Abstract
To persist in the presence of an active immune system, viruses encode proteins that decrease expression of major histocompatibility complex class I molecules by using a variety of mechanisms. For example, murine gamma-2 herpesvirus 68 expresses the K3 protein, which causes the rapid turnover of nascent class I molecules. In this report we show that certain mouse class I alleles are more susceptible than others to K3-mediated down regulation. Prior to their rapid degradation, class I molecules in K3-expressing cells exhibit impaired assembly with β2-microglobulin. Furthermore, K3 is detected predominantly in association with class I molecules lacking assembly with high-affinity peptides, including class I molecules associated with the peptide loading complex TAP/tapasin/calreticulin. The detection of K3 with class I assembly intermediates raises the possibility that molecular chaperones involved in class I assembly are involved in K3-mediated class I regulation.
The outcome of virus infection is affected by the status of the host's immune system, and therefore, to coexist with their hosts, viruses express proteins specifically designed to compromise antiviral host defense mechanisms. These immune evasion strategies improve opportunities for viral infection in naive individuals and facilitate virus persistence in the presence of active immune responses. One target of these viral proteins implicated in immune evasion is antigen presentation to CD8+ T cells (17, 27, 36). More specifically, viruses have evolved diverse and elaborate mechanisms to prevent the expression of class I/peptide complexes on the cell surface and thus block detection of infected cells by cytolytic CD8+ T cells. Recent studies have characterized several interesting and distinct mechanisms whereby viral proteins block class I/peptide expression. These mechanisms include (i) interfering with antigen processing (12, 22), (ii) blocking peptide transport into the endoplasmic reticulum (ER) (1, 10, 18, 49), (iii) dislocation of assembled class I from the ER (21, 25, 46, 47), (iv) retrieval of class I to the ER (2, 5, 20), and (v) targeted degradation of class I from post-ER compartments (31, 35, 39, 51, 52).
Although mechanistically diverse, several of the pathways by which viral proteins impair class I expression share common features. For example, many exploit host cell proteins and normal degradation pathways. In this regard, studies of viral proteins have led to unique insights into class I biogenesis under physiologic as well as pathogenic conditions. Although not well documented to date, a logical target of virus proteins might be ER chaperone proteins implicated in class I assembly. Several molecular interactions associated with class I assembly in the ER have been defined. Prior to assembly with β2-microglobulin (β2m), class I heavy chains are detected in association with calnexin (8, 29). After β2m assembly, the heavy-chain/β2m heterodimers are associated with the complex of TAP/tapasin/calreticulin/ERp57, commonly referred to as the peptide loading complex (34). Once peptide binds to the heavy-chain/β2m complex to complete the assembly, the fully assembled class I heterotrimer dissociates from the peptide loading complex (32). Subsequent to dissociation from the loading complex, class I molecules transit through the Golgi to the cell surface for display of peptides to CD8+ T cells. Although their individual roles are not completely defined, collectively these ER proteins facilitate class I assembly. Thus, viral proteins could block association of ER chaperones with class I and impair full assembly, or virus proteins could use these ER molecules to target class I molecules for early destruction prior to full assembly.
A recent report on the K3 protein of murine gamma herpesvirus 68 (γHV68) suggests that it down regulates class I by a novel pathway and thus is likely to provide insights into class I assembly (44). Stevenson et al. reported that K3 protein is sufficient to down regulate class I expression and abolish CTL recognition (44). Interestingly, K3 was found to inhibit both mouse and human class I molecules, but only in mouse and not human cells. Thus, K3 requires a species-specific interaction with a host protein(s) other than class I. This species-specific protein required for K3-mediated down regulation of class I has not been identified. In addition, Stevenson et al. demonstrated that K3 induces the rapid ER degradation of the mouse class I molecule Db (44). However, no physical association between K3 and class I was demonstrated. In contrast to γHV68 K3, the homologous gene products of Kaposi's sarcoma-associated herpesvirus (KSHV), namely, KSHV K3 and KSHV K5, were reported not to affect assembly and transport of class I (7, 19). Alternatively, they down regulated class I by increasing their cell surface turnover by inducing rapid endocytosis. Thus, the fact that two genetically related and highly homologous K3/5 gene products promote class I down regulation by such dramatically different mechanisms suggests distinct pathways exploiting different host proteins.
In this report we demonstrate that γHV68 K3 down regulates certain mouse alleles more efficiently than others. Analyses of the susceptible class allele, Ld, indicated that K3 is in steady-state association predominantly with class I molecules lacking high-affinity peptide ligands. Furthermore, in the presence of K3, nascent Ld heavy chains exhibit impaired assembly with β2m, an event that could be detected prior to the onset of class I heavy-chain turnover. Surprisingly, K3 was also detected with Ld bound to the peptide loading complex of TAP/tapasin/calreticulin/ERp57, and K3 association did not prevent peptide binding to Ld in cell lysates. The association of K3 with various class I assembly intermediates provides a unique opportunity to investigate the role of ER molecular chaperones in the targeting of class I molecules for destruction by a viral protein.
