Abstract
Sendai virus encodes an RNA-dependent RNA polymerase which is composed of the L and P proteins. Site-directed mutagenesis of the N terminus of L has identified amino acids important for binding P. Seven of nine mutants in amino acids 1 to 350 of Sendai L lost the ability to bind to Sendai P, although they were still able to bind the viral C protein. Loss of P binding correlated with the loss of all RNA synthesis activities. Two L mutants gave limited P-L complex formation and limited viral transcription and replication.
The viruses belonging to the Paramyxovirinae family contain a negative-sense (−), unsegmented RNA genome of about 15 kb, which is encapsidated by the nucleocapsid protein (NP) in a helical nucleocapsid that serves as the template for all viral RNA synthesis (12). The virion-associated phosphoprotein (P) and the L protein are the two subunits of the viral RNA-dependent RNA polymerase. Transcription initiates at the 3′ end of the genome RNA, giving the sequential synthesis of (+) strand leader (le+) RNA and then the NP, P/C/V, M, F, HN, and L mRNAs. During genome replication the synthesis of full-length viral RNA is coupled to its encapsidation by NP protein, utilizing an NP0-P protein complex as the source of NP. For Sendai virus, formation of the RNA polymerase requires the cotranslation of the P and L proteins, in which P binding stabilizes the L protein, probably by facilitating the correct folding of L (8, 9). The Sendai virus P protein is a tetramer forming a coiled coil (18, 19), and the L binding site on P mapped to amino acids (aa) 412 to 479 (6, 17).
The L protein is thought to contain all the catalytic functions required for RNA synthesis. Alignment of the L proteins of viruses of the order Mononegevirales showed six domains of relatively high conservation, designated I to VI from the N terminus to the C terminus of the protein, which were proposed to specify the essential activities common to all L proteins (14, 15). Recent characterization of Sendai virus L mutants in each of the six domains suggests that multiple domains contribute to the different steps in viral RNA synthesis, since mutants in different domains gave the same defective phenotypes (2, 3, 7, 11, 16). Viral RNA synthesis is downregulated by the C proteins encoded from the P gene (12). In the case of Sendai virus, the C proteins were shown to bind to the L polymerase subunit to inhibit RNA synthesis (9).
The L proteins of paramyxoviruses are all over 2,000 aa, and studies to begin mapping the P binding site on L showed a general location in the N-terminal quarter or half of the protein (2, 10, 13). Of the mutants characterized in the various domains in the N-terminal half of Sendai virus L only one, S368R in domain I, abolished binding to Sendai virus P (2, 7, 16). Together these data suggested that the P binding site may reside from aa 1 to 400 of L encompassing the N terminus and domain I. In these studies site-directed mutagenesis identified residues within the N-terminal 350 aa of the L protein of Sendai virus that mediate binding to P protein and the activity of the viral RNA polymerase.
Identification of the P binding site on Sendai virus L protein
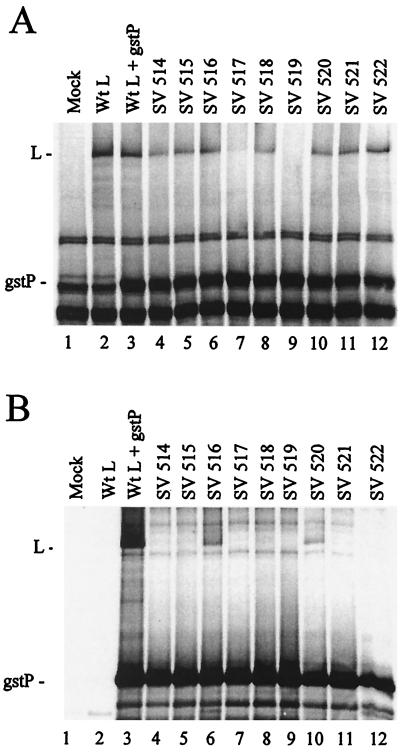
