Abstract
The Autographa californica nucleopolyhedrovirus (AcMNPV) lef-11 gene was previously identified by transient late expression assays as a gene important for viral late gene expression. The lef-11 gene was not previously identified as necessary for DNA replication in transient origin-dependent plasmid DNA replication assays. To examine the role of lef-11 in the context of the infection cycle, we generated a deletion of the lef-11 gene by recombination in an AcMNPV genome propagated as a BACmid in Escherichia coli. The resulting AcMNPV lef-11-null BACmid (vAclef11KO) was unable to propagate in cell culture, although a “repair” AcMNPV BACmid (vAclef11KO-REP), which was generated by transposition of the lef-11 gene into the polyhedrin locus of the vAclef11KO BACmid, was able to replicate in a manner similar to wild-type or control AcMNPV viruses. Thus, the lef-11 gene is essential for viral replication in Sf9 cells. The vAclef11KO BACmid was examined to determine if the defect in viral replication resulted from a defect in DNA replication or from a defect in late transcription. The lef-11-null BACmid and control BACmids were transfected into Sf9 cells, and viral DNA replication was monitored. The viral DNA genome of the lef-11-null BACmid (vAclef11KO) was not amplified, whereas replication and amplification of the genomes of the repair BACmid (vAclef11KO-REP), wild-type AcMNPV, and a nonpropagating gp64-null control BACmid (vAcGUSgp64KO) were readily detected. Northern blot analysis of transcripts from selected early, late, and very late genes showed that late and very late transcription was absent in cells transfected with the lef-11-null BACmid. Thus, in contrast to prior studies using transient replication and late expression assays, studies of a lef-11-null BACmid indicate that LEF-11 is required for viral DNA replication during the infection cycle.
Baculoviridae is a family of large double-stranded DNA viruses of invertebrates. Baculovirus genomes are circular and range in size from approximately 80 to 180 kbp. Members of the family infect a large number of insect species, with most hosts found in the insect order Lepidoptera (5). The Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) is the most intensively studied baculovirus. The AcMNPV genome is approximately 134 kbp in size and encodes 154 predicted genes (2). After entry into the host cell, AcMNPV nucleocapsids are transported to the nuclei of infected cells, where viral transcription and DNA replication occur. The infection cycle can be subdivided into three major phases of gene expression: early, late, and very late. Genes expressed in the early phase are transcribed by the host RNA polymerase II (10). Some of the early genes encode products necessary for DNA replication and late gene expression. The late phase of gene expression begins concomitant with or shortly after replication of the viral genome, and viral DNA replication appears to be a necessary prerequisite to late gene transcription. Inhibitors of viral DNA replication (such as aphidicolin) also block late gene expression in infected cells (19). Late genes are transcribed by a viral RNA polymerase (13, 14) that recognizes distinct late promoters. Most, if not all, late gene promoters contain the conserved sequence 5′-TAAG-3′ (33) at the late transcription start site. The conserved core TAAG sequence plus nonconserved flanking sequences of at least 8 to 12 nucleotides (nt) appear to comprise the late promoter sequences that are necessary for wild-type levels of late transcription (11, 24, 31). Except for the conserved core TAAG motif, little is known regarding the sequence specificity requirements for late promoter recognition and activation. The very late phase of gene expression follows late gene expression and is characterized by the hyperexpression of two genes, polyhedrin and p10. Very late promoters appear to be similar to late promoters in that they also include the conserved core TAAG motif and flanking sequences, but differ in that they also require an additional sequence called a “burst” sequence. The exceptionally high levels of transcription from the polyhedrin and p10 genes appear to be regulated or mediated by binding of viral protein VLF-1 to the “burst” DNA sequence, downstream of the transcription start site. The burst sequence is so called because it appears to regulate the burst of very late transcription (22, 26, 34, 36-38).
The transcription of late genes requires a number of early gene products. In previous studies using a transient late expression assay (29) to identify genes necessary for transient transcription from an AcMNPV late promoter, 19 late expression factor (lef) genes were identified (16, 20, 32). Because DNA replication is a necessary prerequisite for late gene transcription, a transient origin-dependent DNA replication assay was used to identify a subset of lef genes necessary for DNA replication, and these genes were proposed to constitute the subset of lef genes associated with viral DNA replication (15, 20). Transient assays for late gene expression and DNA replication have proved to be extremely important tools for the identification of genes associated with DNA replication and late gene expression. However, because viral proteins are expressed transiently from plasmid constructs in these assays, it is likely that the regulation of expression of each protein differs substantially from its expression in the context of a viral infection. Thus, it is possible that artifacts may arise from over- or underexpression of various LEF proteins. It is therefore critical to examine the effects of LEF proteins in the context of the AcMNPV infection cycle. Because many lef genes are likely essential for viral replication, only a few viruses have been generated with knockout or null mutations in lef genes (12). Recently, the p143 gene of AcMNPV was deleted and replaced with the p137 gene from the Trichoplusia ni granulovirus (TnGV), by recombination in Escherichia coli with an AcMNPV genome propagated as a bacterial artificial chromosome (BACmid) (4). That study demonstrated the utility and convenience of manipulating essential AcMNPV genes in an E. coli-based system.
The lef-11 gene is located immediately upstream of and overlapping the pp31 open reading frame (ORF). lef-11 is expressed as an early gene, and the LEF-11 protein is localized to the nuclei of infected cells (18). lef-11 was initially identified as a gene necessary for efficient transcription from a late promoter in a transient late expression assay (35). Several studies showed that omission of lef-11 in transient late expression assays resulted in only approximately 1 to 10% of the reporter expression that was observed when plasmids containing all 19 lef genes were present (20, 32, 35). In addition, lef-11 was not identified as necessary for transient origin-dependent plasmid DNA replication in two studies using that technique (15, 20).
We initially attempted to delete the lef-11 gene in the AcMNPV genome by recombination in an insect cell line that was stably transfected with the lef-11 gene, but repeated attempts were unsuccessful. Therefore, for the present study, we used a commercially available AcMNPV BACmid to delete the lef-11 gene by recombination in E. coli. After transfection into Sf9 cells, a lef-11-null AcMNPV BACmid (vAclef11KO) was unable to propagate in cell culture. However, a “repair” BACmid (vAclef11KO-REP) generated by transposition of the lef-11 gene into the polyhedrin locus of the vAclef11KO BACmid was able to replicate in a manner similar to wild-type and control AcMNPV. Thus, we found that the lef-11 gene was essential for viral replication in Sf9 cells. The lef-11-null BACmid was subsequently examined to determine if the defect in viral replication resulted from a defect in DNA replication or from a defect in transcription. In transfected Sf9 cells, the viral DNA genome of the lef-11-null BACmid (vAclef11KO) was not amplified, whereas the repair BACmid's genome was amplified in a manner similar to that of either the wild-type or control virus. Repair of the vAclef11KO BACmid confirmed that the defect in DNA replication in vAclef11KO was due to the loss of lef-11. Thus, the lef-11-null virus was deficient in viral DNA replication. Northern blot analysis of early, late, and very late genes showed that late transcription and very late transcription were absent in cells transfected with the lef-11-null BACmid. In contrast to prior studies suggesting a role for lef-11 in late transcriptional regulation, our data obtained with a lef-11-knockout virus indicate that lef-11 is necessary for viral DNA replication in Sf9 cells and that effects on late transcription may represent only secondary effects of the lef-11 knockout.
MATERIALS AND METHODS
Cells and virus.
Spodoptera frugiperda Sf9 cells and cells stably transfected with the lef-11 gene were maintained in TNMFH medium at 27°C as described earlier (23). Infection of cells with AcMNPV or other viruses was performed as described previously (28).
lef-11-knockout BACmid.