MATERIALS AND METHODS
Antibodies and peptides.
The monoclonal antibody (MAb) 64-3-7 (immunoglobulin G2b [IgG2b]) is specific for open forms of Ld and other molecules tagged with this epitope (40, 41, 50). MAb 30-5-7 (IgG2a) is specific for folded forms of Ld (23, 40, 41), and MAb 28-14-8 detects Ld independently of α1/α2 conformation (23). MAbs 11-4-1 (30) and 15-5-5 (33) were used for detection of Kk and Dk, respectively, and were obtained from the American Type Culture Collection (Rockville, Md.). MAbs B8-24-3 and B22-249 (13) were used for the detection of Kb and Db, respectively, and were also obtained from the American Type Culture Collection. Armenian hamster MAb 5D3 to mouse tapasin and rabbit antibodies to mouse TAP (503) have been described (14). Antibodies to K3 (3209) were produced in rabbits immunized with the N-terminal γHV68 K3 peptide SMDSTGEFCWICHICHQPEGPLKRFCGCK. For secondary blotting reagents, the following antibodies were used: goat anti-mouse γ2b-biotin (Caltag Laboratories, San Francisco, Calif.), donkey anti-rabbit biotin, and goat anti-Armenian hamster biotin (both from Jackson ImmunoResearch, West Grove, Pa.). The human cytomegalovirus (HCMV) peptide YPHFMPTNL (38) binds Ld, and CW3 (RYLKNGKETL, derived from HLA-Cw3) (26) was used as a nonbinding control. Peptides were synthesized on an Applied Biosystems (Foster City, Calif.) model 432A peptide synthesizer. Purity was assessed by reverse-phase high-pressure liquid chromatography and mass spectrometry.
Cell lines and flow cytometry.
The mouse L-cell line L-Ld (Kk, Dk, Ld) (27) and B6/WT-3 (Kb, Db) (37) were maintained in RPMI 1640 or Dulbecco's modified Eagle medium (Life Technologies, Gaithersburg, Md.). Media were supplemented with 10% bovine calf serum or fetal calf serum (HyClone Laboratories, Logan, Utah) plus 2 mM l-glutamine, 0.1 mM nonessential amino acids, 1.25 mM HEPES, 1 mM sodium pyruvate, and 100 U of penicillin and streptomycin per ml (all from the Tissue Culture Support Center at Washington University School of Medicine). Where appropriate, Geneticin (Life Technologies) was added to a final concentration of 0.6 mg/ml. To analyze cells for surface expression of class I molecules, cells were stained with 20 μl of culture supernatant containing the appropriate MAb by standard methods. Staining was visualized with fluorescein isothiocyanate (ICN, Costa Mesa, Calif.)-conjugated or phycoerythrin (Pharmingen)-conjugated goat anti-mouse IgG. The data collected on a FACSCalibur (Becton Dickinson, San Jose, Calif.) were expressed in the form of histograms as fluorescence values (x axis) plotted against cell numbers (y axis) by using CellQuest Software (Becton Dickinson).
DNA constructs and transfections.
To create a K3 expression vector, the K3 gene was PCR amplified from a γHV68 genomic subclone (coordinates 24733 to 25335 [45]). The PCR product was cloned into pIRESneo (Clontech, Palo Alto, Calif.). DNA sequence analysis of the complete open reading frame confirmed the correct sequence. Transfections were performed with FuGene 6 (Roche Diagnostics, Indianapolis, Ind.). Stable transfectants were cloned by limiting dilution. The eYFP.K3 construct was made by subcloning a SalI/BamHI fragment of K3 into the SalI/BamHI sites of peYFP.C1 (Clontech). It encodes a fusion protein with the N terminus being eYFP and the C terminus being K3 separated by a 29-residue linker (SGLRSRAQASNSAVDLQAAALVIQHEEVS).
Immunoprecipitations and Western blots.