Nine clustered hydrophobic-to-alanine or charged-to-alanine scanning mutants were constructed in the N-terminal 350 aa of Sendai virus L (Table 1). In vitro transcription and translation of the plasmids showed that most of the L mutants were synthesized at levels comparable to those of wild-type (wt) L with the exception of Sendai virus (SV) 517 and SV 519, which were greatly reduced (data not shown). When initially tested by long-term labeling in transient transfections of mammalian cells, all these L mutants were synthesized at low levels even in the presence of the Sendai virus glutathione-S-transferase P fusion protein (gstP) (data not shown), suggesting that these mutants may not be able to bind to P and were unstable, so pulse-labeling was performed. Cells were infected with a vaccinia virus recombinant encoding T7 RNA polymerase (VVT7), transfected with the Sendai virus plasmids encoding gstP and wt or mutant L (2, 4, 5) under the control of the T7 promoter, and pulse-labeled with Express-35S. Analysis of total protein showed that wt L alone and wt L with gstP were expressed at about the same levels under these conditions (Fig. 1A, lanes 2 and 3). The synthesis of SV 517 and that of SV 519 with gstP were very low and undetectable, respectively (Fig. 1A, lanes 7 and 9), suggesting that these two proteins were intrinsically unstable and degraded immediately in mammalian cells, although small amounts were detected by in vitro translation. The remaining L mutants even in a pulse-label were not synthesized nearly as well as wt L (Fig. 1A), these results being similar to the data obtained with long-term labeling (not shown). The gstP protein migrated like a background band. P-L complex formation was determined by the cobinding of the L proteins with gstP to glutathione-Sepharose beads. No protein bound the beads in the absence of viral proteins or when wt L was expressed in the absence of gstP as expected, but when the gstP and wt L proteins were coexpressed L cobound with gstP (Fig. 1B, lanes 1 to 3). Only SV 516 and SV 520 cobound with gstP to the beads (lanes 6 and 10) in amounts approximately equal to their level of expression. The remaining seven mutants gave no detectable polymerase complex formation (Fig. 1B) identifying amino acids essential for L binding to P.
TABLE 1.
Amino acid changes in the Sendai virus L mutants
| Mutant | Amino acid changesa | P bindingb |
|---|---|---|
| SV 514 | L20A, S22A, I24A, V25A | − |
| SV 515 | T29A, Q31A, L32A, H33A | − |
| SV 516 | Q77A, R78S, T79A, K81A | ± |
| SV 517 | F173A, L174A, T175A, F177A | − |
| SV 518 | Y209A, L210E, T211A, V212A, T213A | − |
| SV 519 | L235A, V236A, L237A, M238A | − |
| SV 520 | L262A, L263A, V264G, K265R, G266A | ± |
| SV 521 | V287A, I288A, L290A, L291A | − |
| SV 522 | I345A, F346A, H347A | − |
Changes are indicated as the amino acid position in the Sendai virus L protein. The letter preceding the position is the wt sequence, and the amino acid to which it was changed appears after the number. Domain I in Sendai virus L extends from aa 225 to 416.
FIG. 1.
P-L complex formation with the Sendai virus L mutants. A549 cells in 35-mm-diameter dishes were infected with VVT7 at a multiplicity of infection of 2.5 PFU/ml and transfected with no plasmids (Mock) or Sendai virus gstP (0.2 μg) and the indicated wt or mutant Sendai virus L (1.67 μg) plasmids. The cells were incubated for 10 h and then labeled for 30 min using Express-35S (100 μCi/ml), and cytoplasmic extracts were prepared. Samples of the extracts were analyzed directly by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) for total protein expression (A) or incubated with glutathione-Sepharose beads, after which the bound proteins were separated by SDS-PAGE (B). The positions of the proteins are indicated.