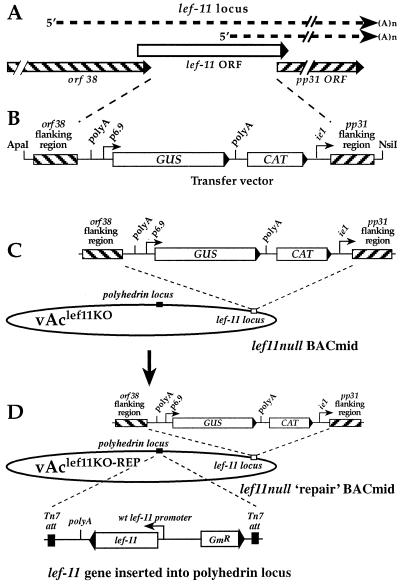
To delete the lef-11 gene in an AcMNPV BACmid, we first generated a transfer vector in which the lef-11 gene was replaced with (i) a chloramphenical acetyltransferase (CAT) cassette for antibiotic selection in E. coli, and (ii) a β-glucuronidase (GUS) reporter cassette. Because lef-11 overlaps both ORF38 (upstream) and pp31 (downstream) (18), removal of the lef-11 ORF also removes the pp31 promoter and sequences immediately downstream of ORF38. Therefore, we also inserted an early/late promoter (the AcMNPV ie1 promoter) to substitute for the pp31 promoter, and a poly(A) signal (derived from the OpMNPV GP64 gene) downstream of ORF38. The cassette designated “polyA-GUS-CAT-ie1 promoter” was cloned between sequences that flank the lef-11 ORF. First, plasmid p166-BRNX (17) was digested with EcoRI and ClaI, and the 205-bp fragment containing the poly(A) signal from the OpMNPV GP64 gene was purified and cloned into vector pGEM72(f+) to generate a recombinant plasmid named “pGEM72(f+)polyA.” Second, a 621-bp fragment containing the AcMNPV ie1 promoter (AcMNPV nt 126600 to 127197) was excised as a BamHI-BglII fragment from plasmid pie1-CAT (8) and ligated into pGEM72(f+)polyA, which also was digested with BamHI, to generate a recombinant plasmid named pGEM72(f+)polyAie1P. Third, a 1-kbp fragment from the pp31 gene region (nt 29052 to 30069) was PCR amplified from AcMNPV genomic DNA, digested with KpnI, and then ligated with pGEM72(f+)polyAie1P, which was digested with KpnI to generate a recombinant plasmid named pGEM72(f+)polyAie1Ppp31. The orientation was determined by SacI digestion. The primers used for amplification of the pp31 region were 5′ pp31 flank primer (5′-GGGGTACCGCCGATAAAGAAGGTGTGCCCG-3′) and 3′ pp31 flank primer (5′-GGGGTACCATGGTAAACGTGCCGGAGC-3′) (KpnI sites are underlined). Fourth, plasmid pAcGP64/GUS, which contains a p6.9 promoter-driven GUS reporter cassette in a BglII fragment (see pΔSmaΔ-GUS [27]), was digested with BglII, and the 2.6-kbp GUS cassette was ligated into plasmid pGEM72(f+)polyAie1Ppp31, which was digested with BamHI to generate a recombinant plasmid named “pGEM72(f+)polyAie1Ppp31GUS.” Fifth, a 1-kbp region of the orf38 ORF (nt 30364 to 31419) was PCR amplified from the AcMNPV genome, and XhoI sites were added to the ends of the PCR product. The PCR product was digested with XhoI and cloned into the XhoI site of plasmid pGEM72(f+)polyAie1Ppp31GUS to generate a recombinant plasmid named pGEM72(f+)polyAie1Ppp31GUSorf38. The orientation was confirmed by a HindIII digestion. The primers used for amplification of the orf38 region were 5′ orf38 flank primer (5′-TCAGTCCGCTCGAGTCACACCCGCCTAAGTGC-3′) and 3′ orf38 flank primer (5′-ATGTGCCCTCGAGATTGAAGTTCCGCTATACG-3′). (XhoI sites are indicated by underlined nucleotides.) Last, a 935-bp CAT gene cassette for antibiotic selection in E. coli cells was PCR amplified from plasmid pRE112 (9), and SmaI sites were added to the ends of the PCR product. The PCR product was digested with SmaI and ligated with SmaI-digested pGEM72(f+)polyAie1Ppp31GUSorf38, to generate a final plasmid named pGEM72(f+)polyAie1Ppp31GUSorf38CAT. The primers used for amplification of the CAT cassette were 5′ SmaI CAT (5′-GCCCGGGTAAATACCTGTGACGGAAGAT-3′) and 3′ SmaICAT (5′-GCCCGGGTATCACTTATTCAGGCGTAGC-3′). (SmaI sites are indicated by underlined nucleotides.) The orientation and structure of the insert in plasmid pGEM72(f+)polyAie1Ppp31GUSorf38CAT are shown in Fig. 1B.
FIG. 1.
Strategy for construction of a lef-11-null BACmid containing a deletion of the AcMNPV lef-11 gene and rescue by reinsertion of the wild-type (wt) lef-11 gene. (A) Relative locations and orientations of overlapping ORFs in the lef-11 locus of AcMNPV. The relative locations of lef-11 and pp31 transcripts are indicated by dashed lines. (B) Organization of the transfer vector DNA used to generate the lef-11 knockout BACmid by recombination in E. coli. A linear DNA fragment containing a poly(A) site, a p6.9 promoter-driven GUS gene, a chloramphenicol resistance gene cassette (CAT), and an ie1 promoter, are flanked by 1,034- and 1,026-bp regions from the orf38 and pp31 genes, as indicated. The linear DNA fragment was excised from plasmid pGEM72(f+)polyAie1Ppp31GUSorf38CAT, as described in Materials and Methods, and cotransfected with BACmid bMON14272 into E. coli strain BJ5183. (C) The organization of the lef-11-null BACmid (vAclef11KO) is shown. vAclef11KO was derived from BACmid bMON14272 and contains a chloramphenicol resistance gene cassette and a p6.9 promoter-driven GUS reporter gene in the lef-11 locus. The majority of the lef-11 ORF was removed. (D) Structure of the lef-11-null repair BACmid (vAclef11KO-REP). vAclef11KO-REP was derived from BACmid vAclef11KO by insertion of the wild-type lef-11 gene (under the control of the lef-11 promoter) into the polyhedrin locus by transposition with plasmid pFastBAClef11-REP.
To generate a recombinant AcMNPV BACmid containing a lef-11 knockout, we used a modification of a method described by Bideshi and Federici (4) for recombination in E. coli. The AcMNPV BACmid genome used in these studies was originally described as bMON14272 by Luckow and coworkers (21) and is commercially available (Invitrogen Life Technologies). Transfer vector pGEM72(f+)polyAie1Ppp31GUSorf38CAT was digested with ApaI-NsiI. The resulting purified linear 6.558-kbp fragment containing the polyA-GUS-CAT-ie1 promoter cassette plus flanking regions from the lef-11 locus was cotransformed with the bMON14272 BACmid DNA into E. coli strain BJ5183 (Stratagene, Inc.). After overnight incubation in SOC, cells were plated onto Luria-Bertani (LB) agar containing 50 μg of kanamycin and 30 μg of chloramphenicol per ml. Plates were incubated at 37°C for a minimum of 24 h. Colonies that were resistant to kanamycin and chloramphenicol were selected. The presence of the polyA-GUS-CAT-ie1 promoter cassette and the absence of the lef-11 ORF were confirmed by PCR analysis.
lef-11 repair BACmid.
To generate a lef-11 repair transfer vector, we modified plasmid pFastBac1 by removing the polyhedrin promoter and replacing it with a fragment containing the lef-11 promoter region and the lef-11 ORF. First, a 478-bp fragment containing the AcMNPV lef-11 promoter was PCR amplified then cloned into vector pCR-Blunt II-TOPO (Invitrogen) to generate plasmid pTOPO-lef11-p. The primers used to amplify the lef-11 promoter were 3′ lef11 promoter (5′-GTACGTACGCTGCAGGATTGTTTATGATAATCG-3′) and 5′ lef11 promoter (5′-GGGATCCTAATTAAAGTATTTGTTGAGCGGCACGATG-3′) (478 bp). The lef-11 promoter fragment was excised from plasmid pTOPO-lef11-p with EcoRI, the EcoRI ends were filled in with Klenow DNA polymerase, and then the fragment was digested with BamHI and cloned into SnaBI-BamHI-digested pFastBac1 plasmid, replacing the polyhedrin promoter. Next, the lef-11 ORF was PCR amplified from the AcMNPV genome. The primers used for amplification of the lef-11 ORF (5′ lef11 EcoRI, 5′-TTTGAATTCTTGCACACGGCCGCAGTCT-3′; 3′ lef11 BamHI, 5′-GCGGATCCAGGACTTTTTCTACGCCACT-3′) added BamHI and EcoRI restriction sites to the ends of the plasmid. The amplified fragment was digested with BamHI and EcoRI and was cloned into the BamHI-EcoRI sites of the pFastBac plasmid containing the lef11 promoter to generate the final recombinant plasmid containing lef-11 under the control of its own promoter, named pFastBAClef11-REP.
