Abstract
Here we report the identification of inbred miniature swine that failed to produce human-tropic replication-competent porcine endogenous retroviruses (HTRC PERVs), using in vitro coculture assays. When HTRC PERVs were isolated from transmitting animals, all were recombinant viruses, with the receptor-binding domain of PERV-A combining with PERV-C-related sequences.
The number of patients awaiting transplantation increases every year due to a lack of suitable donors. Although the xenotransplantation of porcine tissues has the potential to alleviate this shortage (5), there remain certain risks associated with the use of pigs as donor animals. In particular, the inadvertent transmission of porcine microorganisms to the recipient of a xenograft is a concern (1, 7, 14, 17, 20). Because porcine endogenous retroviruses (PERVs) are unaffected by barrier derivation technologies and can infect human cells in vitro (13), these viruses are perceived as a major safety concern.
Three replication-competent classes of PERVs (PERV-A, -B, and -C) have been identified in the genomic DNA of pigs and porcine cell lines (9, 18). The PERV-A and PERV-B classes can infect human and pig cells in vitro. PERV-C is not able to infect human cells but can replicate in porcine cell lines (13, 18). Although the majority of PERV loci in porcine DNA are most likely noninfectious (3, 4, 8, 9), human-tropic viruses have been isolated from porcine cell lines as well as primary tissues (2, 11-13, 16, 18, 21). Interestingly, all clones of replication-competent PERV-A and PERV-B classes with significant infectious titers have been isolated from PK15 cells or cell lines infected by viruses released from PK15 cells (6, 8, 19). Conversely, all PERV clones derived from primary pig cells have had extremely low titers and limited replication competence (6, 8, 19).
We investigated the PERV transmission characteristics of a unique inbred herd of miniature swine by using in vitro coculture assays. Three independent swine leukocyte antigen (SLA) haplotypes (a, c, and d) and three classes of recombinant SLA haplotypes (g, h, and k) are maintained within this herd. Due to its proven sensitivity, the coculture of porcine peripheral blood mononuclear cells (PBMC) with the human 293 cell line (6, 8, 10, 21, 22) and the porcine cell line ST-IOWA (10, 13, 21) has been used to compare the relative infectivity of PERVs produced from different pigs and the replication competence of PERV. In addition to these systems, we varied both the producer cells and target cell lines used in the coculture assays in an attempt to identify conditions under which additional human-tropic replication-competent (HTRC) PERVs could be identified. Pig aortic endothelial cells were investigated as an alternative primary cell source of PERV because single polynucleotide polymorphisms in PERV long terminal repeats (LTRs) have been reported that might confer differential locus responsiveness to activation by phytohemagglutinin and phorbol myristate acetate or tumor necrosis factor alpha (15). Production of PERVs from pig aortic endothelial cells was lower than that from PBMC and resulted in less-reliable coculture assays (data not shown). Therefore, only those assays using PBMC as the primary producer cells are reported below.
Because of the presence of PERV-C in miniature swine, we used the porcine cell line ST-IOWA and the human cell line HT1080 as target cell lines for the transmission assays. The HT1080 cell line is of particular interest because it is the only human cell line that has been reported to express a receptor for PERV-C (18). Although all ST-IOWA coculture assays became infected by replication-competent PERV-C released from the PBMC, infection was not detected with the HT1080 cell line (Table 1). Although it cannot be excluded that PERV-C may have required an extended adaptation period for its replication to become productive in the HT1080 cells, tissue culture-adapted classes of PERV-A and PERV-B, but not PERV-C, were capable of replication in this cell line, albeit at low levels (data not shown). In addition, it has recently been observed that retrovirus pseudotype particles lacking all env proteins can also infect HT1080 cells (Y. Takeuchi and F.-L. Cosset, personal communication). This suggests that the PERV-C infectivity reported for this human cell line is the result of nonspecific uptake mechanisms and therefore that no independent human-tropic capacity can currently be associated with PERV-C.
TABLE 1.
