Abstract
We previously showed that the intrahepatic induction of cytokines such as alpha/beta interferon (IFN-α/β) and gamma interferon (IFN-γ) inhibits hepatitis B virus (HBV) replication noncytopathically in the livers of transgenic mice. The intracellular pathway(s) responsible for this effect is still poorly understood. To identify interferon (IFN)-inducible intracellular genes that could play a role in our system, we crossed HBV transgenic mice with mice deficient in IFN regulatory factor 1 (IRF-1), the double-stranded RNA-activated protein kinase (PKR), or RNase L (RNase L) (IRF-1−/−, PKR−/−, or RNase L−/− mice, respectively), three well-characterized IFN-inducible genes that mediate antiviral activity. We showed that unmanipulated IRF-1−/− or PKR−/− transgenic mice replicate HBV in the liver at slightly higher levels than the respective controls, suggesting that both IRF-1 and PKR individually appear to mediate signals that modulate HBV replication under basal conditions. These same animals were responsive to the antiviral effects of the IFN-α/β inducer poly(I-C) or recombinant murine IFN-γ, suggesting that under these conditions, either the IRF-1 or the PKR genes can mediate the antiviral activity of the IFNs or other IFN-inducible genes mediate the antiviral effects. Finally, RNase L−/− transgenic mice were undistinguishable from controls under basal conditions and after poly(I-C) or IFN-γ administration, suggesting that RNase L does not modulate HBV replication in this model.
Hepatitis B virus (HBV) is a noncytopathic, enveloped virus that causes acute and chronic hepatitis and hepatocellular carcinoma (4). It was previously shown that the intrahepatic induction of alpha/beta interferon (IFN-α/β) that occurs in the livers of HBV transgenic mice after injection of poly(I-C) or infection with unrelated hepatotropic viruses, such as lymphocytic choriomeningitis virus and adenovirus (9, 21), downregulates HBV replication noncytopathically (21). The contribution of IFN-α/β to this process was demonstrated by showing that antiviral activity is completely blocked in HBV transgenic mice that were either genetically deficient for the IFN-α/β receptor (21) or treated with antibodies to IFN-α/β (10). Recent studies have shown that the mechanism whereby IFN-α/β inhibits HBV replication in the transgenic mouse liver relies on the inhibition of formation and/or the destabilization of immature HBV RNA-containing capsids (29).
The intrahepatic induction of gamma interferon (IFN-γ) also inhibits HBV replication noncytopathically; this effect is achieved by injecting HBV transgenic mice with HBV-specific cytotoxic T lymphocytes (10, 21) or interleukin 12 (3) or by infecting them with mouse cytomegalovirus (2). Experiments with antibodies to IFN-γ or mice genetically deficient for IFN-γ have demonstrated the importance of this cytokine as a mediator for the antiviral activity of these stimuli.
Upon binding to specific surface receptors, IFN-α/β and IFN-γ activate a variety of IFN-inducible genes, some of which trigger common intracellular antiviral pathways (25). A large variety of IFN-inducible genes have been identified to date; most of these are activated by the JAK-STAT signal transduction cascade (6, 7, 14, 25, 34). However, how these genes exert their intracellular antiviral activities is still poorly understood. Among the interferon (IFN)-inducible genes, those for the IFN regulatory factor 1 (IRF-1), RNase L, and double-stranded RNA-activated protein kinase (PKR) systems are some of the best characterized. IRF-1 is an IFN-inducible transcription factor that regulates nitric oxide production (1, 19) and cytokine signaling (22) and mediates antiviral activities against several viruses, including coxsackievirus (19). RNase L, a cellular RNase activated by 2′,5′-oligoadenylates produced by IFN-induced, double-stranded RNA-dependent synthetase (2′,5′-OAS), degrades viral and cellular RNAs (5, 15, 31). This pathway has been shown to selectively reduce the intracellular RNA content of viruses such as human immunodeficiency virus (20), encephalomyocardarditis virus (EMCV) (18), and vaccinia virus (8). The inhibition of viral protein synthesis initiation by IFN-inducible PKR has been shown to suppress certain viral infections, including those with EMCV (14) and reovirus (23). Recently, the interaction between the hepatitis C virus NS5 and E2 proteins and PKR was suggested to reduce the sensitivity of this virus to IFN-α/β (13, 26, 28).
