). In contrast, replication of KyAΔ49.5 on complementing RK49.5 cells was morphologically indistinguishable from that shown for parental KyA or the wild-type RacL11 virus (10, 27). After primary envelopment by budding at the inner lamella of the nuclear membrane, nucleocapsids were released into the cytoplasm and received their final envelope by budding at membranes in the Golgi region (Fig. 8A and B). Virus-containing vesicles (Fig. 8C) released their content by fusion at the cell membrane, and a large number of virions were visible in the extracellular space (Fig. 8D and E). Taken together, the electron microscopic studies demonstrated that the impairment of KyAΔ49.5 in virus egress was caused by a defect in secondary envelopment of virions. This defect appeared to be ultrastructurally identical to that of a gM-negative KyA (27) and could be reversed by growth of the virus on a complementing cell line.
FIG. 7.
Electron micrographs of RK13 cells infected with KyAΔ49.5 at 16 h p.i. Budding of nucleocapsids of the UL49.5-negative mutant at the inner lamella of the nuclear membrane and the presence of enveloped virions in the perinuclear space (A), as well as de-envelopment at the outer lamella of the nuclear membrane (B), were visible. Cytoplasmic nucleocapsids released from infected-cell nuclei accumulated in the vicinity of Golgi membranes (C). Higher magnification demonstrated fuzzy material, probably representing tegument proteins, around cytoplasmic nucleocapsids (D). An overview of a section of an infected cell with only few virions in secretory vesicles or the extracellular space is also shown (E). Bars, 250 nm (A, B, and D) and 1 μm (C and E).
FIG. 8.
Electron micrographs of RK13 cells infected with KyAΔ49.5 on complementing RK49.5 cells at 16 h p.i. Various stages of normal EHV-1 morphogenesis can be seen. These stages included budding of nucleocapsids at the inner lamella of the nuclear membrane and de-envelopment at the outer lamella of the nuclear membrane (A), naked nucleocapsids in the cytoplasm (B), and secondary envelopment of cytoplasmic virions at Golgi membranes leading to enveloped mature virions in secretory vesicles (C) Particles in secretory vesicles were finally released into the extracellular space (D). A lower-magnification overview of a section of an infected cell is also shown (E); note the high number of released virions at the cell surface. Bars, 500 nm (A to C) or 1 μm (D and E).
DISCUSSION
Experiments to elucidate the function of the EHV-1 UL49.5 protein and its cooperation with gM were conducted. It could be demonstrated that maturation of gM to the fully glycosylated and functional form of the glycoprotein requires the product of the UL49.5-homologous ORF of EHV-1, gene 10. This conclusion is drawn from the results of cotransfection experiments using gM- and UL49.5-expressing plasmids, analysis of glycosylation patterns of gM in cells infected with a UL49.5-negative EHV-1, the growth properties of a UL49.5-negative mutant in cultured cells, and RIPA experiments which showed that a 10,000-Mr protein was coprecipitated with gM in infected cells and mature extracellular virions. A functional impairment of the larger complex partner gM or the gM/UL49.5 complex was shown for the first time to be dependent on the presence of the smaller complex partner, UL49.5.
The Alphaherpesvirinae encode at least six (glyco)proteins which form hetero-oligomers in the virus envelope. These complexes comprise the gE/gI complex, the gH/gL complex, and a complex between gM and the UL49.5 product which is held together by disulfide bonds (11, 28, 31). The UL49.5 product is O glycosylated in the case of PRV, in which the complex formed between the two highly conserved ORFs, present in all herpesviruses identified to date, was first described. The UL49.5 product does not appear to carry O-glycans in BHV-1 but does so in the members of the Betaherpesvirinae (HCMV) and Gammaherpesvirina (EBV) (15, 17, 31). Our previous studies and the results reported here may suggest that the UL49.5 product encoded by gene 10 (29) is not glycosylated in EHV-1 but forms a disulfide-linked complex with gM, as evidenced by altered electrophoretic mobilities of immunoprecipitates obtained with anti-gM antibodies under nonreducing conditions (24) and by analysis of immunoprecipitates by using gM antibodies, which contained a 10,000-Mr protein which was absent in both gM- and UL49.5-negative viruses. Attempts to unanimously identify this protein as the UL49.5 product by using a rabbit polyclonal antibody have failed, and our laboratory currently focuses on the generation of anti-UL49.5 MAbs to define the 10,000-Mr polypeptide coimmunoprecipitated with gM.