Cells were lysed in 10 mM Tris-buffered saline, pH 7.4 (TBS), containing 1% digitonin (Wako, Richmond, Va.) with 20 mM iodoacetamide (IAA) and 0.2 mM phenylmethylsulfonyl fluoride (Sigma). Saturating amounts of the primary antibody were added to the lysis buffer and subjected to immunoprecipitations as previously described (16). Briefly, lysates were centrifuged to remove cell debris and nuclei, supernatants were added to protein A-Sepharose CL-4B (Amersham Pharmacia, Uppsala Sweden) for 60 min on ice, and protein A-bound material was washed in 0.1% digitonin in TBS with 20 mM IAA. Immunoprecipitates were eluted from protein A by boiling for 5 min in lithium dodecyl sulfate sample buffer (Invitrogen, Carlsbad, Calif.). Samples were electrophoresed on either Tris-glycine or Tris-acetate prepoured polyacrylamide gels (Invitrogen) and transferred to Immobilon-P transfer membranes (Millipore, Bedford, Mass.). After overnight blocking in 10% milk-phosphate-buffered saline (PBS)-0.05% Tween 20, membranes were incubated in a dilution of antibody for 2 h, washed three times with PBS-0.05% Tween 20, and incubated for 1 h with the appropriate biotin-conjugated secondary reagents (described above). Following three washes with PBS-0.05% Tween 20, membranes were incubated for 1 h with streptavidin-conjugated horseradish peroxidase (Zymed, San Francisco, Calif.), washed three times with PBS-0.3% Tween 20, and incubated with enhanced chemiluminescence reagents (Amersham Pharmacia Biotech, Piscataway, N.J.). Western blots were then developed on BioMax-MR film (Eastman Kodak, Rochester, N.Y.).
Pulse-chase experiments.
After a 30-min preincubation in Met/Cys-free medium (Dulbecco's modified Eagle medium with 5% dialyzed fetal calf serum), L-Ld or L-Ld+K3 cells (at ∼20 × 106 cells/ml) were pulse-labeled with Express [35S]Met/Cys labeling mix (Perkin Elmer Life Sciences, Boston, Mass.) at 250 to 300 μCi/ml for 10 min. Cells were then washed extensively, an aliquot was removed for the zero time point, and the remaining cells were placed back in culture at 37°C for various times. In certain experiments, labeling and chase were performed in the presence of lactacystin (Alexis Biochemical, San Diego, Calif.) and N-acetyl-Leu-Leu-Nle (ALLN) (Calbiochem, La Jolla, Calif.). For immunoprecipitations, labeled cells were lysed in 1% NP-40 (Sigma) dissolved in TBS with 20 mM IAA and 5 mM phenylmethylsulfonyl fluoride. Postnuclear lysates were precleared over protein A-Sepharose CL-4B for 30 min on ice. Lysates were then transferred to protein A-Sepharose pellets containing the appropriate prebound MAbs. After binding for 45 min on ice, protein A pellets were washed 4 times with 0.1% NP-40 in TBS, and bound proteins were eluted by boiling in 10 mM Tris-Cl, pH 6.8, with 0.5% sodium dodecyl sulfate (SDS) and 1% 2-mercaptoethanol. Eluates were mixed with an equal volume of 100 mM sodium acetate, pH 5.4, and digested (or mock digested) overnight with 1 mU of endoglycosidase H (Endo H; ICN) that was reconstituted in 50 mM sodium acetate, pH 5.4. Samples were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) on 4 to 20% Tris-glycine gradient gels (Invitrogen). Gels were treated with Amplify (Amersham), dried, and exposed to BioMax-MR film.
RESULTS
Down regulation of class I by γHV-68 protein K3 is allele specific.
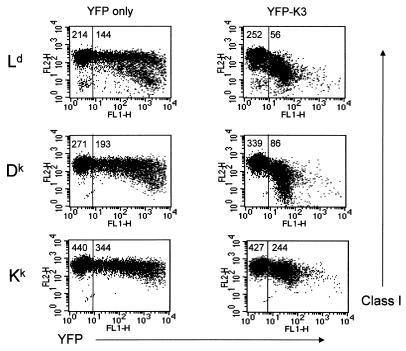
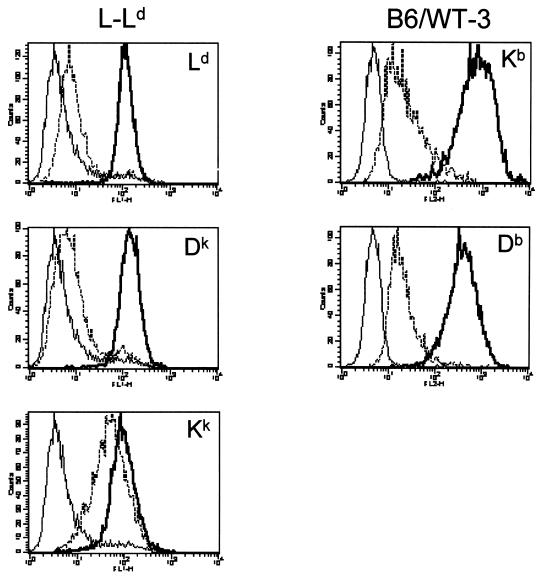
To determine whether different mouse class I alleles are variably affected by K3, we expressed K3 in L-Ld cells. L-Ld is a mouse fibroblast cell line expressing the endogenous Kk and Dk alleles as well as Ld, which was introduced by transfection. To initially test K3-mediated class I regulation, a YFP-K3 construct encoding a fusion protein with YFP linked to the N terminus of K3 was produced for transient-transfection assays. As shown in Fig. 1, transient expression of K3 resulted in a >2-fold reduction of expression of Ld and Dk but only a 29% reduction of Kk expression. This preliminary evidence suggested that class I was down regulated over a range of K3 expression levels and that this regulation was class I allele specific. To confirm and extend these findings, we also produced L-Ld cell lines that stably express K3. As shown in Fig. 2, stable expression of K3 resulted in a 15- to 20-fold reduction in expression of Ld and Dk proteins, whereas only a 2-fold reduction of Kk protein expression was observed. Indeed, in some assays no difference in Kk expression was observed in the presence and absence of K3 (data not shown). These assays provide evidence that Ld and Dk proteins are more susceptible to K3 downregulation than Kk proteins. It is important to note that surface Ld molecules expressed in the presence or absence of K3 were found to be comparably inducible with exogenous peptide and displayed similar levels of peptide occupancy (data not shown). Thus, K3 sharply reduces the level of surface Ld molecules without affecting the overall quality of their peptide loading.