Based on the aforementioned studies, it is possible that the IRF-1, RNase L, or PKR genes represent intracellular candidate genes that mediate the antiviral activities of IFN-α/β and/or IFN-γ in our system. To test this hypothesis, we crossed transgenic mice that replicate HBV with mice that are genetically deficient for IRF-1, RNase L, or PKR (IRF-1−/−, RNase L−/−, or PKR−/− mice, respectively), and we monitored the contributions of these gene products to the antiviral effects of the systemic administration of poly(I-C) or IFN-γ.
MATERIALS AND METHODS
Mice.
The HBV transgenic mouse lineage 1.3.32 (inbred C57BL/6) used in this study (official designation, Tg[HBV 1.3 genome]Chi32) was described previously (11). These mice replicate HBV at high levels in the liver without any evidence of cytopathology. Lineage 1.3.32 was crossed with IRF-1−/− (16, 27), RNase L−/− (32), or PKR−/− (30) mice. IRF-1−/− mice were obtained from Jackson Laboratory (Bar Harbor, Maine). Heterozygous mice from lineage 1.3.32 were repeatedly backcrossed with homozygous mice from each of the three knockout lineages to yield progeny that were screened for hepatitis B e antigen (HBeAg) in the serum (by using a commercially available kit from Abbott Laboratories, Abbott Park, Ill.). HBeAg-positive progeny were screened for homozygosity of the null mutations by PCR as described previously (16, 30, 32). Mice found either homozygous or heterozygous for the null mutation were matched for age (8 to 10 weeks), sex (male), and levels of HBeAg in their serum before experimental manipulations. All animals were housed in pathogen-free rooms under strict barrier conditions.
Poly(I-C) and IFN-γ treatments.
Mice were injected intravenously with a single dose of either poly(I-C) complex (Sigma Chemical Co., St. Louis, Mo.) (200 μg/mouse) or recombinant murine IFN-γ (Genentech, Inc., San Francisco, Calif.) (200,000 U/mouse) and sacrificed 24 h later. Their livers were processed for histological analysis or snap frozen in liquid nitrogen and stored at −80°C for subsequent molecular analyses (see below).
Tissue DNA and RNA analyses.
Frozen liver tissue was mechanically pulverized under liquid nitrogen, and total genomic DNA and RNA were isolated exactly as previously described (11). Analyses of HBV DNA by Southern blotting and various cytokine and 2′,5′-OAS mRNAs by an RNase protection assay (RPA) were performed exactly as previously described (10, 11). The relative abundances of specific DNA and RNA molecules were quantitated by phosphorimaging analysis with Optiquant image analysis software (Packard, Meriden, Conn.).
Biochemical and histological analyses.
The extent of hepatocellular injury was monitored by measuring serum alanine aminotransferase (sALT) activity at multiple time points after treatment with saline, poly(I-C), or IFN-γ. sALT activity was measured with a Paramax chemical analyzer (Baxter Diagnostics Inc., McGaw Park, Ill.) exactly as previously described (10). For histological analysis, liver tissue samples were fixed in 10% zinc-buffered formalin (Anatech, Battle Creek, Mich.), embedded in paraffin, sectioned (3 μm), and stained with hematoxylin and eosin exactly as described elsewhere (10).
Statistical analysis.
A two-tailed nonparametric Wilcoxon test was used to assess the statistical significance of the experimental variation in the intrahepatic content of HBV replicative forms among transgenic mice that were either heterozygous or homozygous for the IRF-1 or PKR null mutation. A P value of <0.05 was considered significant.