The absence of the EHV-1 UL49.5 product had profound effects on the maturation of its complex partner, gM, and also on virus growth in cultured cells. Using EHV-1 strain KyA, which lacks the gE and gI genes (9), the critical and yet partially redundant functions of gM or the gM/UL49.5 complex in virus egress and cell-to-cell spread could be demonstrated (27). The more pronounced effect of a deletion of gM in a gE/gI-negative background is obviously caused by partially overlapping although distinct functions of the glycoproteins in virus egress and cell-to-cell spread, which has been shown not only for EHV-1 but also for PRV (2, 3, 27). Using the same system of a concomitant absence of gM or the gM/UL49.5 complex and the gE/gI complex, the consequences of a deletion of UL49.5 in strain KyA were analyzed. Coexpression of UL49.5 and gM resulted in complete processing of the multiply hydrophobic gM, whereas absence of the UL49.5 product led to an incomplete trimming of sugar moieties of gM in infected and stably or transiently transfected cells. These findings and the absence of gM processing and transport to the TGN in cells infected with the UL49.5-negative mutant demonstrated that expression of the UL49.5 product is both necessary and sufficient for functional maturation of gM to the endo H-resistant glycoprotein. To our knowledge, incomplete gM maturation in the absence of its complex partner is unprecedented in any other member of the Herpesviridae. It was previously shown, however, that processing of the smaller complex partner gN was dependent on gM expression in EBV and HCMV (15) and that EBV mutants having a gN deletion exhibited reduced growth in cultured cells (14). In contrast, deletion of the UL49.5 product in PRV did not lead to altered growth properties of the mutant virus in vitro, whereas deletion of gM led to an approximately 50-fold decrease of virus titers and reduced plaque diameters. Moreover, gM appeared to be fully processed and was incorporated into extracellular virus particles, whereas gN was absent from virions which were also lacking gM (6, 11). From these results it appears that gM but not gN (UL49.5) or the gM/gN complex of PRV is the functional entity. Because EHV-1 gM does not reach the TGN and secretory vesicles in the absence of its complex partner, it is consequently absent from extracellular virus particles. Therefore, the situation in EHV-1 is fundamentally different from that observed in PRV with regard to the UL49.5 protein, gM, and the complex between the two envelope constituents. Besides the structural consequences, deletion of either gM or the UL49.5 product, at least in a gE/gI-negative virus, led to essentially identical phenotypes in cultured cells. These findings leave several ways for interpretation, but it appears that the function of the EHV-1 complex partners is more likely to reside in the formation of the complex than in action of the individual proteins, as is the case in PRV. This interpretation is also based on the ultrastructural investigations, which revealed a block of virus egress at the stage of secondary envelopment in the trans-Golgi region and virus egress for both KyAΔ49.5 and KyAΔgM. In addition, cell-to-cell spread defects of the UL49.5- and gM-negative KyA mutants were virtually identical, suggesting that fully processed gM is also needed for direct transmission of infectivity from an infected cell to a neighboring uninfected cell. Further studies will focus on the physical interaction between the EHV-1 UL49.5 protein and its complex partner by targeted mutagenesis of cysteine residues that are predicted to be involved in the complex formation between the two virion components. These studies will be necessary to investigate whether physical interaction is necessary for the function of the individual proteins or the complex formed between these highly conserved proteins at various stages of EHV-1 maturation and egress.