Sendai virus L mutants did not bind with P to nucleocapsids
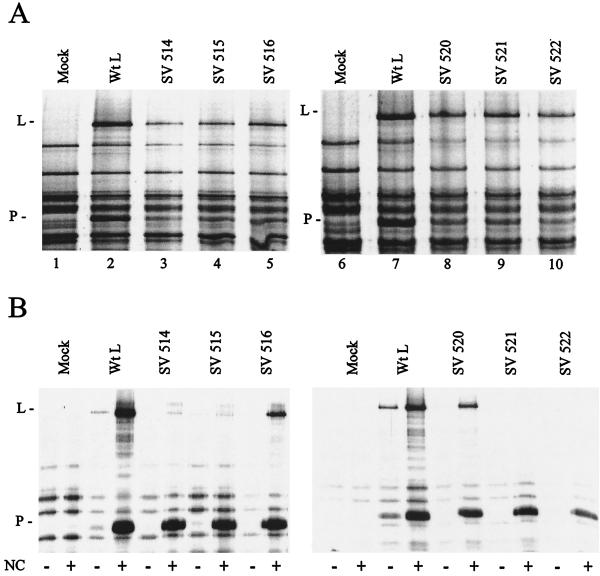
The L protein itself is unable to bind to the nucleocapsid template but is bound through the P subunit of the P-L complex (11). We have identified L mutants that were unable to bind to gstP in a pull-down assay (Fig. 1) but wanted to confirm this by another method that does not rely on a P fusion protein. We therefore measured the binding of mutant L with P to the nucleocapsid template by a cosedimentation assay. VVT7-infected cells were transfected with the P and the wt or mutant L plasmids, except for SV 517 and SV 519, which were not expressed, and labeled with Express-35S. Analysis of total protein synthesis showed the same low levels of synthesis of the L mutants (Fig. 2A) observed with expression with gstP (Fig. 1A). Samples of the extracts were incubated with or without polymerase-free NC and sedimented through glycerol. Analysis of the pellets showed some host or vaccinia virus background proteins in the absence of Sendai virus proteins and small amounts of P and wt L in the absence of NC, which were probably due to some aggregation (Fig. 2B, mock and wt L). In the presence of NC greatly increased levels of both P and wt L bound NC, showing polymerase complex formation. For the L mutants, SV 516 and SV 520 cobound, at levels approximately equal to their degree of synthesis, with the P protein to NC, confirming polymerase complex formation with these mutants (Fig. 2B). However, none of the other L mutants sedimented with NC, even though P bound in each case, confirming the results with the gstP experiments.
FIG. 2.
Mutant virus Sendai polymerase binding to nucleocapsids. VVT7-infected A549 cells were transfected with no plasmids (Mock) or the Sendai virus P and the indicated wt or mutant L plasmids. The cells were incubated overnight with Express-35S, and extracts were prepared. (A) Total samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). (B) Samples were incubated in the absence (−) and presence (+) of Sendai virus nucleocapsids (NC; 1 μg) and then pelleted through glycerol as described previously (11). The NC-associated proteins were analyzed by SDS-PAGE. The positions of the proteins are indicated.
In these studies the mutations in L that abolish P binding are spread from aa 20 to 347, suggesting a site with nonlinear amino acids. Together with the previously identified L mutant at S368 that did not bind P (2), the P binding site on L encompasses aa 20 to 368. In Sendai virus L, of the total eight mutants unable to bind to P four were constructed by changing primarily hydrophobic amino acids to alanine, while the remainder changed various other amino acids. These data suggest that while hydrophobic interactions are important, other interactions also contribute to the P binding site on L.