RESULTS AND DISCUSSION
Higher levels of HBV replication in the livers of IRF-1−/− or PKR−/− mice.
HBV transgenic mice from lineage 1.3.32 were crossed with IRF-1−/−, RNase L−/−, or PKR−/− mice. Groups (six mice per group) of age (8 to 10 weeks)-, sex (male)-, and serum HBeAg-matched animals that were either heterozygous (plus/minus) or homozygous (minus/minus) for the respective null mutation were sacrificed, and their livers were harvested. Following extraction, total hepatic DNAs were pooled for each group and analyzed for HBV replication by Southern blot analysis.
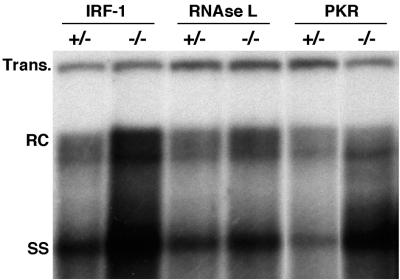
As shown in Fig. 1, IRF-1−/− or PKR−/− mice replicated HBV in the liver at levels about 2.2- or 1.5-fold higher than the respective heterozygous control littermates or RNase L−/− mice (as measured by phosphorimaging analysis with the transgene band for normalization). As shown in Table 1, the experimental variation in the levels of HBV replication between individual IRF+/− and IRF−/− mice and individual PKR+/− and PKR−/− mice was statistically significant (P values of 0.035 and 0.030, respectively), while no significant difference was detected between RNase L+/− and RNase L−/− mice (data not shown). The levels of HBV replication observed in all groups of heterozygous control mice were comparable to those observed in wild-type mice from lineage 1.3.32 (data not shown). The livers from the mice shown in Fig. 1 were also tested for the expression of IFN-γ and 2′,5′-OAS (a known IFN-α/β-inducible enzyme that produces an activator of RNase L) (25) by an RPA. As expected, neither message was detected in these uninflamed livers (data not shown), and similar results were obtained for saline-injected controls from all lineages (Fig. 2 and 3). Nonetheless, the higher content of HBV replicative forms in IRF-1−/− and PKR−/− mice suggests that both of these pathways (perhaps induced by undetectable amounts of interferons) appear to contribute individually to the control of HBV replication in uninflamed livers of wild-type mice.
FIG. 1.
Higher levels of HBV replication in the livers of IRF-1−/− or PKR−/− mice. Six age (8 to 10 weeks)-, sex (male)-, and serum HBeAg-matched mice that were either heterozygous or homozygous for the indicated null mutation were sacrificed, and the livers were harvested. Following extraction, total DNAs were pooled for each group and analyzed for HBV replication by Southern blot analysis. Bands corresponding to the integrated transgene (Trans.), relaxed circular (RC), and single-stranded (SS) linear HBV DNA replicative forms are indicated. The integrated transgene can be used to normalize the amount of DNA bound to the membrane. The filter was hybridized with a 32P-labeled HBV-specific DNA probe.
TABLE 1.
Statistically significant experimental variation of the intrahepatic content of HBV replicative forms in mice heterozygous (plus/minus) or homozygous (minus/minus) for the IRF-1 or PKR null mutationa
| Mouse | Ratio for the following mice:
|
|||
|---|---|---|---|---|
| IRF-1+/− | IRF-1−/− | PKR+/− | PKR−/− | |
| 1 | 78 | 104 | 47 | 68 |
| 2 | 52 | 91 | 58 | 47 |
| 3 | 67 | 88 | 36 | 54 |
| 4 | 48 | 102 | 34 | 71 |
| 5 | 56 | 107 | 61 | 78 |
| 6 | 44 | 77 | 74 | |
| 7 | 48 | |||
| Mean ± SD | 56.1 ± 12 | 94.8 ± 11 | 47.2 ± 12 | 63.6 ± 11 |
Data represent the ratio between the values obtained by phosphorimaging analysis for HBV replicative forms and the transgene.