Acknowledgments
The skillful technical assistance of Kerstin Wink and Petra Meyer is gratefully acknowledged. E. Zorn and H. Stephan helped with the artwork. We thank Dennis J. O'Callaghan for providing strain KyA and anti-gD 20-mer antibody, and we thank Lindsey Day and Richard A. Killington for the generous gift of anti-gM MAbs.
This study was supported by DFG grant Os 143/2 to N.O.
REFERENCES
- 1.Adams, R., C. Cunningham, M. D. Davison, C. A. MacLean, and A. J. Davison. 1998. Characterization of the protein encoded by gene UL49A of herpes simplex virus type 1. J. Gen. Virol. 79:813-823. [DOI] [PubMed] [Google Scholar]
- 2.Brack, A. R., J. M. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J. Virol. 73:5364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brack, A. R., B. G. Klupp, H. Granzow, R. Tirabassi, L. W. Enquist, and T. C. Mettenleiter. 2000. Role of the cytoplasmic tail of pseudorabies virus glycoprotein E in virion formation. J. Virol. 74:4004-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Csellner, H., C. Walker, J. E. Wellington, L. E. McLure, D. N. Love, and J. M. Whalley. 2000. EHV-1 glycoprotein D (EHV-1 gD) is required for virus entry and cell-cell fusion, and an EHV-1 gD deletion mutant induces a protective immune response in mice. Arch. Virol. 145:2371-2385. [DOI] [PubMed] [Google Scholar]
- 5.Day, L. 1999. Characterisation of selected glycoproteins of equine herpesvirus-1. Ph.D. thesis. University of Leeds, Leeds, United Kingdom.
- 6.Dijkstra, J. M., N. Visser, T. C. Mettenleiter, and B. G. Klupp. 1996. Identification and characterization of pseudorabies virus glycoprotein gM as a nonessential virion component. J. Virol. 70:5684-5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorange, F., B. K. Tischer, J.-F. Vautherot, and N. Osterrieder. 2002. Characterization of Marek's disease virus serotype 1 (MDV-1) deletion mutants that lack UL46 to UL49 genes: MDV-1 UL49, encoding VP22, is indispensable for virus growth. J. Virol. 76:1959-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flowers, C. C., and D. J. O'Callaghan. 1992. Equine herpesvirus 1 glycoprotein D: mapping of the transcript and a neutralization epitope. J. Virol. 66:6451-6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flowers, C. C., and D. J. O'Callaghan. 1992. The equine herpesvirus type 1 (EHV-1) homolog of herpes simplex virus type 1 US9 and the nature of a major deletion within the unique short segment of the EHV-1 KyA strain genome. Virology 190:307-315. [DOI] [PubMed] [Google Scholar]
- 10.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 75:3675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jöns, A., J. M. Dijkstra, and T. C. Mettenleiter. 1998. Glycoproteins M and N of pseudorabies virus form a disulfide-linked complex. J. Virol. 72:550-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jöns, A., H. Granzow, R. Kuchling, and T. C. Mettenleiter. 1996. The UL49.5 gene of pseudorabies virus codes for an O-glycosylated structural protein of the viral envelope. J. Virol. 70:1237-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kyhse-Andersen, J. 1984. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J. Biochem. Biophys. Methods 10:203-209. [DOI] [PubMed] [Google Scholar]
- 14.Lake, C. M., and L. M. Hutt-Fletcher. 2000. Epstein-Barr virus that lacks glycoprotein gN is impaired in assembly and infection. J. Virol. 74:11162-11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lake, C. M., S. J. Molesworth, and L. M. Hutt-Fletcher. 1998. The Epstein-Barr virus (EBV) gN homolog BLRF1 encodes a 15-kilodalton glycoprotein that cannot be authentically processed unless it is coexpressed with the EBV gM homolog BBRF3. J. Virol. 72:5559-5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang, X., M. Tang, B. Manns, L. A. Babiuk, and T. J. Zamb. 1993. Identification and deletion mutagenesis of the bovine herpesvirus 1 dUTPase gene and a gene homologous to herpes simplex virus UL49.5. Virology 195:42-50. [DOI] [PubMed] [Google Scholar]