Transmission of PERV from lines of inbred miniature swinea
| Miniature swine line haplotype | No. of positive transmission assays/no. of animals tested with indicated cell line
|
||
|---|---|---|---|
| ST-IOWA | HT1080 | 293 | |
| a/a | 7/7 | NT | 2/7 |
| c/c | 8/8 | 0/1 | 4/8 |
| d/d | 28/28 | 0/6 | 3/28 |
| g/g | 10/10 | 0/1 | 1/10 |
| h/h | 4/4 | NT | 1/4 |
| k/k | 4/4 | NT | 1/4 |
Infection of porcine and human cell lines following exposure to miniature swine PBMC. Prior to coculture, PBMC were stimulated with phytohemagglutinin P (2.5 μg/ml) and phorbol 12-myristate-13-acetate (1 ng/ml). Infection of cells was measured by the presence of RT activity in the culture supernatants (HS-kit Mn2+ RT; Cavidi Tech AB) and, for transmitting animals, was typically detectable after 10 to 14 or 20 to 30 days in ST-IOWA and 293 cultures, respectively. 293 and HT1080 cell cocultures were maintained for a minimum of approximately 60 days before being classified as negative for PERV replication. PERV-C RNA expression was detected in ST-IOWA cells concurrently with the development of RT activity in the culture supernatants. NT, not tested.
Animals representing all lines of miniature swine were identified whose PBMC transmitted HTRC PERVs to 293 cells (Table 1). However, the frequency of transmission within the haplotype lines varied markedly (Table 1). In particular, two lines of miniature swine (SLAd/d and SLAg/g) showed low incidences of transmission. It is noteworthy that the three SLAd/d animals showing transmission are closely related (one sow and two offspring) and, similarly, that the single transmission-positive animal identified within the SLAg/g subline is from a line of animals distinct from the nontransmitting SLAg/g animals. To assess the reliability of the coculture assays, and therefore the significance of negative transmission results with 293 cells, repeat cocultures for individual animals were initiated using PBMC isolated from additional blood samples. Although a low false-negative rate in the assay system was identified, animals were identified that consistently produced transmitting or nontransmitting phenotypes, following repeat testing on up to four occasions (Table 2). The identification of transmitting animals within all of the lines of SLA-defined miniature swine indicates that critical PERV proviruses are unlikely to be closely linked to the SLA on chromosome 7.
TABLE 2.
Summary of 293 cell transmission assays from repeat blood draws of individual miniature swine
| No. of negative assays/no. of positive assays | No. of animals tested |
|---|---|
| 1/0 | 42 |
| 2/0 | 4 |
| 3/0 | 1 |
| 4/0 | 2 |
| 0/1 | 6 |
| 0/2 | 2 |
| 0/3 | 1 |
| 1/1 | 2 |
| 1/2 | 1 |
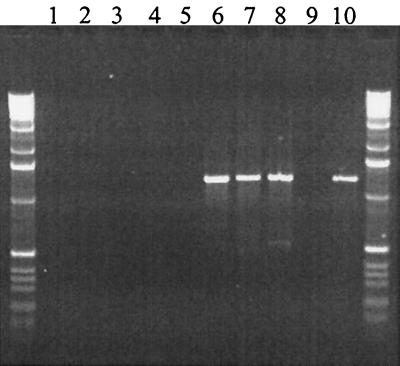
To gain information regarding the genomic organization of the HTRC PERVs produced by transmitting miniature swine, we examined the PERV proviruses present in eight independently infected 293 cultures using LTR-to-LTR PCR. In total, 127 PERV clones were analyzed by sequencing regions of their gag, pol, and env genes. All clones possessed gag, pol, and env transmembrane (TM) sequences most closely related to PERV-C and the env receptor-binding domain of PERV-A. To determine whether recombination events were required to generate the PERV-A-PERV-C hybrid sequences detected in the HTRC PERVs, we analyzed the sequence of PERV loci present in the genomic DNA of miniature swine. PERV-A clones were generated using LTR-to-LTR PCR, and regions of the gag, pol, and env genes of nominally full-length clones were sequenced. Analysis indicated that many of these PERV-A clones possessed gag and pol genes most closely related to PERV-C, suggesting that the PERV-C gag-pol and PERV-A env hybrid sequences were derived directly from genomic sequences. Alignment of the pol- env gene sequences of approximately 30 HTRC-PERV clones indicated that three distinct families of PERV-C pol and PERV-A env sequences were present and is suggestive of only a limited number of PERV-A loci having contributed toward the formation of HTRC PERVs in the samples tested (Fig 1). In contrast, PERV-A variable region A (VRA)-PERV-C TM hybrid sequences were not detected in the genomic DNA of miniature swine by PCR (Fig. 2), indicating that they were being generated by recombination during the in vitro coculture period. Interestingly, although PERV-A loci with long env open reading frames (ORFs) were identified in miniature swine DNA, their env TM ORFs were disrupted within a few residues of the carboxy terminus. In HTRC PERVs, this region was replaced by in-frame PERV-C sequences that repaired the TM ORF, suggesting a growth advantage of the recombinant viruses over the truncated PERV-A viruses or rescue of defective sequences during the in vitro coculture period. Further experiments are required to identify the interactions driving these recombination events. In this regard, it is noteworthy that an env TM truncation has also been identified in a PERV-A clone derived from PK15 cells (8). Cells infected by this virus produced only transient reverse transcriptase (RT) activities at levels approximately 1,000-fold below those infected by tissue culture-adapted PERVs (6, 8, 19) and lower than those identified with porcine cells that behave as if uninfected by PERVs (13, 18, 21, 22). While such minimal replication competence can be demonstrated using cloned viral DNA in combination with highly sensitive RT detection systems, it remains to be determined if such clones can be isolated directly from primary pig cells using coculture systems.