FIG. 1.
Class I allele-specific down regulation by K3. Surface expression of Kk, Dk, and Ld on L-Ld cells after transient transfection with YFP-K3 was assessed. The vertical line through each panel separates transfected from nontransfected cells, and the numbers represent the mean fluorescence of class I expression. Ld, Dk, and Kk molecules were detected with MAbs 30-5-7, 15-5-5, and 11-4-1, respectively.
FIG. 2.
Class I expression in cells with or without stable expression of K3. L-Ld cells and their K3 transfectants and B6/WT-3 cells and their K3 transfectants were analyzed for surface class I expression. Expression of the indicated class I allele with (dashed line) or without (solid thick line) stable expression of K3 is shown in each panel. Background staining is shown with solid thin lines. Ld, Dk, Kk, Kb, and Db molecules were detected with MAbs 30-5-7, 15-5-5, 11-4-1, B8-24-3, and B22-249, respectively.
To further probe the nature of the class I allele-specific regulation by K3, the H2b-derived fibroblast cell line B6/WT-3 was transfected with K3. As shown in Fig. 2, expression of both Kb and Db was sharply reduced in the presence of K3. Importantly, the level of K3 expression in this cell line was comparable to that expressed in the L-Ld+K3 line (data not shown). Thus, of the class I alleles tested thus far, Kk appears to be the most resistant to K3-mediated downregulation. Interestingly, Kk molecules assemble with peptide and transit to the Golgi exceedingly fast compared with other mouse class I molecules (4, 43, 48). This finding suggests a kinetic model, whereby relative resistance to K3 down regulation of alleles such as Kk could be explained by their rapid assembly, thus escaping K3 targeted destruction. Alternatively, structural disparities between class I alleles could also influence targeting by K3.
The K3 protein is associated with Ld heavy chains lacking a high-affinity peptide.
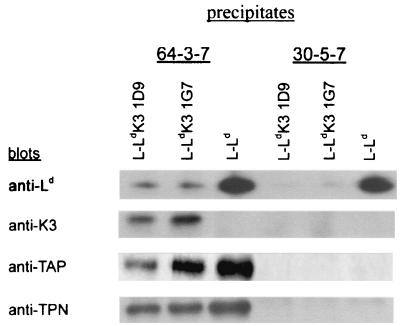
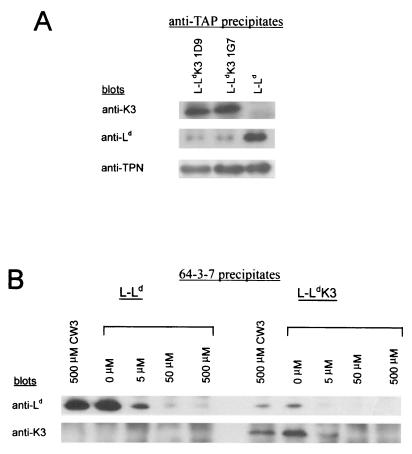
To determine whether K3 is associated with Ld, lysates of L-Ld cells with and without K3 expression were analyzed by immunoprecipitation and Western blotting. Ld was selected for these studies because it is highly susceptible to K3 regulation, and MAbs 30-5-7 and 64-3-7, which distinguish Ld heavy chains with and without a bound high-affinity peptide, respectively, are available (23, 40, 41, 50). As shown in Fig. 3 approximately equal amounts of both conformers of Ld were detected in lysates of L-Ld cells lacking K3 expression. By contrast, in lysates from two independent clones of L-Ld+K3 (1D9 and 1G7), a 6- to 10-fold reduction of 30-5-7+ conformers (with high-affinity peptide) and 64-3-7+ conformers (lacking a high-affinity peptide) was detected (Fig. 3). Thus, K3 expression severely reduces the steady-state level of Ld detected in cell lysates. To determine whether K3 was in physical association with these class I molecules, precipitates of Ld were blotted with a rabbit serum to K3. The results shown in Fig. 3 demonstrate that K3 was readily detected in anti-Ld precipitates in both L-Ld+K3 cell lines. Interestingly, K3 was found to be predominantly associated with open forms of Ld as detected with MAb 64-3-7.