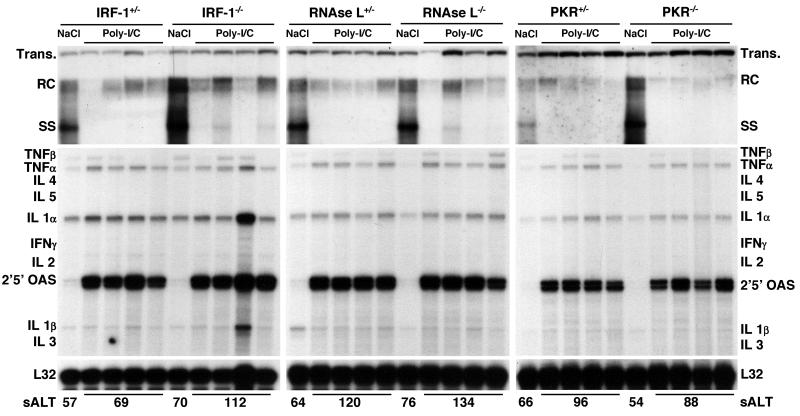
FIG. 2.
Role of IRF-1, RNase L, or PKR in antiviral activity induced by poly-I/C. Groups of age-, sex-, and serum HBeAg-matched transgenic mice (four mice per group) that were either heterozygous or homozygous for the indicated null mutation were injected intravenously with a single dose of poly(I-C) (200 μg/mouse) and sacrificed 24 h later. Total hepatic DNA was analyzed for HBV replication by Southern blot analysis. Bands corresponding to integrated transgene (Trans.), relaxed circular (RC), and single-stranded (SS) linear HBV DNA replicative forms are indicated. The integrated transgene can be used to normalize the amount of DNA bound to the membrane. Total hepatic RNA was analyzed for the expression of cytokine- and 2′,5′-OAS-specific transcripts by an RPA. TNF, tumor necrosis factor; IL, interleukin. The RNA encoding ribosomal protein L32 was used to normalize the amount of RNA loaded in each lane. The results were compared with those observed for livers pooled from 10 age-, sex-, and serum HBeAg-matched transgenic littermates injected with saline (NaCl). The mean sALT activity, measured at the time of autopsy, is indicated for each group and is expressed in units per liter.
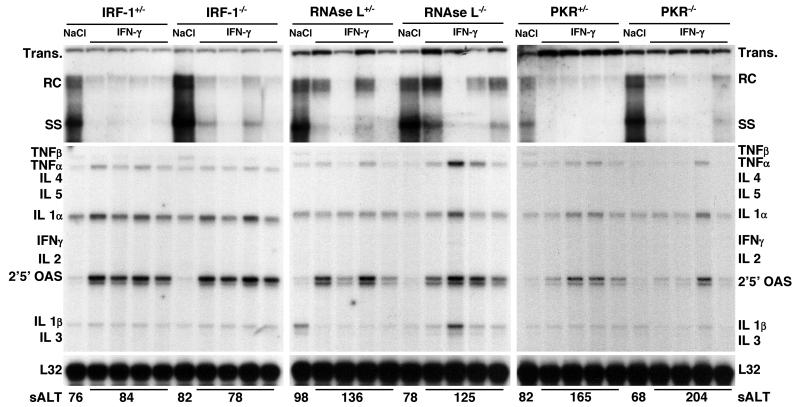
FIG. 3.