FIG. 1.
Alignment of pre-VRA (A) and VRA (B) regions of the env genes of HTRC PERV. Only those nucleotides that vary between the PERV-A (shaded boxes) and PERV-C (unshaded boxes) reference sequences are indicated. HTRC PERV loci were PCR amplified from PERV-infected 293 cells using oligonucleotides designed to match LTR sequences conserved between the three infectious classes of PERV (5′-CCTGGTGGTCTCCTACTGTCG-3′ and 5′-GCTTTTATGGGGTTCACAACAAA-3′) with TaKaRa DNA polymerase (Intergen). The sequences were divided into three families as defined by the position at which their nucleotide sequence underwent transition between PERV-C and PERV-A. Representative members of each family are presented. A nucleotide substitution that did not correlate with either reference sequence was identified at base 192 in sequence T6E5 (black box).
FIG. 2.
PCR analysis of miniature swine genomic DNA indicating that PERV-A-PERV-C TM recombinants are not present in miniature swine genomic DNA but develop during tissue culture. The sense primer specific for PERV-A is located in the VRA region, and the PERV-C specific antisense primer is located in the TM region. Samples are as follows: water control (lane 1), DNA from miniature swine (nontransmitting animals, lanes 2 and 3; transmitting animals, lanes 4 and 5), 293 cells infected with HTRC-PERV (lanes 6 to 8), 293 cells (lane 9), and a plasmid-positive control of a cloned HTRC-PERV env gene (lane 10). Control amplifications of cytochrome b sequences were successful and comparable from each sample (data not shown).
In summary, we report the in vitro PERV transmission characteristics of a unique herd of inbred miniature swine and demonstrate that animals can be identified that consistently do not transmit HTRC PERVs. When HTRC PERVs were isolated from transmitting miniature swine, they were shown to be recombinant viruses derived from PERV-A and PERV-C. It will be important to determine whether a transmitting phenotype is inherited in a strictly Mendelian manner or whether additional factors such as exogenous PERV transmission between animals affects the production of HTRC PERV by pigs.
Nucleotide sequence accession numbers
Representative members of full-length HTRC PERV env sequences (accession no. AF417227 through AF417232), HTRC PERV-A and genomic PERV locus gag sequences (accession no. AF417210 through AF417221), and near-full-length genomic PERV loci env sequences (accession no. AF417222 through AF417226) have been deposited at GenBank.
Acknowledgments
We thank Jay Fishman and Gillian Langford for expert discussions, Gabi Cruz for excellent technical assistance, and Leanne Duncan for administrative support.
This research was supported in part by a Small Business Innovation Research program grant (1R43AI48349-01).