FIG. 3.
K3 displays steady-state association with open forms of class I. Precipitates were formed with MAb 64-3-7 (open Ld conformers) or MAb 30-5-7 (folded Ld conformers). Blotting antibodies were anti-Ld (MAb 64-3-7), anti-γHV68 K3 (3209); anti-TAP (503), and anti-tapasin (MAb 5D3).
The K3 protein impairs β2m assembly and induces the rapid turnover of nascent Ld heavy chains.
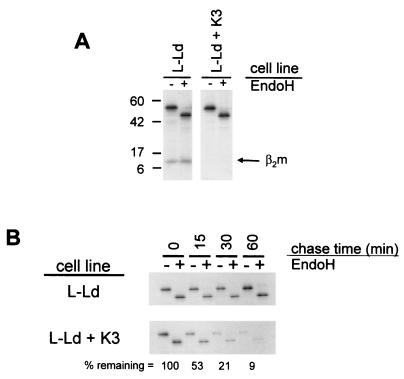
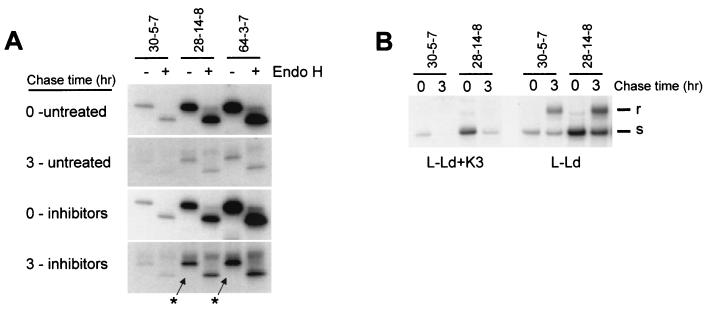
To monitor the effect of K3 on class I assembly and turnover, pulse-chase analyses were performed with L-Ld and L-Ld+K3 cells. Samples were initially compared to determine the amount of assembly with β2m in the presence or absence of K3, since β2m association is one of the earliest events in class I assembly. The comparisons shown in Fig. 4A were from precipitates of cells pulsed for 10 min followed by no chase period (the zero time point). As shown in Fig. 4A, Ld heavy chains in the presence of K3 displayed sharply reduced β2m association, compared with substantial β2m association in the absence of K3. In data not shown, β2m association with Ld was maximal after the 10-min pulse and did not increase at later time points. Furthermore after the 10-min pulse, comparable amounts of Ld heavy chain were detected in both L-Ld and L-Ld+K3 cells (Fig. 4A). Thus, there was no appreciable K3-induced degradation within the first 10 min. The implication of these combined findings is that K3 impairs assembly with β2m prior to inducing degradation of the Ld class I heavy chains.
FIG. 4.
K3 effects on class I assembly and stability. (A) L-Ld and L-Ld+K3 cells were pulse-labeled with [35S]methionine for 10 min. Cell lysates were precipitated with MAb 28-14-8, and precipitates were treated with Endo H and subjected to SDS-PAGE and autoradiography. Results from 28-14-8 precipitations are shown here, since this MAb yields a stronger β2m signal than 64-3-7. However, the same disparity in β2m assembly between L-Ld and L-Ld+K3 cells was also observed with 64-3-7 precipitates (not shown). (B) Pulse-chase analysis of newly synthesized Ld molecules in L-Ld and L-Ld+K3 cells. Cells were pulse-labeled with [35S]methionine for 10 min and then chased for the indicated times. Cell lysates were precipitated with MAb 64-3-7, incubated overnight with or without Endo H, and then separated by SDS-PAGE and exposed to X-ray film. The percent remaining is the fraction of H-chain signal at each time point, relative to time zero, for the L-Ld+K3 samples. Band quantitation was performed with ImageQuant software (Molecular Dynamics, Sunnyvale, Calif.).