Role of IRF-1, RNase L, or PKR in antiviral activity induced by IFN-γ. Groups of age-, sex-, and serum HBeAg-matched transgenic mice (four mice per group) that were either heterozygous or homozygous for the indicated null mutation were injected intravenously with a single dose of IFN-γ (200,000 U/mouse) and sacrificed 24 h later. Total hepatic DNA was analyzed for HBV replication by Southern blot analysis. Bands corresponding to integrated transgene (Trans.), relaxed circular (RC), and single-stranded (SS) linear HBV DNA replicative forms are indicated. The integrated transgene can be used to normalize the amount of DNA bound to the membrane. Total hepatic RNA was analyzed for the expression of cytokine- and 2′,5′-OAS-specific transcripts by an RPA. TNF, tumor necrosis factor; IL, interleukin. The RNA encoding ribosomal protein L32 was used to normalize the amount of RNA loaded in each lane. The results were compared with those observed for livers pooled from 10 age-, sex-, and serum HBeAg-matched transgenic littermates injected with saline (NaCl). The mean sALT activity, measured at the time of autopsy, is indicated for each group and is expressed in units per liter.
Role of IRF-1, RNase L, or PKR in antiviral activity induced by poly-I/C.
To test the role of IRF-1, RNase L, or PKR in the antiviral activity of IFN-α/β induced by poly(I-C) in IRF-1−/−, RNase L−/−, or PKR−/− animals, groups of age-, sex-, and serum HBeAg-matched transgenic mice that were either heterozygous or homozygous for the respective null mutation were injected intravenously with a single dose of poly(I-C) (200 μg/mouse). Mice were bled and sacrificed, and livers were harvested 24 h later, when the antiviral activity of poly(I-C) is maximal (21, 29). Results were compared with those observed for livers pooled from six age-, sex-, and serum HBeAg-matched transgenic controls that were sacrificed 24 h after injection with saline.
As shown in Fig. 2, the hepatic content of HBV DNA replicative forms was profoundly reduced in all groups of IRF-1, RNase L, or PKR heterozygous or homozygous transgenic mice after poly(I-C) injection compared to the respective saline-injected control mice (as measured by phosphorimaging analysis, the average reduction of HBV DNA replicative forms was over 10-fold in all groups of mice). The most significant reduction involved the single-stranded DNA forms, while the more mature, high-molecular-weight relaxed circular double-stranded DNA forms remained detectable, albeit at levels lower than those detected in control mice (Fig. 2). The relative resistance of the mature HBV DNA forms to the antiviral effect of poly(I-C) is consistent with previous studies that showed that poly(I-C) either destabilizes or inhibits the formation of HBV RNA-containing capsids (29). Under these circumstances, single-stranded DNA-containing capsids disappear after RNA-containing capsids but before mature capsids, which are cleared from hepatocytes with slower kinetics (29).
As also shown in Fig. 2, the signal transduction pathway(s) required for the poly-I/C-dependent induction of 2′,5′-OAS RNA was intact in all groups of mice. Furthermore, the absence of detectable levels of IFN-γ RNA in these same livers (Fig. 2) suggests that this cytokine did not mediate the antiviral effect of poly-I/C. This result is consistent with previous results showing that the entire inhibitory effect of poly(I-C) in this system is mediated by IFN-α/β (21).
In keeping with the notion that poly(I-C) inhibits HBV replication noncytopathically (21), little or no liver disease was observed either histologically (data not shown) or biochemically (Fig. 2, bottom), as indicated by the very modest elevation in the level of sALT (a hepatocellular enzyme that is released into the circulation by injured hepatocytes) at the time of autopsy.
Role of IRF-1, RNase L, or PKR in antiviral activity induced by IFN-γ.
The intrahepatic induction of IFN-γ inhibits HBV replication in transgenic mice (3, 10), and this effect occurs independently of IFN-α/β (21). IFN-γ and IFN-α/β bind to distinct surface receptors and activate multiple intracellular antiviral pathways, some of which are common and involve IRF-1, RNase L, or PKR (25). To monitor the role of IRF-1, RNase L, or PKR in the direct antiviral activity of IFN-γ, groups of animals (four mice per group) from the same lineages as those used in the experiment shown in Fig. 1 were injected intravenously with a single dose of murine recombinant IFN-γ (200,000 U/mouse) and sacrificed 24 h after injection.