- 17.Mach, M., B. Kropff, P. Dal Monte, and W. Britt. 2000. Complex formation by human cytomegalovirus glycoproteins M (gpUL100) and N (gpUL73). J. Virol. 74:11881-11892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsumura, T., T. Kondo, S. Sugita, A. M. Damiani, D. J. O'Callaghan, and H. Imagawa. 1998. An equine herpesvirus type 1 recombinant with a deletion in the gE and gI genes is avirulent in young horses. Virology 242:68-79. [DOI] [PubMed] [Google Scholar]
- 19.Meindl, A., and N. Osterrieder. 1999. The equine herpesvirus 1 Us2 homolog encodes a nonessential membrane-associated virion component. J. Virol. 73:3430-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neubauer, A., B. Braun, C. Brandmüller, O.-R. Kaaden, and N. Osterrieder. 1997. Analysis of the contributions of the equine herpesvirus 1 glycoprotein gB homolog to virus entry and direct cell-to-cell spread. Virology 227:281-294. [DOI] [PubMed] [Google Scholar]
- 21.Osterrieder, N., O.-R. Kaaden, and A. Neubauer. 1999. Structure and function of equine herpesvirus glycoproteins—a review, p. 111-118. In Proceedings of the 8th International Conference on Equine Infectious Diseases. R&W Publications, Newmarket, United Kingdom.
- 22.Osterrieder, N., A. Neubauer, C. Brandmüller, B. Braun, O.-R. Kaaden, and J. D. Baines. 1996. The equine herpesvirus 1 glycoprotein gp21/22a, the herpes simplex virus type 1 gM homolog, is involved in virus penetration and cell-to-cell spread of virions. J. Virol. 70:4110-4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osterrieder, N., A. Neubauer, C. Brandmüller, O.-R. Kaaden, and D. J. O'Callaghan. 1998. The equine herpesvirus 1 IR6 protein that colocalizes with nuclear lamins is involved in nucleocapsid egress and migrates from cell to cell independently of virus infection. J. Virol. 72:9806-9817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osterrieder, N., A. Neubauer, B. Fakler, C. Brandmüller, C. Seyboldt, O.-R. Kaaden, and J. D. Baines. 1997. Synthesis and processing of the equine herpesvirus 1 glycoprotein M. Virology 232:230-239. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 26.Schumacher, D., B. K. Tischer, S. M. Reddy, and N. Osterrieder. 2001. Glycoproteins E and I of Marek's disease virus serotype 1 are essential for virus growth. J. Virol. 75:11307-11318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seyboldt, C., H. Granzow, and N. Osterrieder. 2000. Equine herpesvirus 1 (EHV-1) glycoprotein M: effect of deletions of transmembrane domains. Virology 278:477-489. [DOI] [PubMed] [Google Scholar]
- 28.Spear, P. G. 1993. Entry of alphaherpesviruses into cells. Semin. Virol. 4:167-180. [Google Scholar]
- 29.Telford, E. A., M. S. Watson, K. McBride, and A. J. Davison. 1992. The DNA sequence of equine herpesvirus-1. Virology 189:304-316. [DOI] [PubMed] [Google Scholar]
- 30.van Regenmortel, M. H. V., C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, and C. R. Pringle, and R. B. Wickner (ed.). 1999. Virus taxonomy. Seventh Report of the International Committee on Taxonomy of Viruses. Academic Press, New York, N.Y.
- 31.Wu, S. X., X. P. Zhu, and G. J. Letchworth. 1998. Bovine herpesvirus 1 glycoprotein M forms a disulfide-linked heterodimer with the U(L)49.5 protein. J. Virol. 72:3029-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]