To compare the rate of turnover of Ld heavy chains in the presence and absence of K3, L-Ld+K3 and L-Ld cells were pulsed for 10 min and chased for various amounts of time. These comparisons were done on 64-3-7+ forms of Ld, since they are the primary form of Ld detected in the presence of K3 (Fig. 3). Furthermore, in the absence of K3, 64-3-7+ Ld molecules are known to bind β2m prior to peptide (41). As shown in Fig. 4B, Ld turnover (half-life [t1/2]) in the presence of K3 was about 15 min, compared with Ld in the absence of K3, which displayed no detectable turnover during the 1-h time course. As also shown in Fig. 4B, the fast degradation of Ld in the presence of K3 occurred with immature (Endo H-sensitive) forms, indicating that K3 affects ER-resident class I molecules. Indeed, published studies have demonstrated that the rate of oligosaccharide maturation of Ld in the absence of K3 (t1/2) is about 3 h (4, 43). Thus, subsequent to inhibiting assembly (or promoting disassembly) with β2m, K3 induces the rapid turnover of nascent Ld molecules, an event that occurs before peptide binding and prior to their expected transit from the ER.
The K3 protein is associated with the peptide loading complex.
After assembly with β2m and prior to binding high-affinity peptides, class I molecules are detected in association with the peptide loading complex of TAP/tapasin/calreticulin/ERp57 (6, 32). Since the residual Ld molecules detected in the presence of K3 lack evidence of high-affinity peptide binding, we next asked whether K3 might affect the association of Ld with the loading complex. To assess loading complex association, Ld precipitates were blotted with antibodies to TAP and tapasin. As shown in Fig. 3, the Ld molecules expressed in the presence of K3 displayed high levels of TAP/tapasin association. Indeed, the data in Fig. 3 suggest that a higher percentage of Ld molecules is associated with the TAP/tapasin complex in the presence than in the absence of K3. This observation is consistent with K3 preferentially targeting class I molecules before they associate with the peptide loading complex. To determine whether K3 remains associated with Ld while bound to TAP/tapasin, TAP was precipitated and then these precipitates were blotted with antibodies to K3. As shown in Fig. 5A, significant levels of K3 were detected in association with TAP. To determine whether K3 affected the interaction of TAP with tapasin, the anti-TAP precipitates from K3-positive and -negative cells were blotted with antibody to tapasin. As shown in Fig. 5A, K3 expression had little if any affect on TAP/tapasin association or the steady-state level of TAP and tapasin in these cells (data not shown). Also in data not shown, K3 was found to be associated with calreticulin and tapasin. Thus, K3 is detected in association with TAP/tapasin/calreticulin, three components of the class I peptide loading complex. The implication of these findings is that K3 association with open forms of class I does not prevent their binding to the loading complex.
FIG. 5.
(A) K3 displays steady-state association with TAP/tapasin. Precipitates were formed using anti-TAP antibody 503, and blotting antibodies were anti-K3 (3209), anti-Ld (MAb 64-3-7), and anti-tapasin (MAb 5D3). (B) K3 association does not prevent peptide binding to Ld. Cell lysates from L-Ld and L-Ld+K3 were incubated with the indicated dilution of the HCMV peptide (YPHFMPTNL) or the CW3 peptide (RYLKNGKETL) for 2 h on ice prior to precipitation with MAb 64-3-7. Blotting antibodies were anti-Ld (MAb 64-3-7) and anti-K3 (3209).
K3 association does not prevent peptide-induced folding of class I in cell lysates.
Class I molecules normally bind peptide while associated with the peptide loading complex. Thus, the association of K3 with class I while it is bound by TAP/tapasin raises the possibility that K3 association prevents the binding of high-affinity peptides. To test this theory, a high-affinity Ld peptide ligand was added to cell lysates to induce the folding of Ld as detected by the loss of 64-3-7+ conformers. Indeed, previous studies using this in vitro assay have shown that nascent (Endo H-sensitive), β2m-assembled, TAP/tapasin-associated class I molecules undergo preferential peptide-induced folding (6, 15, 24, 28, 40, 41). Thus, this in vitro assay mimics several known features of peptide-induced folding of class I in the ER. To determine whether K3 prevents peptide binding to Ld, peptide was added to cell lysates of L-Ld and L-Ld+K3 cells, and then lysates were precipitated with MAb 64-3-7. For these studies, the HCMV peptide YPHFMPTNL was used, since it is known to bind Ld with a high affinity (38), and the peptide RYLKNGKETL was used as a control, since it does not bind Ld (26, 28). As shown in Fig. 5B, peptide addition resulted in the loss of Ld as detected by MAb 64-3-7, which is specific for the open conformer, in both the presence and absence of K3. Thus, peptide-induced Ld folding was not prevented in K3-expressing cells. To determine whether K3-associated Ld molecules specifically underwent folding, these precipitates were blotted with antibodies to K3. As shown in Fig. 5B, K3-associated 64-3-7+ Ld molecules also disappeared with the addition of peptide. Thus, these data provide evidence that K3 association with class I does not prevent peptide binding.
K3 does not induce the accumulation of deglycosylated intermediates of Ld even in the presence of proteasome inhibitors.