The lef-11 repair BACmid was generated by moving the lef-11 gene into the lef-11-null BACmid (described above) by transposition as described by Luckow and coworkers (21) (Fig. 1D). DH10B cells were transformed with helper plasmid pMON7124 (a plasmid containing Tn7 transposition functions). Competent DH10B cells containing the pMON7124 helper plasmid were cotransformed with both pFastBAClef11-REP DNA and vAclef11KO DNA. After a 6-h incubation at 37°C in SOC, transformed cells were plated onto media containing 50 μg of kanamycin, 30 μg of chloramphenicol, 7 μg of gentamicin, 10 μg of tetracycline, 100 μg of Bluo-gal, and 40 μg of isopropyl-β-d-thiogalactopyranoside (IPTG) per ml. Plates were incubated at 37°C for a minimum of 24 h. White colonies resistant to kanamycin, chloramphenicol, gentamicin, and tetracycline were selected, streaked onto fresh plates to verify the phenotype, and then confirmed by PCR.
Analysis of recombinant BACmids.
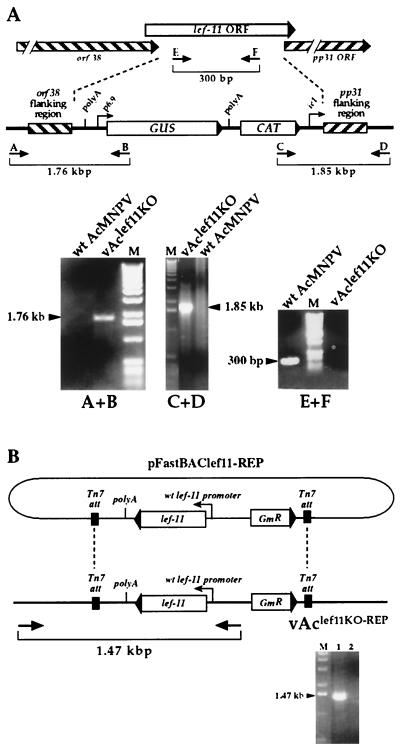
PCR analysis was used to confirm the lef-11-null BACmid. To confirm the insertion of the polyA-GUS-CAT-ie1 promoter cassette into the lef-11-null BACmid, two specific pairs of PCR primers (Fig. 2A) were used. For each primer pair, one primer corresponded to sequences within the inserted sequence (the GUS or CAT ORF), and the second primer was from the baculovirus genome, just outside of the flanking sequence included for recombination. The first set of primers consisted of GUS primer (5′-CGCGCTTTCCCACCAACGCTGATCAATTCC-3′) and lef11-3′ detect primer (5′-GTCAGGCGATTGTTAATCAAGCATGAAATGCTGC-3′). The second primer set consisted of 3036 primer (5′-CAAGGCGACAAGGTGCTGATGC-3′) and lef11- 5′detect primer (5′-AAACGGAACCCGCAGAGACGGTGGCCGATCTTAAGC-3′). To confirm the absence of lef-11 in the lef-11-null BACmid, two primers within the lef-11 ORF were used for PCR analysis: lef11 inside5′ (5′-CTTCTTGTAAACCTTTGAAACAACA-3′) and lef11inside3′ (5′-GACTGCTTGACTCGCAGCGAAATACAAG-3′). A similar strategy was used to confirm the reinsertion of the lef-11 gene into the polyhedrin locus of the lef-11-null BACmid. A primer pair consisting of one primer from the inserted sequence (between the two Tn7 att sites within pFastBAClef11-REP) and another primer from sequence within the BACmid (Fig. 2B). These primers consisted of M13 reverse (5′-CAGGAAACAGCTATGAC-3′) and 5′FastBac (5′-GGACTCTAGCTATAGTTCTAGTGG-3′).
FIG.2.
Confirmation of BACmid constructs vAclef11KO and vAclef11KO-REP by PCR analysis. (A) The strategy for PCR analysis of the lef-11 locus in BACmid vAclef11KO is indicated by the positions of primer pairs (arrows and brackets). The top diagram shows the structure of the wild-type (wt) lef-11 locus, and the lower diagram shows the structure of the BACmid vAclef11KO. To confirm the insertion of the polyA-GUS-CAT-ie1 cassette in BACmid vAclef11KO, primer pairs A+B and C+D were used to examine the recombination junctions by PCR analysis. For each primer pair, one primer corresponded to sequences within the inserted sequence (the GUS or CAT ORF), and the second primer was from the baculovirus genome, just outside the homologous flanking sequences used for recombination. Primer pair E+F was designed to amplify a fragment from within the lef-11 ORF and was used to confirm the absence of the lef-11 ORF in vAclef11KO. The sizes of expected PCR amplification products are shown below each primer pair on the diagram, and the panels below show the agarose gel electrophoresis results of each PCR, with the sizes of PCR products indicated beside an arrowhead. Primer pairs used for each PCR analysis are indicated below each panel and template DNAs are indicated above the panels. M, DNA size markers. (B) Analysis of the polyhedrin locus in BACmid vAclef11KO-REP. PCR analysis was used to confirm the insertion of a cassette containing the lef-11 ORF under the control of the wild-type lef-11 promoter from plasmid pFastBAClef11-REP, into BACmid vAclef11KO. The relative location of the primer pair used to confirm the insertion of the lef-11 gene is shown below the diagram of the resulting BACmid (vAclef11KO-REP). The panel below shows an ethidium bromide-stained agarose gel with the expected 1.47-kbp DNA product of PCR amplification from vAclef11KO-REP (lane 1). A similar PCR amplification from a negative control (wild-type AcMNPV) is also shown (lane 2).
Control BACmid DNAs.
For a control AcMNPV BACmid that contained a wild-type lef-11 locus, we used a BACmid containing a deletion of the gp64 gene at the gp64 locus and reinsertion of the gp64 gene and a p6.9 promoter-driven GUS reporter gene at the polyhedrin locus. This BACmid, named vAcgp64−/Acgp64+gus, will be referred to as vAc64−/+GUS for the purposes of the current study. A second control BACmid was used for DNA replication studies. In this case, the BACmid contained a wild-type lef-11 locus, but contained a deletion of the gp64 gene and a p6.9-GUS reporter inserted into the polyhedrin locus. This BACmid, named vAcgp64−/gus, will be referred to as vAcGUSgp64KO in the current study. The two control BACmids described above were kindly provided by Oliver Lung, and their construction and characterization will be described elsewhere.
Transfection of BACmid DNAs.
For transfections of insect cells, BACmid DNAs were prepared as follows. Each BACmid DNA was isolated and transformed into DH10B cells and then selected on kanamycin (in the absence of tetracycline), and colonies were screened for sensitivity to tetracycline and for the absence of the helper plasmid pMON7124 by analysis of DNAs on agarose gels. Each resulting E. coli strain carrying the helper-free BACmid was used to prepare BACmid DNA for transfections. DNA was prepared from 0.5- to 3-liter cultures and purified by CsCl gradient purification. Transfections of BACmid DNAs into insect cells were performed with cationic liposomes prepared as described previously (7). BACmid DNA (5 μg) was mixed with cationic liposomes (20 μl) and added to 2 × 106 insect cells. After a 5-h incubation period, the medium was removed and replaced with TNMFH containing 10% fetal bovine serum (FBS). Transfection efficiency was monitored by parallel transfections of cells with a BACmid containing a knockout of the gp64 gene (vAcGUSgp64KO) followed by staining for GUS activity.
Detection of GUS expression.
For analysis of viral infectivity and late gene expression, Sf9 or Sf9-derived cells were transfected with BACmids (5 μg of BACmid DNA per 2 × 106 cells, in six-well plates) and then stained for GUS activity. At 4 days posttransfection, cells were washed with 2 ml of phosphate-buffered saline (PBS) and then fixed in 1 ml of PBS containing 2% formaldehyde and 0.05% gluteraldehyde for 5 min at room temperature. The fixed cells were washed twice with 2 ml of PBS, and then cells were stained with an X-Gluc solution (PBS containing 1 mg of X-glucuronide per ml, 5 mM potassium ferricyanide, and 2 mM MgCl2) for approximately 1 to 2 h. The cells were rinsed with 2 ml of PBS and observed on an inverted microscope. The percentage of GUS-positive (blue) cells was determined.