Sendai virus L mutants bound the viral C protein
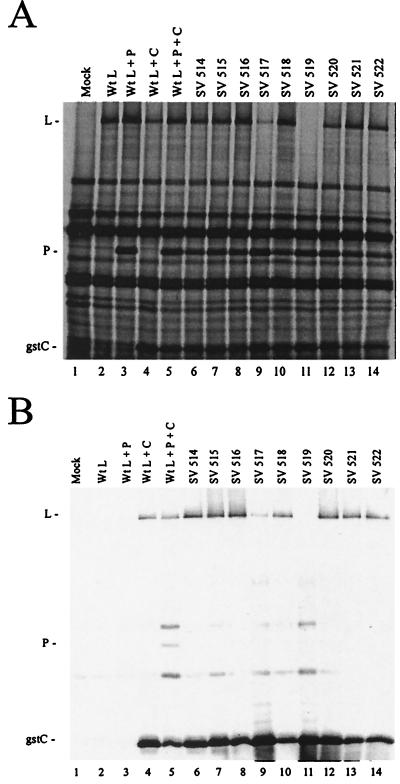
There are at least two explanations for the inability of the L mutants to bind to P. First, their structures may be globally disrupted and the proteins may be degraded as quickly as they are synthesized, as appears to be the case for SV 517 and SV 519. Alternatively, the P binding site on L may be specifically disrupted such that binding cannot occur and the L protein is unstable as a result. To differentiate between these two possibilities the Sendai virus L mutants were tested for their ability to bind to a different viral protein, the C protein, which binds to L at a site distinct from the P binding site (9). VVT7-infected cells were transfected with the gstC plasmid (9) together with P and the wt or mutant L plasmids. Analysis of total protein synthesis in a pulse-label showed that wt L expressed alone or with the P or gstC proteins or all together gave good expression of L (Fig. 3A, lanes 2 to 5). SV 517 and SV519 coexpressed with both the gstC and P proteins, however, were still barely or not detectable (Fig. 3A, lanes 9 and 11) and thus did not appear to be stabilized by gstC. Interestingly, the remaining L mutants synthesized with gstC and P were all better expressed, some even at levels close to that of wt (Fig. 3A), in contrast to their reduced expression with gstP (Fig. 1A). In the analysis of P-L complex formation wt L was cobound to beads only when expressed with gstC or gstC and P (Fig. 3B, lanes 2 to 5) as expected (9). A small amount of the SV 517 but no SV 519 coexpressed with both P and gstC cobound to the beads (Fig. 3B, lanes 9 and 11). The remaining L mutants were all able to bind gstC at close to or better than the level of wt L; still, however, no P binding was observed (Fig. 3B). Apparently the low level of P binding to SV 516 and SV 520 observed previously (Fig. 1B) was below the limit of detection in this assay. These data show that the C binding site on most of these mutant L proteins was intact and binding stabilized the protein, which in turn suggested that the overall structure of these proteins was not disrupted by these mutations. The L mutations, therefore, specifically abolished the P binding site on the L protein.
FIG. 3.
Complex formation between Sendai virus gstC and the L mutants. A549 cells in 35-mm-diameter dishes were infected with VVT7 and transfected with no plasmids (Mock) or with the Sendai virus gstC (0.2 μg), P (1.67 μg), and the indicated wt or mutant L (1.67 μg) plasmids. At 10 h the cells were incubated with Express-35S for 30 min, and cytoplasmic extracts were prepared. Samples were analyzed directly by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (A) or incubated with glutathione-Sepharose beads after which the bound proteins were separated by SDS-PAGE as described previously (B) (9). The positions of the proteins are indicated.
Sendai virus L mutants are defective in RNA synthesis in vitro
The Sendai virus L mutants were then tested for their ability to synthesize various viral RNAs in vitro and in vivo. We predicted that the L mutants which could not bind P would be inactive in all RNA synthesis. For transcription, VVT7-infected cells were transfected with the P and wt or mutant L plasmids and cell extracts were prepared as described previously (7, 8). The extracts were incubated with a wt polymerase-free RNA-NP template in the presence of [α-32P]CTP, and the products were analyzed by gel electrophoresis. The wt L and P proteins gave good transcription, while no mRNA product was detected in the absence of viral proteins (Fig. 4A, lanes 1 and 2). In this overexposure of the gel none of the L mutants gave mRNA synthesis except SV 520, which showed 7% of the activity of wt L (Fig. 4A, lane 9). While both SV 516 and SV 520 gave a small amount of binding to P (Fig. 1), only SV 520 had limited activity. Western analysis of samples showed that P protein expression was similar in all of the extracts (Fig. 4B).
FIG. 4.
In vitro transcription with the Sendai virus L mutants. VVT7-infected A549 cells were transfected with no plasmids (Mock) or with the Sendai virus P (1.5 μg) and the indicated wt or mutant L (0.5 μg) plasmids. (A) Cytoplasmic cell extracts were incubated with polymerase-free wt Sendai virus RNA-NP (1 μg) and [α-32P]CTP (200 μCi/ml) as described previously (7, 8). Total RNA was isolated and separated on an agarose-urea gel. The position of the NP and P transcripts which comigrate is indicated. (B) Samples of the extracts were immunoblotted with α-P antibody. The position of the P protein is indicated. (C) For leader RNA synthesis the extracts were incubated with polymerase-free wt RNA-NP and unlabeled nucleotides, and the leader RNA was detected by Northern analysis with a 32P-labeled oligonucleotide probe complementary to le+ RNA as described previously (7). The position of the le+ product is indicated.