To compare K3-mediated degradation with class I degradation mediated by HCMV proteins US2 and US11, inhibitors of the proteasome were included in a pulse-chase experiment. Previous studies demonstrated that US2 and US11 proteins induce the accumulation of deglycosylated, 40-kDa breakdown intermediates of class I heavy chains in the presence of proteasome inhibitors (46, 47). To determine whether K3 also induces the accumulation of deglycosylated forms of class I, synthesis of Ld was compared in the presence or absence of proteasome inhibitors. For the experiment whose results are shown in Fig. 6A, a mixture of lactacystin, a specific proteasome inhibitor (3, 9), and ALLN, a proteasome inhibitor with broader activity, was used. As shown in Fig. 6A, comparison between the 0- and 3-h chase times demonstrated the expected loss of Ld as detected by MAb 30-5-7+ (folded Ld), MAb 28-14-8 (total Ld), or MAb 64-3-7 (open Ld) in L-Ld+K3 cells. Furthermore, treatment with the mixture of lactacystin and ALLN partially prevented turnover of Ld in the presence of K3. Similar partial reversals of K3-mediated Ld turnover were also observed when cells were treated with only lactacystin (data not shown). Thus, K3 induced degradation of class I appears to be mediated, at least in part, by the proteasome. More conclusively, however, no deglycosylated Ld heavy chains were detected in the presence or absence of proteasome inhibitors, even after 3 h (Fig. 6A). Therefore, unlike HCMV proteins US2 and US11, γHV68 protein K3 does not induce the accumulation of deglycosylated heavy chains in the presence of proteasome inhibitors. Indeed, after 3 h Ld molecules remained predominantly Endo H sensitive in the presence of K3 (Fig. 6A). In comparison, Ld molecules synthesized in the absence of K3 attained about 50% Endo H-resistant forms after 3 h (Fig. 6B), consistent with their previously reported ER-to-Golgi transit times (4, 43). Thus, in the presence of K3, most remaining Ld molecules, including ones that accumulate in the presence of proteasome inhibitors, retain high levels of mannose glycans, indicating that within 3 h they do not transit to the Golgi and are not deglycosylated.
FIG. 6.
Effect of K3 on oligosaccharide maturation of Ld after 3 h in the presence or absence of proteasome inhibitors. (A) L-Ld+K3 cells were pulse-labeled for 10 min and then chased for 3 h. For proteasome inhibitor-treated samples, cells were preincubated in 80 μM lactacystin plus 250 μM ALLN for 30 min prior to labeling and then chased in the continuous presence of inhibitors. This dose of inhibitors was determined to be optimal (data not shown). Cell lysates were then precipitated with the indicated MAb and digested or mock digested with Endo H. Arrows indicate expected migration of deglycosylated Ld heavy chains. (B) L-Ld and L-Ld+K3 cells were pulse-labeled for 10 min and then chased for 3 h. After immunoprecipitation, all samples were treated with Endo H prior to SDS-PAGE.
DISCUSSION
We demonstrate here that γHV68 protein K3 targets Ld class I heavy chains and induces their rapid degradation (t1/2 = 15 min). This observation is in complete agreement with previous observations of K3-mediated turnover of Db molecules studied by Stevenson and colleagues (44). Here we also show that prior to degradation, Ld heavy chains lack appreciable β2m assembly in the presence of K3 but display a high level of β2m assembly in the absence of K3. Thus, β2m disassembly occurs prior to detectable degradation of heavy chains. This block of β2m assembly is dramatic and could be explained by either preventing assembly in the ER or removing Ld from the ER prior to assembly. If the former model is correct, then K3 could either sterically block or dislodge β2m from the heavy chain. Alternatively, K3 might block heavy chain interaction with calnexin or calreticulin, ER chaperones implicated in oligomeric assembly of glycoproteins (16). However, if assembly is prevented by K3 purging of heavy chains from the ER, then this event must occur very rapidly, because β2m assembles with Ld in less than 10 min. Interestingly, these features of γHV68 K3-mediated degradation are similar to those of HLA class I regulation mediated by the US2 protein of HCMV (21, 47). Indeed, in comparison with previously described viral proteins, the mechanism by which γHV68 K3 downregulates class I would appear to be most similar to that of the US2 protein of HCMV.