Virus growth curves.
Sf9 cells (3 × 105) were transfected with 2 μg of DNA from each BACmid or virus (AcMNPV, vAclef11KO, vAclef11KO-REP, and vAc64−/+GUS). After a 5-h transfection period, cells were washed three times with TNMFH and then incubated for various periods of time before supernatants were collected. Data from each time point represent accumulated infectivity from transfection through the indicated time. The titers of all supernatants were determined by 50% tissue culture infective dose (TCID50) on Sf9 cells (28).
Dot blot analysis of DNA replication.
To quantify viral DNA replication in cells transfected with various BACmids, Sf9 cells were transfected as described above, and viral DNAs were detected and quantified at various times by Southern dot blot hybridization assays (12, 30). Sf9 cells (2 × 106 cells per well in six-well plates) were transfected with 5 μg of BACmid DNA or viral DNA (AcMNPV, vAclef11KO, vAclef11KO-REP, vAcGUSgp64KO or vAc64−/+GUS). At 0, 6, 12, 24, 48, 72, and 96 h posttransfection, cells were removed and pelleted by centrifugation at 10,000 × g for 10 min. Note that vAclef11KO-transfected cells were collected through 10 days posttransfection and then processed as described above. The cell pellet was washed twice with PBS, and 1 × 104 cells were resuspended in 500 μl of a 0.4 M NaOH-10 mM EDTA solution, incubated at 100°C for 10 min, and then blotted onto Magnacharge nylon transfer membrane (MSI Micron SEPERATION, Inc.) with a dot blot apparatus (Bio-Dot SF; Bio-Rad, Inc.). Samples were hybridized with 32P-labeled AcMNPV DNA, which was labeled to 4 × 108 cpm/μg by random priming (DECAprimeII random priming DNA labeling kit; Ambion, Inc.). The blot was visualized, and the bound probe was quantified with a PhosphorImager (Molecular Dynamics, Inc.).
RNA isolation and Northern blot analysis.
For RNA isolation from BACmid-transfected cells, Sf9 cells were plated in six-well plates (2 × 106 cells/well), and each well of cells was transfected with 5 μg of DNA from BACmids vAclef11KO, vAclef11KO-REP, and vAc64−/+GUS. Total RNAs from transfected cells were isolated at 12, 18, 48, 72, and 96 h postinfection with Trizol reagent (Gibco BRL, Inc.). Cells from each well were lysed in 1 ml of Trizol reagent at room temperature for 5 min, an additional 0.2 ml of chloroform was added, and lysates were vigorously agitated. Samples were incubated at room temperature for 2 min and then centrifuged at 12,000 × g for 15 min at 4°C. RNA was precipitated from the aqueous phase with 2 volumes of ethanol. RNA pellets were washed with 1 ml of 70% ethanol, dried, and then resuspended in 50 μl of water.
To examine the effect of the lef-11 knockout on gene transcription, several gene-specific probes were used to examine steady-state transcript levels from early, late, and very late genes. The ie1 gene was used to examine early transcription. Genes p6.9, p24, and gp16 were chosen to examine late transcription, and p10 was selected to examine very late transcription. For Northern blot analyses, 5 μg of total RNA isolated from BACmid-transfected cells at each selected time point was electrophoresed on a formaldehyde-1.2% agarose gel essentially as described previously (6). RNA was blotted onto Magnacharge nylon transfer membrane. To generate labeled single-stranded cRNA riboprobes for hybridization, fragments from the genes p6.9 (190 bp), p10 (303 bp), and ie1 (1.35 kb) and a single 1.5-kb DNA fragment that hybridizes to RNAs from the five genes (25) in the p24-to-alk-exo region (p24 capsid, gp16, pp34-pep, 132-orf, and alk-exo) were PCR amplified. In each primer pair, the downstream PCR primer included a terminal T7 RNA polymerase promoter sequence that was later used to generate a negative-sense cRNA riboprobe. The following PCR primers were used to generate the different probes: 5′IE1 (5′-CCAACCATCGGCAACTGGAACTAAACGGAAGC-3′), 3′IE1 (5′-CTAATACGACTCACTATAGGGCCGCAAACGTTATAGCG-3′), 5′VP39 (5′-CAATATGGCGCTAGTGCCCGTGGGTATGGC-3′), 3′VP39 (5′-CTAATACGACTCACTATAGGGTCCTCCACCTGCTTCGCCTGC-3′), 5′p74 (5′-GTCCAACACGACGCCGTTCATGTACATGCAG-3′), 3′p74 (5′-CTAATACGACTCACTATAGGGCTCCATGCGAGTGTATAGCGAGC-3′), 5′p6.9 (5′-CATGGTTTATCGTCGCCGTCGCCGTTCTTC-3′), 3′p6.9 (5′-CTAATACGACTCACTATAGGGTTAATAGTAGCGTGTTCTG-3′), 5′p10 (5′-TCAAAGCCTAACGTTTTGACGCAAATTTTAGAC-3′), 3′p10 (5′-CTAATACGACTCACTATAGGGTTACTTGGAACTGCGTTTAC-3′), 5′orf133 (5′-GACGTATCCCATGGCCTATTTTGTCAATACCG-3′), and 3′orf133 (5′-CTAATACGACTCACTATAGGGCGTTTAAATGATCGTGTTTGG-3′).
The underlined sequences represent the optimal T7 promoter sequence (3). The PCR products were used to generate a labeled cRNA probe by in vitro transcription with T7 RNA polymerase (MAXIscript; Ambion, Inc.) by using [α-32P]UTP (approximately 3,000 Ci/mmol; NEN Life Science Products, Inc.) according to the supplied protocols. The labeled riboprobes were purified on G-50 spin columns (Princeton Separations, Inc.) and used for Northern blot hybridization. For Northern blots, membranes were prehybridized, hybridized, and processed as described previously (18).
Stable Sf9 cell line expressing LEF-11.
To generate a cell line expressing the LEF-11 protein, the AcMNPV lef-11 gene was first amplified from the AcMNPV genome by using the following PCR primers: 5′lef11EcoRI (5′-TTTGAATTCTTGCACACGGCCGCAGTCT-3′) and 3′lef11BamHI (5′-GCGGATCCAGGACTTTTTCTACGCCACT-3′). The PCR product, which contained EcoRI and BamHI sites engineered onto the ends (underlined above), was digested with EcoRI and BamHI and ligated with insect cell expression plasmid vector p166-BRNX (17), which was also digested with EcoRI and BamHI. The resulting recombinant LEF-11 expression plasmid was designated p166-lef11. By a modification of the method described previously (17), 2 × 106 Sf9 cells were cotransfected with 5 μg of p166-lef11, 2 μg of p166-EGFP (8), and 1 μg of pIE1-Neo (23). After 48 h posttransfection, cells were selected by culture in medium containing 1 mg of G418 per ml for 3 to 4 weeks, until no control Sf9 cells survived under the same selection conditions. Cells were then propagated as an uncloned cell line.
GUS activity assay.
To examine activity from the p6.9-GUS reporter gene in recombinant viruses, GUS activity was determined with a GUS detection kit according to the manufacturer's instructions (Sigma, Inc.). Cell monolayers (3 × 105 cells) in each well of a 24-well plate were infected or transfected with AcMNPV, vAclef11KO, vAclef11KO-REP, and vAc64−/+GUS. Sf9 cells or LEF-11-expressing cells that were transfected or infected with BACmids were collected at various time points postinfection or posttransfection. Cell pellets were lysed in 200 μl of 1× extraction buffer (50 mM sodium phosphate, 10 mM EDTA, 10 mM β-mercaptoethanol [pH 7.0]). For each reaction, 10 μl of (4-methylumbelliferyl β-d-glucuronide (4-MUG) substrate was mixed with 5 μl of 1× extraction buffer and preincubated for 1 to 2 min at 37°C and then added to 5 μl of cell extract and incubated at 37°C for 1 h. After incubation, 10 μl of stop solution (1 M sodium carbonate) was added. 4-Methylumbelliferone (4-MU) was used to generate a standard curve and to confirm that the GUS assay result from each sample was within the linear range of the assay.