As shown in Fig. 3, HBV DNA replication was profoundly inhibited in the different groups of IRF-1, RNase L, or PKR heterozygous or homozygous transgenic mice after IFN-γ injection compared to the respective saline-injected controls (as measured by phosphorimaging analysis, the average reduction of HBV DNA replicative forms was about 10-fold in all groups of mice, with the exception of RNase L−/− mice, in which the reduction was about 6-fold). As in the experiment with poly-I/C, the inhibitory effect of IFN-γ on HBV replication was associated with the intrahepatic induction of 2′,5′-OAS RNA (Fig. 3), although this effect was less pronounced than that observed after poly(I-C) injection (Fig. 2), particularly for PKR−/− mice (Fig. 3). The notion that IFN-γ induces lower levels of 2′,5′-OAS RNA than IFN-α/β was previously demonstrated (24). Moreover, it was previously shown that the lack of the PKR gene results in a defect in IFN-γ-dependent signaling (17). Again, little or no liver disease was observed either histologically (data not shown) or biochemically (Fig. 3, bottom).
In summary, the results reported here showed that unmanipulated RNase L−/− transgenic mice replicate HBV in the liver at levels similar to those in the respective controls. This result, coupled with the fact that the deletion of RNase L activity does not block the antiviral effect of poly(I-C) or IFN-γ, suggests that RNase L is not likely to mediate the ability to inhibit HBV replication. The results also showed that unmanipulated IRF-1−/− or PKR−/− transgenic mice replicate HBV in the liver at levels slightly higher than those in the respective controls, suggesting that, under basal conditions, IRF-1 or PKR appears to mediate signals that modulate HBV replication. Follow-up experiments showed that the antiviral effect of poly(I-C) or IFN-γ was fully operative in the absence of either IRF-1 or PKR. Although there was a defect in IFN-γ induction of 2′,5′-OAS in the PKR−/− mice, the antiviral activity of IFN-γ was intact. Since 2′,5′-OAS is upstream of RNase L, these results are in accord with the lack of constitutive or induced anti-HBV activity in the RNase L−/− mice. Collectively, the data suggest either that high local concentrations of IFN-α/β or IFN-γ inhibit HBV replication by activating IRF-1 or PKR or that other IFN-inducible pathways mediate their antiviral effects. Future experiments with both IRF-1−/− and PKR−/− HBV transgenic mice will attempt to discriminate between these two hypotheses.
In keeping with the possibility that other IFN-inducible genes are involved, it is noteworthy that the advent of oligonucleotide arrays has enabled investigators to identify many novel genes that are induced or repressed by IFN-γ or IFN-α/β (6, 7). Furthermore, it was recently shown that although EMCV is susceptible to the antiviral activity of either RNase L or PKR, the simultaneous disruption of both gene products is still associated with residual IFN-dependent antiviral activity (33); this result indicates that as-yet-undefined IFN-inducible antiviral pathways are operative in the control of EMCV. It is also worth mentioning that the Mx protein, an IFN-induced GTPase that selectively inhibits influenza viruses and bunyaviruses (12), is not involved in our system, since the genetic backgrounds (C57BL/6 and 129/Sv) of the mice used here are deficient for this particular protein (25). Future research aimed at further defining IFN-induced intracellular molecular events that control HBV is clearly warranted.
Acknowledgments
We thank Monte Hobbs for providing the cytokine gene probes used in the RPA experiments and Margie Chadwell for excellent technical assistance.
This work was supported by grants AI40696 (to L.G.G.), CA40489 (to F.V.C.), AI34039 (to B.R.G.W.), and CA44059 (to R.H.S.) from the National Institutes of Health.
Footnotes
Manuscript 14438-MEM from The Scripps Research Institute.