REFERENCES
- 1.Birmingham, K. 1999. FDA subcommittee finds no evidence of PERV transmission. Nat. Med. 5:855.. [DOI] [PubMed] [Google Scholar]
- 2.Blusch, J. H., C. Patience, Y. Takeuchi, C. Templin, C. Roos, K. Von Der Helm, G. Steinhoff, and U. Martin. 2000. Infection of nonhuman primate cells by pig endogenous retrovirus. J. Virol. 74:7687-7690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boeke, J. D., and J. P. Stoye. 1997. Retrotransposons, endogenous retroviruses, and the evolution of retroelements, p. 343-436. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 4.Bösch, S., C. Arnauld, and A. Jestin. 2000. Study of full-length porcine endogenous retrovirus genomes with envelope gene polymorphism in a specific-pathogen-free Large White swine herd. J. Virol. 74:8575-8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cozzi, E., S. Masroor, B. Soin, C. Vial, and D. J. White. 2000. Progress in xenotransplantation. Clin. Nephrol. 53:13-18. [PubMed] [Google Scholar]
- 6.Czauderna, F., N. Fischer, K. Boller, R. Kurth, and R. R. Tonjes. 2000. Establishment and characterization of molecular clones of porcine endogenous retroviruses replicating on human cells. J. Virol. 74:4028-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fishman, J. A. 2000. Xenotransplantation from swine: making a list, checking it twice. Xenotransplantation 7:93-95. [DOI] [PubMed] [Google Scholar]
- 8.Krach, U., N. Fischer, F. Czauderna, and R. R. Tonjes. 2001. Comparison of replication-competent molecular clones of porcine endogenous retrovirus class A and class B derived from pig and human cells. J. Virol. 75:5465-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le Tissier, P., J. P. Stoye, Y. Takeuchi, C. Patience, and R. A. Weiss. 1997. Two sets of human-tropic pig retrovirus. Nature 389:681-682. [DOI] [PubMed] [Google Scholar]
- 10.Martin, U., V. Kiessig, J. H. Blusch, A. Haverich, K. von der Helm, T. Herden, and G. Steinhoff. 1998. Expression of pig endogenous retrovirus by primary porcine endothelial cells and infection of human cells. Lancet 352:692-694. [DOI] [PubMed] [Google Scholar]
- 11.Martin, U., M. E. Winkler, M. Id, H. Radecke, L. Arseniev, R. Groteluschen, A. R. Simon, and G. Steinhoff. 2000. Transmission of pig endogenous retrovirus to primary human cells. Transplant. Proc. 32:1157.. [DOI] [PubMed] [Google Scholar]
- 12.Martin, U., M. E. Winkler, M. Id, H. Radeke, L. Arseniev, Y. Takeuchi, A. R. Simon, C. Patience, A. Haverich, and G. Steinhoff. 2000. Productive infection of primary human endothelial cells by pig endogenous retrovirus (PERV). Xenotransplantation 7:138-142. [DOI] [PubMed] [Google Scholar]
- 13.Patience, C., Y. Takeuchi, and R. A. Weiss. 1997. Infection of human cells by an endogenous retrovirus of pigs. Nat. Med. 3:282-286. [DOI] [PubMed] [Google Scholar]
- 14.Platt, J. L. 2000. Xenotransplantation. New risks, new gains. Nature 407:27, 29-30. [DOI] [PubMed]
- 15.Quinn, G., and G. Langford. 2001. The porcine endogenous retrovirus long terminal repeat contains a single nucleotide polymorphism that confers distinct differences in estrogen receptor binding affinity between PERV A and PERV B/C subtypes. Virology 286:83-90. [DOI] [PubMed] [Google Scholar]
- 16.Specke, V., S. Rubant, and J. Denner. 2001. Productive infection of human primary cells and cell lines with porcine endogenous retroviruses. Virology 285:177-180. [DOI] [PubMed] [Google Scholar]
- 17.Takeuchi, Y. 2000. Risk of zoonosis in xenotransplantation. Transplant. Proc. 32:2698-2700. [DOI] [PubMed] [Google Scholar]
- 18.Takeuchi, Y., C. Patience, S. Magre, R. A. Weiss, P. T. Banerjee, P. Le Tissier, and J. P. Stoye. 1998. Host range and interference studies of three classes of pig endogenous retrovirus. J. Virol. 72:9986-9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tonjes, R. R., F. Czauderna, N. Fischer, U. Krach, K. Boller, P. Chardon, C. Rogel-Gaillard, M. Niebert, G. Scheef, A. Werner, and R. Kurth. 2000. Molecularly cloned porcine endogenous retroviruses replicate on human cells. Transplant. Proc. 32:1158-1161. [DOI] [PubMed] [Google Scholar]
- 20.Weiss, R. A., S. Magre, and Y. Takeuchi. 2000. Infection hazards of xenotransplantation. J. Infect. 40:21-25. [DOI] [PubMed] [Google Scholar]
- 21.Wilson, C. A., S. Wong, J. Muller, C. E. Davidson, T. M. Rose, and P. Burd. 1998. Type C retrovirus released from porcine primary peripheral blood mononuclear cells infects human cells. J. Virol. 72:3082-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson, C. A., S. Wong, M. VanBrocklin, and M. J. Federspiel. 2000. Extended analysis of the in vitro tropism of porcine endogenous retrovirus. J. Virol. 74:49-56. [DOI] [PMC free article] [PubMed] [Google Scholar]