RESULTS
The lef-11 gene was originally identified as a gene essential for transient late gene expression in a plasmid-based transient expression assay. The lef-11 ORF overlaps the upstream orf38 and the downstream pp31 ORFs (Fig. 1A). Previous transcriptional mapping of lef-11 and pp31 showed that lef-11 transcripts initiate within the upstream orf38 reading frame and that pp31 transcripts initiate within the lef-11 reading frame (18). To examine the role of lef-11 in the context of the AcMNPV genome and in a viral infection, we generated a modified AcMNPV genome containing a lef-11 knockout. In our initial attempts to generate a lef-11 knockout, we constructed a transfer vector containing sequences from the lef-11 region, but with the majority of the lef-11 ORF removed and replaced by a p6.9-GUS reporter cassette. Because the lef-11 ORF is small and removal of the lef-11 ORF also removed the pp31 promoter, an AcMNPV ie1 promoter was inserted immediately upstream of the pp31 ORF so as to reconstitute transcription of the pp31 gene. A poly(A) signal was also inserted downstream of orf38 to terminate transcription. This transfer vector construct was used in attempts to generate a lef-11-null virus by recombination with wild-type AcMNPV in a stably transfected cell line expressing LEF-11 (described below). Although recombinant viruses containing the expected modification at the lef-11 locus were generated, all constructs also rapidly reacquired the lef-11 gene by homologous or nonhomologous recombination during viral growth in the cell line, and we were unable to isolate and clone a lef-11-null virus in this manner. We therefore used an AcMNPV genome propagated as a bacterial artificial chromosome (BACmid) to generate a lef-11-null BACmid.
Generation of lef-11-null and repair AcMNPV BACmids.
To generate a lef-11-null AcMNPV BACmid by recombination in E. coli, we used a modification of a method described by Bideshi and Federici (4). For this procedure, the above transfer vector plasmid was modified by the addition of a chloramphenicol resistance gene (cat) cassette for selection of recombinants in E. coli (Fig. 1B). A linear DNA fragment containing the polyA-p6.9GUS-CAT-ie1-promoter insertion shown in Fig. 1B was cotransformed with the commercially available AcMNPV BACmid genome (bMON14272) into E. coli BJ5183 cells. BACmids resulting from recombination and deletion of the lef-11 ORF were selected by growth on medium containing kanamycin and chloramphenicol. A single cloned BACmid containing the lef-11 deletion and insertion of the polyA-p6.9GUS-CAT-ie1-promoter cassette at the lef-11 locus was selected and named vAclef11KO. Insertion of the polyA-p6.9GUS-CAT-ie1-promoter cassette (Fig. 1B) and knockout of lef11 were confirmed by PCR analysis (Fig. 2). Primer pairs flanking the predicted recombination sites resulted in PCR amplification of predicted fragments of 1.76 and 1.85 kbp (Fig. 2A, A+B and C+D, respectively). In addition, a PCR amplification product of 300 bp from a region from within the lef11 ORF was negative when BACmid vAclef11KO DNA was used as template, whereas wild-type AcMNPV DNA yielded the expected 300-bp fragment (Fig. 2A, E+F). Thus, PCR analysis confirmed the absence of the lef-11 ORF and the insertion of the polyA-p6.9GUS-CAT-ie1-promoter cassette.
For rescue or confirmation of the phenotype resulting from the lef-11 knockout, a repair BACmid was constructed from vAclef11KO by reinserting the lef-11 gene under the control of its own promoter into the polyhedrin locus of vAclef11KO. The lef-11 gene was inserted into the polyhedrin locus by transposition, using the BAC-to-BAC system (Fig. 1D and 2B). This was accomplished by first removing the polyhedrin promoter from plasmid pFastbac-1 (Life Technologies) and then inserting the lef-11 ORF under the control of its own promoter. The resulting plasmid construct (pFastBAClef11-REP) was used for Tn7-based transposition as described previously (21). Insertion of the lef-11 cassette into the polyhedrin locus of vAclef11KO was confirmed by PCR analysis with a primer pair that flanked a transposition site and amplified the inserted lef-11 gene (Fig. 2B). The repaired vAclef11KO BACmid was named vAclef11KO-REP.
Analysis of viral replication in Sf9 cells.
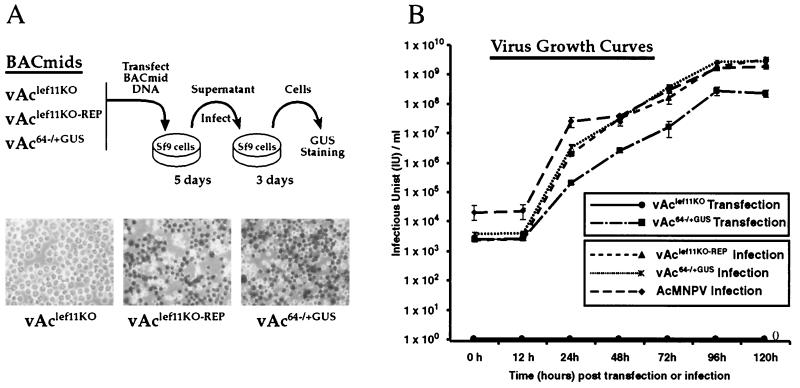
To determine whether the lef-11 gene was necessary or important for viral replication, Sf9 cells were transfected with either the vAclef11KO BACmid, the vAclef11KO-REP BACmid, or a control BACmid, vAc64−/+GUS, and examined for viral propagation. The vAc64−/+GUS BACmid contains a wild-type lef-11 locus and a GUS reporter under the control of a p6.9 promoter (vAc64−/+GUS was provided by O. Lung). In initial experiments, the BACmid constructs described above were examined for viral propagation in cell culture by passage through Sf9 cells, followed by staining for GUS activity (Fig. 3A). At 5 days posttransfection, supernatants were removed and added to freshly plated Sf9 cells and then incubated for 3 days to detect infection by virus generated from cells transfected with these BACmids. At each step, cells were examined for visible signs of cytopathic effects (CPE) and stained with X-Gluc to detect expression of GUS from the late gene reporter construct (p6.9-GUS). No GUS staining or CPE was observed from Sf9 cells transfected with the vAclef11KO BACmid or from cells incubated in the supernatant from that transfection (Fig. 3A, vAclef11KO). In contrast, Sf9 cells infected with supernatants from cells transfected with vAclef11KO-REP or the vAc64−/+GUS control BACmid stained positive for GUS activity, and infected cells showed typical CPE (not shown). Thus, initial viral passaging experiments suggested that deletion of the lef-11 gene resulted in a viral genome unable to propagate infection in Sf9 cells.
FIG. 3.
Analysis of viral replication by a lef-11-null BACmid. (A) A transfection-infection assay was used to examine lef-11-null BACmids for viral replication in Sf9 cells. BACmid DNAs from the indicated constructs were used to transfect Sf9 cells, and cells were incubated for 5 days. Supernatants from transfected cells were transferred to a second group of Sf9 cells, which were subsequently incubated for 3 days and then stained for GUS expression from the p6.9 late promoter-GUS reporter. The results of GUS staining are shown in the three lower panels. (B) Virus growth curves were determined from either transfected cells or infected cells. For transfections, Sf9 cells were transfected in triplicate with either vAclef11KO or vAc64−/+GUS, and then supernatants were removed at the indicated times posttransfection, and the TCID50 in Sf9 cells was determined. For infections, infectious budded virions were prepared from wild-type AcMNPV or BACmids vAclef11KO-REP or vAc64−/+GUS. Infections were performed at an MOI of 5 in triplicate, and supernatants were collected and assayed for production of infectious virus by TCID50.
To refine our initial analysis and to better characterize the rescue of the lef-11-knockout virus, a viral growth curve experiment was performed (Fig. 3B). Two separate growth curve experiments are shown in Fig. 3B. In one experiment, cells were transfected with BACmids, and in the second experiment, cells were infected with virions. In the first experiment, Sf9 cells were transfected with DNA from either the lef-11-knockout BACmid (vAclef11KO) or the control BACmid (vAc64−/+GUS). No infectivity was detected from cells transfected with the lef-11-knockout BACmid at any time examined through 120 h postinfection. In contrast, a steady increase in virus production was detected from cells transfected with the control BACmid (vAc64−/+GUS). In the second experiment, cells were infected at a multiplicity of infection (MOI) of 5 with virions prepared from either the repair BACmid (vAclef11KO-REP) or the vAc64−/+GUS control BACmid or wild-type AcMNPV. We found that virus production from the lef-11 repair virus was similar to that from the control virus (vAc64−/+GUS) and from wild-type AcMNPV (Fig. 3B). Thus, the defect in viral replication in the lef-11-null BACmid (vAclef11KO) was rescued by reinsertion of the lef-11 gene into the polyhedrin locus of the repair BACmid (vAclef11KO-REP). In addition, replication of the repair BACmid (vAclef11KO-REP) was similar to that of a control BACmid or wild-type AcMNPV. These data therefore indicate that the AcMNPV lef-11 gene is required for virus replication.