REFERENCES
- 1.Bachmaier, K., N. Neu, C. Pummerer, G. S. Duncan, T. W. Mak, T. Matsuyama, and J. M. Penninger. 1997. iNOS expression and nitrotyrosine formation in the myocardium in response to inflammation is controlled by the interferon regulatory transcription factor 1. Circulation 96:585-591. [PubMed]
- 2.Cavanaugh, V. J., L. G. Guidotti, and F. V. Chisari. 1998. Inhibition of hepatitis B virus (HBV) replication during adenovirus and cytomegalovirus infections in HBV transgenic mice. J. Virol. 72:2630-2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavanaugh, V. J., L. G. Guidotti, and F. V. Chisari. 1997. Interleukin-12 inhibits hepatitis B virus (HBV) replication in HBV transgenic mice. J. Virol. 71:3236-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chisari, F. V., and C. Ferrari. 1995. Hepatitis B virus immunopathogenesis. Ann. Rev. Immunol. 13:29-60. [DOI] [PubMed] [Google Scholar]
- 5.Clemens, M. J., and B. R. Williams. 1978. Inhibition of cell-free protein synthesis by pppA2′p5′A2′p5′A: a novel oligonucleotide synthesized by interferon-treated L cell extracts. Cell 13:565-572. [DOI] [PubMed] [Google Scholar]
- 6.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Veer, M. J., M. Holko, M. Frevel, E. Walker, S. Der, J. M. Paranjape, R. H. Silverman, and B. R. Williams. 2001. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 69:912-920. [PubMed] [Google Scholar]
- 8.Diaz-Guerra, M., C. Rivas, and M. Esteban. 1997. Inducible expression of the 2-5A synthetase/RNase L system results in inhibition of vaccinia virus replication. Virology 227:220-228. [DOI] [PubMed] [Google Scholar]
- 9.Guidotti, L. G., P. Borrow, M. V. Hobbs, B. Matzke, I. Gresser, M. B. A. Oldstone, and F. V. Chisari. 1996. Viral cross talk: intracellular inactivation of the hepatitis B virus during an unrelated viral infection of the liver. Proc. Natl. Acad. Sci. USA 93:4589-4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guidotti, L. G., T. Ishikawa, M. V. Hobbs, B. Matzke, R. Schreiber, and F. V. Chisari. 1996. Intracellular inactivation of the hepatitis B virus by cytotoxic T lymphocytes. Immunity 4:25-36. [DOI] [PubMed] [Google Scholar]
- 11.Guidotti, L. G., B. Matzke, H. Schaller, and F. V. Chisari. 1995. High-level hepatitis B virus replication in transgenic mice. J. Virol. 69:6158-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haller, O., M. Frese, and G. Kochs. 1998. Mx proteins: mediators of innate resistance to RNA viruses. Rev. Sci. Tech. Off. Int. Epizoot. 17:220-230. [DOI] [PubMed] [Google Scholar]
- 13.He, Y., S. L. Tan, S. U. Tareen, S. Vijaysri, J. O. Langland, B. L. Jacobs, and M. G. Katze. 2001. Regulation of mRNA translation and cellular signaling by hepatitis C virus nonstructural protein NS5A. J. Virol. 75:5090-5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalvakolanu, D. V., and E. C. Borden. 1996. An overview of the interferon system: signal transduction and mechanisms of action. Cancer Investig. 14:25-53. [DOI] [PubMed] [Google Scholar]
- 15.Kerr, I. M., and R. E. Brown. 1978. pppA2′p5′A2′p5′A: an inhibitor of protein synthesis synthesized with an enzyme fraction from interferon-treated cells. Proc. Natl. Acad. Sci. USA 75:256-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimura, T., K. Nakayama, J. Penninger, M. Kitagawa, H. Harada, T. Matsuyama, N. Tanaka, R. Kamijo, J. Vilcek, T. W. Mak, et al. 1994. Involvement of the IRF-1 transcription factor in antiviral responses to interferons. Science 264:1921-1924. [DOI] [PubMed] [Google Scholar]