The first product of transcription is actually le+ RNA and not NP mRNA. To study le+ RNA synthesis the infected, transfected extracts were incubated with wt RNA-NP in the absence of radiolabeled nucleotides and the le+ RNA was detected by Northern blotting with a specific oligonucleotide probe. While wt L with P gave good le+ synthesis as well as some longer read-through products, compared to no product in the absence of viral proteins, the majority of the L mutants were unable to synthesize detectable le+ RNA (Fig. 4C). SV 520, however, gave very good le+ synthesis (60% of wt L) (Fig. 4C, lane 9), although mRNA synthesis by this mutant was severely impaired (Fig. 4A). SV 516 had only 4% of the activity of the wt L (Fig. 4C, lane 5), whereas it was unable to synthesize mRNA (Fig. 4A). Since the amount of SV 520 protein in the extract was considerably less than wt L, this protein appears to have better than wt L activity for le+ synthesis; however, it has a defect in initiation at the NP gene. For the majority of the L mutants the lack of binding to P protein correlated with the lack of transcription activity.
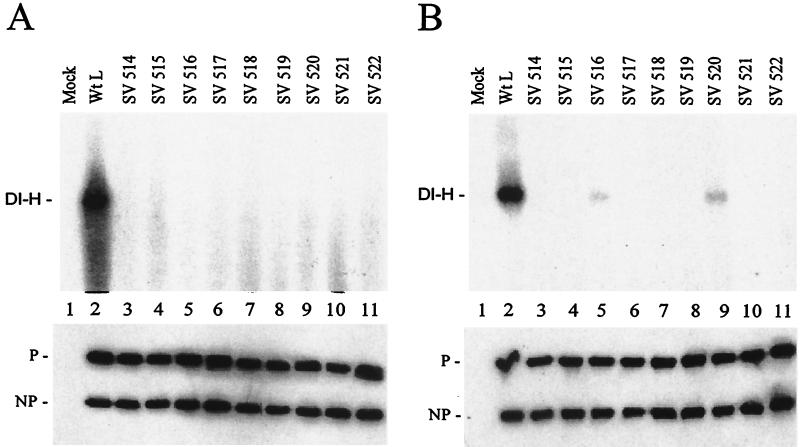
The L mutants were also tested for function in DI-H genome replication under two conditions, in vitro with added template and in vivo with a DI-H clone. For in vitro replication there is a large molar excess of input DI RNA-NP template, so this assay measures the ability of the viral polymerase to carry out a single round of synthesis and encapsidation of genomic length DI RNA. VVT7-infected cells were transfected with the Sendai virus NP, P, and wt or mutant L plasmids. Cell extracts were prepared and incubated with DI-H polymerase-free RNA-NP and [α-32P]CTP as described previously (8). Nuclease-resistant (encapsidated) RNA products were isolated and analyzed by gel electrophoresis. Coexpression of the wt proteins gave good DI-H replication; however, none of the Sendai virus L mutants gave any DI-H synthesis (Fig. 5A). Immunoblot analysis showed that the NP and P proteins were uniformly expressed in the extracts (Fig. 5A, bottom).
FIG. 5.
DI-H replication with the Sendai virus L mutants. (A) A549 cells were infected with VVT7 and transfected with no plasmids (Mock) or the Sendai virus NP, P, and the indicated wt or mutant L plasmids. Cytoplasmic extracts were incubated with polymerase-free DI-H RNA-NP in the presence of [α-32P]CTP, and total RNA was isolated and analyzed on an agarose-urea gel as described previously (8). The position of DI-H RNA is indicated. (B) For in vivo replication infected cells were transfected as above with the addition of pSPDI-H plasmid, and extracts were prepared. The extracts were nuclease treated, and the nuclease-resistant RNA was isolated and then separated on an agarose-urea gel. The RNA was transferred to a nylon membrane, and the blot was probed with a DI-H-specific (+) sense 32P-labeled riboprobe as described previously (8). The position of the DI-H RNA is indicated. (Bottom panels) Samples of the extracts were immunoblotted using α-SV and α-P antibodies. The positions of the proteins are indicated.