We noted that the kinetics of the infection cycle for the control BACmid (vAc64−/+GUS) was somewhat delayed in the first experiment (transfected cells) when compared with the same BACmid used in the second experiment (infected cells). This difference likely results from the fact that the first experiment does not represent a one-step growth curve, since transfections rarely resulted in initial infection efficiencies of >10 to 40%, whereas cells were synchronously infected in the second experiment.
Complementation by Sf9lef-11 cells.
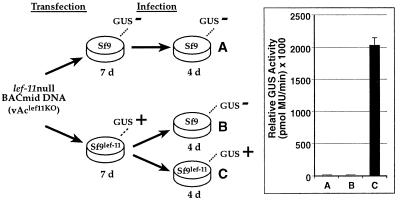
To confirm that lef-11 was necessary for viral replication in Sf9 cells and to further ensure that this initial phenotype resulted from deletion of lef-11 and not from a second site mutation or from effects of a cis-acting element, we also performed a complementation experiment with a cell line, Sf9lef-11, that was stably transfected with the lef-11 gene expressed under the control of an early baculovirus promoter. In this experiment, Sf9 or Sf9lef-11 cells were transfected with the lef-11-null BACmid (Fig. 4). The lef-11-null BACmid (vAclef11KO) was transfected into Sf9 cells, and after 7 days, the supernatant was transferred to a second group of Sf9 cells and incubated for 4 days. No GUS activity was detected from the transfected Sf9 cells or from the second group of Sf9 cells receiving the supernatant from those transfected cells. In parallel, Sf9lef-11 cells were also transfected with the lef-11-null BACmid (vAclef11KO). After 7 days, high levels of GUS activity were detected (not shown). To examine whether the observed GUS activity resulted from complementation by the LEF-11 protein expressed from the cell line, we transferred the supernatant from the transfected Sf9lef11cells to either Sf9 cells or to Sf9lef-11 cells. After 4 days, no GUS activity was detected from the Sf9 cells, but relatively high levels of GUS activity were detected from the infected LEF-11-expressing cells (Sf9lef-11) (Fig. 4). These results show that the lef-11-null BACmid (vAclef11KO) can be rescued by LEF-11-expressing cells (Sf9lef-11) and suggest that the observed complementation is due to LEF-11 protein expressed from the cell line and not from acquisition of the lef-11 gene from the cell line.
FIG. 4.
Complementation of a lef-11-null BACmid by a stable cell line expressing LEF-11. The diagram shows the strategy used to examine a LEF-11-expressing cell line (Sf9lef11) for complementation of the lef-11-null BACmid (vAclef11KO). GUS activity results are indicated above the wells (GUS+ or GUS−). Quantitative measurements from triplicate transfection-infections are shown for the infected wells (A, B, and C) in the graph on the right.
Analysis of DNA replication.
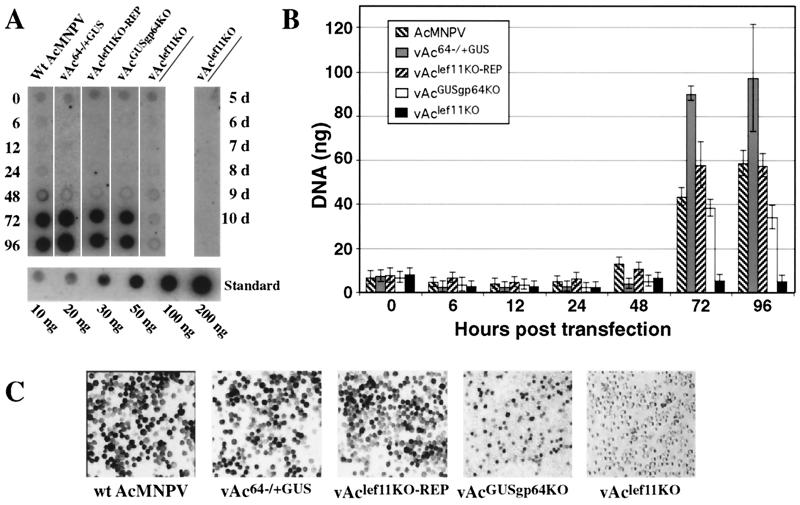
To determine if AcMNPV DNA replication was affected by the lef-11 knockout, we examined DNA replication in cells transfected with the lef-11-null BACmid (vAclef11KO) (Fig. 5A). As controls, cells were also transfected with either wild-type AcMNPV DNA, a control BACmid (vAc64−/+GUS), or the repair BACmid (vAclef11KO-REP). We also used an additional control BACmid (vAcGUSgp64KO) that contains a deletion of the major envelope protein gene, gp64. Deletion of gp64 results in a virus unable to propagate infection from cell to cell (23). This control ensured that we could detect DNA replication from the lower percentage of cells that were initially transfected. The vAcGUSgp64KO BACmid DNA can enter cells by transfection and is DNA replication competent, but it cannot exit from the transfected cells and cannot propagate in culture. After transfection with the BACmid DNAs described above, cells were incubated for the indicated times, and DNA was isolated and used for Southern dot blot analysis. Replication of viral DNA was detected with a labeled wild-type AcMNPV genomic DNA hybridization probe (Fig. 5A), and DNAs were quantified by PhosphorImager analysis (Fig. 5B). At 0 h posttransfection, similar DNA signals were detected from cells transfected with each BACmid or wild-type AcMNPV DNA. The signal from the 0-h time point represents the level of detectable DNA transfected into Sf9 cells. At 72 and 96 h posttransfection, DNA replication was detectable as a strong signal in cells transfected with wild-type AcMNPV DNA and control BACmid DNAs. In contrast, DNA replication was not observed from cells transfected with the lef-11-null BACmid (vAclef11KO), even when these cells were examined as late as 10 days posttransfection (Fig. 5A). DNA replication was readily detected from cells transfected with the BACmid containing a gp64 deletion (vAcGUSgp64KO), although the intensity of the DNA signal was around two- to fourfold lower than that from other positive control BACmids at late times (Fig. 5B, 96 h). This was expected, because the virus containing a gp64 deletion should not spread from cell to cell. Thus, the number of cells in which AcMNPV replication occurs is limited to those cells initially transfected with the gp64-null BACmid. Figure 5B shows a quantitative analysis of the same DNA replication assay represented in Fig. 5A, but incorporating the results of replicate transfections and quantitative DNA analyses. Figure 5C shows GUS staining of transfected Sf9 cells at 96 h posttransfection. GUS staining was detected in all transfected cells, except those transfected with the lef-11-null BACmid (vAclef11KO). In the case of the gp64-null BACmid (vAcGUSgp64KO), the percentage of GUS-positive cells is lower than that from other positive control BACmids, as expected. These results show no detectable DNA replication in cells transfected with the lef-11-null BACmid and therefore indicate that lef-11 is necessary for viral DNA replication.
FIG. 5.
Analysis of AcMNPV DNA replication in Sf9 cells transfected with lef-11-null and control BACmids. (A) Sf9 cells were transfected with either wild-type (Wt) AcMNPV, control BACmid (vAc64−/+GUS), lef-11 repair BACmid (vAclef11KO-REP), gp64-null control BACmid (vAcGUSgp64KO), or lef-11-null BACmid (vAclef11KO) DNA (lanes 1 to 5, respectively), and total cellular DNAs were isolated at various times (0 to 96 h) posttransfection. Viral DNA replication was detected by Southern dot blot hybridization with total AcMNPV DNA as a 32P-labeled hybridization probe. Cells transfected with the lef-11-null BACmid (vAclef11KO) were also examined after an extended period (right lane, 5 to 10 days). A standard curve of AcMNPV DNA is shown below (10 to 200 ng of DNA). (B) Quantitative analysis of BACmid DNA replication by Southern dot blot analysis. Three replicates of each virus and time point were examined as shown in panel A, and DNA was measured by PhosphorImager analysis. Bars represent the average of three dot blot samples, and error bars represent standard deviation. (C) Sf9 cells transfected with each of the indicated DNAs (wild-type AcMNPV, vAc64−/+GUS, vAclef11KO-REP, vAcGUSgp64KO, or vAclef11KO) were stained for GUS activity at 4 days posttransfection.