- 17.Kumar, A., Y. L. Yang, V. Flati, S. Der, S. Kadereit, A. Deb, J. Haque, L. Reis, C. Weissmann, and B. R. Williams. 1997. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-kappaB. EMBO J. 16:406-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, X. L., J. A. Blackford, and B. A. Hassel. 1998. RNase L mediates the antiviral effect of interferon through a selective reduction in viral RNA during encephalomyocarditis virus infection. J. Virol. 72:2752-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, P., J. Penninger, K. Aitken, M. Sole, and T. Mak. 1995. The role of transgenic knockout models in defining the pathogenesis of viral heart disease. Eur. Heart J. 16(Suppl. O):25-27. [DOI] [PubMed] [Google Scholar]
- 20.Maitra, R. K., and R. H. Silverman. 1998. Regulation of human immunodeficiency virus replication by 2′,5′-oligoadenylate-dependent RNase L. J. Virol. 72:1146-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McClary, H., R. Koch, F. V. Chisari, and L. G. Guidotti. 2000. Relative sensitivity of hepatitis B virus and other hepatotropic viruses to the antiviral effects of cytokines. J. Virol. 74:2255-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pitha, P. M., W. C. Au, W. Lowther, Y. T. Juang, S. L. Schafer, L. Burysek, J. Hiscott, and P. A. Moore. 1998. Role of the interferon regulatory factors (IRFs) in virus-mediated signaling and regulation of cell growth. Biochimie 80:651-658. [DOI] [PubMed] [Google Scholar]
- 23.Samuel, C. E. 1998. Reoviruses and the interferon system. Curr. Top. Microbiol. Immunol. 233:125-145. [DOI] [PubMed] [Google Scholar]
- 24.Staeheli, P. 1990. Interferon-induced proteins and the antiviral state. Adv. Virus Res. 38:147-200. [DOI] [PubMed] [Google Scholar]
- 25.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 26.Tan, S. L., and M. G. Katze. 2001. How hepatitis C virus counteracts the interferon response: the jury is still out on NS5A. Virology 284:1-12. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka, N., M. Ishihara, M. Kitagawa, H. Harada, T. Kimura, T. Matsuyama, M. S. Lamphier, S. Aizawa, T. W. Mak, and T. Taniguchi. 1994. Cellular commitment to oncogene-induced transformation or apoptosis is dependent on the transcription factor IRF-1. Cell 77:829-839. [DOI] [PubMed] [Google Scholar]
- 28.Taylor, D. R., S. T. Shi, P. R. Romano, G. N. Barber, and M. M. Lai. 1999. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science 285:107-110. [DOI] [PubMed] [Google Scholar]
- 29.Wieland, S. F., L. G. Guidotti, and F. V. Chisari. 2000. Intrahepatic induction of alpha/beta interferon eliminates viral RNA-containing capsids in hepatitis B virus transgenic mice. J. Virol. 74:4165-4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang, Y. L., L. F. Reis, J. Pavlovic, A. Aguzzi, R. Schafer, A. Kumar, B. R. Williams, M. Aguet, and C. Weissmann. 1995. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 14:6095-6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou, A., B. A. Hassel, and R. H. Silverman. 1993. Expression cloning of 2-5A-dependent RNAase: a uniquely regulated mediator of interferon action. Cell 72:753-765. [DOI] [PubMed] [Google Scholar]
- 32.Zhou, A., J. Paranjape, T. L. Brown, H. Nie, S. Naik, B. Dong, A. Chang, B. Trapp, R. Fairchild, C. Colmenares, and R. H. Silverman. 1997. Interferon action and apoptosis are defective in mice devoid of 2′,5′- oligoadenylate-dependent RNase L. EMBO J. 16:6355-6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou, A., J. M. Paranjape, S. D. Der, B. R. Williams, and R. H. Silverman. 1999. Interferon action in triply deficient mice reveals the existence of alternative antiviral pathways. Virology 258:435-440. [DOI] [PubMed] [Google Scholar]
- 34.Zhu, H., J. P. Cong, and T. Shenk. 1997. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc. Natl. Acad. Sci. USA 94:13985-13990. [DOI] [PMC free article] [PubMed] [Google Scholar]