The activity of the mutant L proteins in DI-H replication in vivo was also determined. Infected cells were transfected overnight with the plasmids for the viral proteins as well as with the pSPDI-H plasmid which encodes a (+) strand DI-H genome RNA. The DI-H RNA is nonspecifically encapsidated by the NP protein, creating a template which can be replicated by the viral polymerase where multiple rounds of amplification are required to detect product. Cell extracts were prepared, nuclease-resistant RNA was isolated, and the (−) sense DI-H RNA was analyzed by Northern blotting with a (+) sense DI riboprobe. The wt polymerase complex gave good DI-H replication; however, again the majority of the L mutants had no activity (Fig. 5B). Surprisingly, SV 516 and SV 520 did replicate low levels of DI-H RNA (6 and 11%, respectively) (Fig. 5B, lanes 5 and 9), whereas in vitro no replication was detected (Fig. 5A). Western blot analysis showed that the NP and P proteins were uniformly synthesized (Fig. 5B, bottom). It may be that replication is more efficient in vivo because this process requires some cellular protein or structure which is disrupted in the cell extracts used for in vitro replication. In previous studies some, but not all, L mutants in other domains also showed a phenotype similar to that of SV 516 and SV 520, whereby in vitro replication was inhibited but in vivo replication occurred (3, 7). Thus, this in vivo replication stimulation is mutant specific and can occur at sites throughout the L protein. In summary, L mutants which were unable to form a polymerase complex were unable to synthesize any viral RNAs, whereas the two mutants which bound some P gave limited transcription and limited in vivo, but not in vitro, replication. Thus, P binding to L is required for RNA synthesis.
The polymerase complex of Sendai virus is thought to be composed of a homotetramer of the P protein and a monomer of the L protein (5, 19). Recently the crystal structure of the portion of Sendai virus P (aa 320 to 433) containing the oligomerization domain and a portion of the L binding site showed a four-stranded parallel coiled coil (18). Within the L binding portion of this region from aa 408 to 433, studies by Bowman et al. (1) identified both charged and hydrophobic amino acids that were essential for polymerase activity. The charged amino acids, K408A/R409A (mutant P408/9) and E412A/K415A/E416A (mutant P1), which were altered by site-directed mutagenesis, are located on the exterior of the coiled coil. Although neither mutant alone abolished P binding to L, both were deficient in transcription in vitro. The L421A and L425A mutations, which are located on the interior of the coiled coil, gave a similar phenotype. Presumably these charged amino acids on P interact directly with the N terminus of the L subunit, while the hydrophobic mutations may alter the coiled coil to affect the overall P-L interaction. While none of the mutants disrupted binding, they did interfere with polymerase activity, possibly by promoting an incorrect, inactive conformation of the L protein. Perhaps binding was not abolished by individual changes due to the rather rigid structure of P extending from the oligomerization domain into the L binding domain. Further understanding of the P-L interaction will require additional mutagenesis analysis of the required regions of both the P and L proteins and would be facilitated by a structural analysis of this portion of P cocrystallized with the bound N-terminal fragment of L.
Acknowledgments
This work was supported by NIH grant AI14594 (S.A.M.).