Analysis of early, late, and very late transcription.
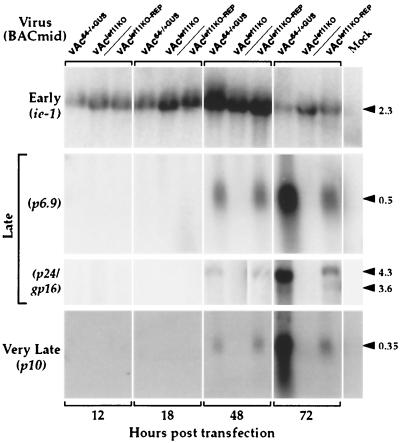
To determine the effects of the lef-11 knockout on gene expression in the context of an AcMNPV infection, the lef-11-null BACmid was used to transfect Sf9 cells, and transcription of several indicator genes was monitored (Fig. 6). Early, late, and very late transcription in cells transfected with the lef-11-null BACmid (vAclef11KO) were compared with that from similar transfections with a control BACmid (vAc64−/+GUS) and the repair BACmid (vAclef11KO-REP). For these analyses, ie1 was used to monitor effects on early transcription; p6.9, p24, and gp16 were used to monitor effects on late transcription; and p10 was selected to monitor effects on very late transcription. Sf9 cells were transfected with either the lef-11-null BACmid or control or repair BACmids, and then total RNAs were isolated at various times posttransfection and used for Northern blot analysis with the indicated early, late, or very late gene-specific probes. Transcription of the ie1 gene appeared similar in cells transfected with the lef-11-null BACmid (vAclef11KO) or with the repair or control BACmids (vAclef11KO-REP or vAc64−/+GUS) (Fig. 6, early). Thus, no effect on early (ie1) gene transcription was detected in the lef-11-null BACmid. In contrast, late transcription and very late transcription were both completely absent from cells transfected with the lef-11-null BACmid. Late transcription and very late transcription were present in the lef-11 repair BACmid (vAclef11KO-REP), in which lef-11 was expressed under its own promoter. The complete absence of late and very late transcription is consistent with prior studies, suggesting that late gene transcription is dependent on prior viral DNA replication. Interestingly, at 48 h posttransfection, late and very late transcription from the repair BACmid appeared similar to that from the control BACmid construct. However, at 72 h posttransfection, the levels of late and very late transcripts from the repair BACmid had increased, but were lower than that from the control vAc64−/+GUS BACmid. The reason for this difference is unclear, but might be related to possible subtle differences in the levels of LEF-11 or PP31 in the repair BACmid construct. Because late and very late transcription are completely absent in cells transfected with the lef-11-null BACmid, but were rescued in the repair BACmid, these data are consistent with our earlier observations indicating that LEF-11 is necessary for viral DNA replication.
FIG. 6.
Northern blot analysis of early, late, and very late transcripts from cells transfected with lef-11-null or control BACmids. Sf9 cells were transfected with either the lef-11-null BACmid (vAclef11KO), a control BACmid (vAc64−/+GUS), or the lef-11 repair BACmid (vAclef11KO-REP). At various times (12, 18, 48, or 72 h) posttransfection, total RNAs were isolated and used for Northern blot analysis with early (ie1), late (p6.9, p24, and gp16), or very late (p10) gene-specific probes. BACmids used for transfections are indicated at the top of the lanes, and gene-specific probes are indicated on the left. The sizes of expected RNAs from each gene-specific probe are indicated in kilobases on the right.
DISCUSSION
Previous studies with transient origin-dependent DNA replication assays showed that AcMNPV lef-1, lef-2, lef-3, p143, p35, and ie1 were necessary for origin-dependent transient DNA replication, and that replication in this assay was stimulated by ie2, lef-7, dnapol, and pe-38 (1, 15, 19, 20, 35). Thus, approximately 10 early gene products were identified as potential replication proteins in transient origin-dependent DNA replication assays. Interestingly, lef-11 was not identified as a DNA replication-associated gene by these transient assay methods. By using traditional methods to generate recombinant BmNPV viruses in insect cells, knockout viruses were generated by interrupting the reading frames of the ie2, lef-7, and p35 genes, demonstrating that these genes were not essential for BmNPV replication in cell culture (12). DNA replication was reduced in each knockout virus, confirming the stimulatory role for each of these genes in viral DNA replication. However, by those methods, those authors were unable to generate viruses containing knockouts in ie1, lef-1, lef-2, lef-3, p143, dnapol, lef-4, lef-8, lef-9, p47, lef-5, lef-6, lef-10, or lef-11 genes.
In the current study, we used a genetic analysis to examine the role of the AcMNPV lef-11 gene in the context of an AcMNPV infection in Sf9 cells. To accomplish this, we used an AcMNPV genome propagated in E. coli to generate a knockout mutation in the lef-11 gene. We used two separate strategies to rescue the lef-11-null phenotype generated by this knockout. First, we inserted a copy of the wild-type lef-11 gene into the polyhedrin locus. Next, we used a stably transfected cell line expressing LEF-11, to complement the defect in the lef-11-null BACmid. We found that the lef-11-null BACmid was unable to propagate after transfection into Sf9 cells, but this defect in replication could be rescued by either the reinsertion of the lef-11 gene into the polyhedrin locus of the same BACmid or by transfection of the lef-11-null BACmid into cells expressing LEF-11. Therefore, our studies indicated that the lef-11 gene is essential for AcMNPV replication in Sf9 cells. We next examined the specific cause of this defect more closely. In studies of DNA replication in cells transfected with lef-11-null or control BACmids, we found that the lef-11-null BACmid was defective in DNA replication, and, as expected, late gene transcription was also absent. Therefore, the lef-11 gene appears to be necessary for DNA replication of the AcMNPV genome in Sf9 cells. Because some lef genes appear to have species-specific roles, it will be of interest to determine whether LEF-11 is required for viral DNA replication in other cell lines or animals that are permissive for AcMNPV replication.
One concern with strategies such as those employed here is the possibility that cis-acting regulatory sequences may be disrupted as the knockout is generated, and that such disruption could have effects on the regulation of adjacent genes or even global effects. To avoid these concerns in the current studies, we used two separate strategies to rescue the defect resulting from the lef-11 knockout. First, we generated a repair virus by reinserting the lef-11 gene at a different locus (the polyhedrin locus). Thus, the modified lef-11 locus is left unchanged, and any effects on adjacent genes or global effects caused by changes at the lef-11 locus should be identical in both the knockout and repair viruses. Second, we rescued the defect by using an Sf9 cell line stably transfected with the lef-11 gene. Since the lef-11 gene sequences are not reinserted into the viral genome in this case, this eliminates the likelihood of cis-acting effects from sequences within the lef-11 ORF.
In this study, we developed and used a new and powerful method for identification and analysis of the roles of essential baculovirus regulatory genes. The methods employed were based on a combination of methods previously described by Bideshi and Federici (4) and Luckow et al. (21). The use of infectious BACmid clones of baculoviruses permits the rapid mutagenesis, modification, and study of many genes and proteins that were previously intractable. In this study of lef-11, we found that, in contrast to prior studies based on transient assays, lef-11 was necessary for viral DNA replication. The development of a lef-11-knockout BACmid system will permit the further refinement and study of lef-11 functions and will permit us to test hypotheses on the roles and functions of various protein domains, in the context of the viral genome and in infected cells.
Acknowledgments
We thank Oliver Lung for providing BACmids vAc64−/+GUS and vAcGUSgp64KO and for thoughtful comments and advice during the course of these studies. We also thank Brian Federici and Dennis Bideshi for providing E. coli strain BJ5183 and Gretchen Hoffmann for expert technical assistance.
This work was supported by USDA grant 99-35302-7952 and NIH grant AI33657.
REFERENCES
- 1.Ahrens, C. H., D. J. Leisy, and G. F. Rohrmann. 1996. Baculovirus DNA replication, p. 855-872. In M. L. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Ayres, M. D., S. C. Howard, J. Kuzio, M. Lopez-Ferber, and R. D. Possee. 1994. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202:586-605. [DOI] [PubMed] [Google Scholar]
- 3.Baklanov, M. M., L. N. Golikova, and E. G. Malygin. 1996. Effect on DNA transcription of nucleotide sequences upstream to T7 promoter. Nucleic Acids Res. 24:3659-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bideshi, D. K., and B. A. Federici. 2000. The Trichoplusia ni granulovirus helicase is unable to support replication of Autographa californica multicapsid nucleopolyhedrovirus in cells and larvae of T. ni. J. Gen. Virol. 81:1593-1599. [DOI] [PubMed] [Google Scholar]
- 5.Blissard, G., B. Black, N. Crook, B. A. Keddie, R. Possee, G. Rohrmann, D. Theilmann, and L. E. Volkman. 2000. Baculoviridae: taxonomic structure and properties of the family, p. 195-202. In M.H.V. van Regenmortel et al. (ed.), Virus taxonomy. The classification and nomenclature of viruses. The seventh report of the International Committee for the Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 6.Blissard, G. W., R. L. Quant-Russell, G. F. Rohrmann, and G. S. Beaudreau. 1989. Nucleotide sequence, transcriptional mapping, and temporal expression of the gene encoding P39, a major structural protein of the multicapsid nuclear polyhedrosis virus of Orgyia pseudotsugata. Virology 168:354-362. [DOI] [PubMed] [Google Scholar]
- 7.Campbell, M. J. 1995. Lipofection reagents prepared by a simple ethanol injection technique. BioTechniques 18:1029-1032. [PubMed] [Google Scholar]
- 8.Chang, M.-J., J. Kuzio, and G. W. Blissard. 1999. Modulation of translational efficiency by contextual nucleotides flanking a baculovirus initiator AUG codon. Virology 259:369-383. [DOI] [PubMed] [Google Scholar]
- 9.Edwards, R. A., L. H. Keller, and D. M. Schifferli. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149-157. [DOI] [PubMed] [Google Scholar]
- 10.Friesen, P. D. 1997. Regulation of baculovirus early gene expression, p. 141-166. In L. K. Miller (ed.), The baculoviruses. Plenum Press, New York, N.Y.
- 11.Garrity, D. B., M.-J. Chang, and G. W. Blissard. 1997. Late promoter selection in the baculovirus gp64 envelope fusion protein gene. Virology 231:167-181. [DOI] [PubMed] [Google Scholar]
- 12.Gomi, S., C. E. Zhou, W. Yih, K. Majima, and S. Maeda. 1997. Deletion analysis of four of eighteen late gene expression factor gene homologues of the baculovirus, BmNPV. Virology 230:35-47. [DOI] [PubMed] [Google Scholar]
- 13.Grula, M. A., P. L. Buller, and R. F. Weaver. 1981. α-Amanitin-resistant viral RNA synthesis in nuclei isolated from nuclear polyhedrosis virus-infected Heliothis zea larvae and Spodoptera frugiperda cells. J. Virol. 38:916-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guarino, L. A., B. Xu, J. Jin, and W. Dong. 1998. A virus-encoded RNA polymerase purified from baculovirus-infected cells. J. Virol. 72:7985-7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kool, M., C. H. Ahrens, R. W. Goldbach, G. F. Rohrmann, and J. M. Vlak. 1994. Identification of genes involved in DNA replication of the Autographa californica baculovirus. Proc. Natl. Acad. Sci. USA 91:11212-11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, L., S. H. Harwood, and G. F. Rohrmann. 1999. Identification of additional genes that influence baculovirus late gene expression. Virology 255:9-19. [DOI] [PubMed] [Google Scholar]
- 17.Lin, G., G. Li, R. R. Granados, and G. W. Blissard. 2001. Stable cell lines expressing baculovirus P35: resistance to apoptosis and nutrient stress, and increased glycoprotein secretion. In Vitro Cell. Dev. Biol. Anim. 37:293-302. [DOI] [PubMed] [Google Scholar]
- 18.Lin, G., J. M. Slack, and G. W. Blissard. 2001. Expression and localization of LEF-11 in Autographa californica nucleopolyhedrovirus infected Sf9 cells. J. Gen. Virol. 82:2289-2294. [DOI] [PubMed] [Google Scholar]
- 19.Lu, A., P. J. Krell, J. M. Vlak, and G. R. Rohrmann. 1997. Baculovirus DNA replication, p. 171-192. In L. K. Miller (ed.), The baculoviruses. Plenum Press, New York, N.Y.
- 20.Lu, A., and L. K. Miller. 1995. The roles of eighteen baculovirus late expression factor genes in transcription and DNA replication. J. Virol. 69:975-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luckow, V. A., S. C. Lee, G. F. Barry, and P. O. Olins. 1993. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol. 67:4566-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLachlin, J. R., and L. K. Miller. 1994. Identification and characterization of vlf-1, a baculovirus gene involved in very late gene expression. J. Virol. 68:7746-7756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monsma, S. A., A. G. P. Oomens, and G. W. Blissard. 1996. The GP64 envelope fusion protein is an essential baculovirus protein required for cell-to-cell transmission of infection. J. Virol. 70:4607-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris, T. D., and L. K. Miller. 1994. Mutational analysis of a baculovirus major late promoter. Gene 140:147-153. [DOI] [PubMed] [Google Scholar]
- 25.Oellig, C., B. Happ, T. Müller, and W. Doerfler. 1987. Overlapping sets of viral RNAs reflect the array of polypeptides in the EcoRI J and N fragments (map positions 81.2 to 85.0) of the Autographa californica nuclear polyhedrosis virus genome. J. Virol. 61:3048-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ooi, B. G., C. Rankin, and L. K. Miller. 1989. Downstream sequences augment transcription from the essential initiation site of a baculovirus polyhedrin gene. J. Mol. Biol. 210:721-736. [DOI] [PubMed] [Google Scholar]
- 27.Oomens, A. G. P., and G. W. Blissard. 1999. Requirement for GP64 to drive efficient budding of Autographa californica multicapsid nucleopolyhedrovirus. Virology 254:297-314. [DOI] [PubMed] [Google Scholar]
- 28.O'Reilly, D. R., L. K. Miller, and V. A. Luckow. 1992. Baculovirus expression vectors: a laboratory manual. W. H. Freeman and Co., New York, N.Y.
- 29.Passarelli, A. L., and L. K. Miller. 1993. Identification and characterization of lef-1, a baculovirus gene involved in late and very late gene expression. J. Virol. 67:3481-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prikhod'ko, E. A., A. Lu, J. A. Wilson, and L. K. Miller. 1999. In vivo and in vitro analysis of baculovirus ie-2 mutants. J. Virol. 73:2460-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rankin, C., B. G. Ooi, and L. K. Miller. 1988. Eight base pairs encompassing the transcriptional start point are the major determinant for baculovirus polyhedrin gene expression. Gene 70:39-49. [DOI] [PubMed] [Google Scholar]
- 32.Rapp, J. C., J. A. Wilson, and L. K. Miller. 1998. Nineteen baculovirus open reading frames, including LEF-12, support late gene expression. J. Virol. 72:10197-10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohrmann, G. F. 1986. Polyhedrin structure. J. Gen. Virol. 67:1499-1513. [DOI] [PubMed] [Google Scholar]
- 34.Todd, J. W., A. L. Passarelli, A. Lu, and L. K. Miller. 1996. Factors regulating baculovirus late and very late gene expression in transient-expression assays. J. Virol. 70:2307-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Todd, J. W., A. L. Passarelli, and L. K. Miller. 1995. Eighteen baculovirus genes, including lef-11, p35, 39K, and p47, support late gene expression. J. Virol. 69:968-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang, S., and L. K. Miller. 1999. Activation of baculovirus very late promoters by interaction with very late factor 1. J. Virol. 73:3404-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang, S., and L. K. Miller. 1998. Control of baculovirus polyhedrin gene expression by very late factor 1. Virology 248:131-138. [DOI] [PubMed] [Google Scholar]
- 38.Yang, S., and L. K. Miller. 1998. Expression and mutational analysis of the baculovirus very late factor 1 (vlf-1) gene. Virology 245:99-109. [DOI] [PubMed] [Google Scholar]