REFERENCES
- 1.Bowman, M. C., S. Smallwood, and S. A. Moyer. 1999. Dissection of individual functions of the Sendai virus phosphoprotein in transcription. J. Virol. 73:6474-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chandrika, R., S. M. Horikami, S. Smallwood, and S. A. Moyer. 1995. Mutations in conserved domain I of the Sendai virus L polymerase protein uncouple transcription and replication. Virology 213:352-363. [DOI] [PubMed] [Google Scholar]
- 3.Cortese, C. K., J. A. Feller, and S. A. Moyer. 2000. Mutations in domain V of the Sendai virus L polymerase protein uncouple transcription and replication and differentially affect replication in vitro and in vivo. Virology 277:387-396. [DOI] [PubMed] [Google Scholar]
- 4.Curran, J., R. Boeck, and D. Kolakofsky. 1991. The Sendai virus P gene expresses both an essential protein and an inhibitor of RNA synthesis by shuffling modules via mRNA editing. EMBO J. 10:3079-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curran, J., R. Boeck, N. Lin-Marq, A. Lupas, and D. Kolakofsky. 1995. Paramyxovirus phosphoproteins form homotrimers as determined by an epitope dilution assay, via predicted coiled coils. Virology 214:139-149. [DOI] [PubMed] [Google Scholar]
- 6.Curran, J., T. Pelet, and D. Kolakofsky. 1994. An acidic activation-like domain of the Sendai virus P protein is required for RNA synthesis and encapsidation. Virology 202:875-884. [DOI] [PubMed] [Google Scholar]
- 7.Feller, J. A., S. Smallwood, S. M. Horikami, and S. A. Moyer. 2000. Mutations in conserved domains IV and VI of the large (L) subunit of the Sendai virus RNA polymerase give a spectrum of defective RNA synthesis phenotypes. Virology 269:426-439. [DOI] [PubMed] [Google Scholar]
- 8.Horikami, S. M., J. Curran, D. Kolakofsky, and S. A. Moyer. 1992. Complexes of Sendai virus NP-P and P-L proteins are required for defective interfering particle genome replication in vitro. J. Virol. 66:4901-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horikami, S. M., R. E. Hector, S. Smallwood, and S. A. Moyer. 1997. The Sendai virus C protein binds the L polymerase protein to inhibit viral RNA synthesis. Virology 235:261-270. [DOI] [PubMed] [Google Scholar]
- 10.Horikami, S. M., S. Smallwood, B. Bankamp, and S. A. Moyer. 1994. An amino-proximal domain of the L protein binds to the P protein in the measles virus RNA polymerase complex. Virology 205:540-545. [DOI] [PubMed] [Google Scholar]
- 11.Horikami, S. M., and S. A. Moyer. 1995. Alternative amino acids at a single site in the Sendai virus L protein produce multiple defects in RNA synthesis in vitro. Virology 211:577-582. [DOI] [PubMed] [Google Scholar]
- 12.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 689-724. In D. M. Knipe and P. M. Howley (ed.), Fundamental virology, 4th ed. Lippincott, Williams & Wilkins, Philadelphia, Pa.
- 13.Parks, G. D. 1994. Mapping of a region of the paramyxovirus L protein required for the formation of a stable complex with the viral phosphoprotein P. J. Virol. 68:4862-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poch, O., B. M. Blumberg, L. Bougueleret, and N. Tordo. 1990. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J. Gen. Virol. 71:1153-1162. [DOI] [PubMed] [Google Scholar]
- 15.Sidhu, M. S., J. P. Menonna, S. D. Cook, P. C. Dowling, and S. A. Udem. 1993. Canine distemper virus L gene: sequence and comparison with related viruses. Virology 193:50-65. [DOI] [PubMed] [Google Scholar]
- 16.Smallwood, S., C. D. Easson, J. A. Feller, S. M. Horikami, and S. A. Moyer. 1999. Mutations in conserved domain II of the Large (L) subunit of the Sendai virus RNA polymerase abolish RNA synthesis. Virology 262:375-383. [DOI] [PubMed] [Google Scholar]
- 17.Smallwood, S., K. W. Ryan, and S. A. Moyer. 1994. Deletion analysis defines a carboxyl-proximal region of Sendai virus P protein that binds to the polymerase L protein. Virology 202:154-163. [DOI] [PubMed] [Google Scholar]
- 18.Tarbouriech, N., J. Curran, R. W. H. Ruigrok, and W. P. Burmeister. 2000. Tetrameric coiled coil domain of Sendai virus phosphoprotein. Nat. Struct. Biol. 7:777-781. [DOI] [PubMed] [Google Scholar]
- 19.Tarbouriech, N., J. Curran, C. Ebel, K. W. H. Ruigrok, and W. P. Burmeister. 2000. On the domain structure and the polymerization state of the Sendai virus P protein. Virology 266:99-109. [DOI] [PubMed] [Google Scholar]