US2, like K3, induces the rapid turnover of class I heavy chains, and in the presence of US2, class I heavy chains display impaired β2m assembly (21, 47). By a mechanism commonly referred to as dislocation, US2 removes class I molecules from the ER to the cytosol. Recently, dislocation was shown to involve the Sec61 complex of proteins, implying that the same complex that translocates nascent proteins from the cytosol to the ER appears to be involved in US2-induced dislocation (47). Furthermore after arrival in the cytosol both the US2 protein and class I heavy chains are deglycosylated, where they accumulate in the presence of proteasome inhibitors. Thus, US2-targeted degradation occurs in the cytosol and is mediated by N-glycanase and proteasome. Interestingly, US2 protein in the ER is detected in physical association with fully assembled class I (W6/32+) heterotrimers (47). Indeed, fully assembled HLA-A2 heterotrimers were used to refold Escherichia coli-expressed US2 protein to obtain cocrystals (11). Analyses of these cocrystals revealed that US2 bound to the class I heavy chain at a junction between the peptide binding region and the α3 domain, a unique interaction site distinct from known ER chaperone binding sites. Furthermore, US2 binding did not significantly alter HLA-A2 conformation, suggesting that it may be an adapter protein that requires other, yet-to-be-defined host proteins involved in the dislocation pathway (11). Although we do not know whether K3 also engages a dislocation pathway, distinctions between K3- and HCMV US2-mediated regulation of class I are noteworthy and suggest important mechanistic differences.
The structural difference between US2 and K3 proteins suggests different modes of interaction with class I molecules.
US2 is a type I, transmembrane, 24-kDa glycoprotein, with N-linked sugars suggesting predominant ER residence (21, 47). Furthermore, the predicted ER luminal domain (residues 1 to 164 of a total of 199) was found to form an Ig-like fold of seven β-strands that directly binds to the fully assembled class I molecule (11). By comparison, γHV68 K3 is a 24-kDa protein predicted to transect the membrane twice, with an overall domain structure unlike that of US2 but very similar to that of KSHV proteins K3 and K5. The γHV68 K3 and the KSHV K3/K5 homologues each have N termini containing a highly conserved C4HC3 zinc finger-like motif, followed by two predicted transmembrane segments connected by a very short (about 16-residue) domain. The C-terminal domains of these viral K3/K5 homologues contain no apparent structural motifs and vary from one another considerably. Furthermore, it has been predicted that the C- and N-terminal K3/K5 domains extend into the cytosol, whereas the short sequence between the transmembrane segments is in the ER lumen. The length of K3 luminal domain would preclude its interaction with the ectodomain of class I similar to US2. In support of this conclusion, we have introduced an epitope tag (of nine additional residues) into the lumen domain of K3 without disrupting its ability to down regulate class I expression (unpublished observation). Alternatively, the K3 protein could interact with class I through its transmembrane or cytosolic domains, or K3 could bind to class I through an intermediary host protein. In any case K3 is very likely to interact with class I in a manner distinct from that defined for US2 (11).
The forms of class I associated with K3 and US2 appear to be different.
Glycosylated forms of US2 are detected predominantly in complex with fully assembled class I heterotrimers in the ER (47). However, US2 is also associated with the Sec61 complex, suggesting that US2 may also bind nascent class I heavy chains and reverse the process by which they are translocated into the ER. Thus, US2 appears to associate with both incompletely and fully assembled class I molecules. By comparison, we show here that K3 is detected in steady-state association with predominantly class I heavy chains lacking peptide or β2m, and K3 is clearly associated with class I molecules bound to TAP and other members of the peptide loading complex. Unfortunately, there are no reports of whether US2-bound class I molecules are detected in association with TAP. However, published results thus far indicate that both US2 and K3 associate with class I molecules in multiple stages of assembly. An unresolved question with both US2 and K3 is that of which class I assembly intermediate(s) is the primary target for destruction. Interestingly, human class I molecules transition through a fully assembled (W6/32+) form before US2-induced degradation (47). By contrast, incompletely assembled Ld (64-3-7+) molecules disappear in K3-positive cells during a time frame when no loss of 64-3-7+ Ld occurs in K3-negative cells (Fig. 4). Thus, K3 appears to be capable of directly targeting incompletely assembled Ld. However, it is important to note that the few folded (30-5-7+) Ld molecules expressed in the presence of K3 are turned over more quickly than folded Ld molecules in the absence of K3 (Fig. 6). Thus, K3 may be capable of also targeting fully assembled class I molecules.
At later time points, deglycosylated forms of US2 are detected in association with deglycosylated free class I heavy chains which accumulate and in the presence of proteasome inhibitors (47). By contrast, we detected no deglycosylated class I molecules in cells expressing K3 as monitored in steady-state comparisons, in pulse-chase experiments, or when cells were treated with proteasome inhibitors (Fig. 3 and 6). Thus, there are potential differences between the US2 and K3 pathways in terms of both the class I interaction in the ER and the targeted degradation in the cytosol. The molecular basis of these differences and the question of whether species-specific factors (human versus mouse) are involved warrant future investigation.
The association of K3 with TAP and the peptide loading complex is intriguing and raises questions of functional significance.
Our data indicate that K3 does not block association of class I with TAP/tapasin as a mechanism to prevent class I assembly. Alternatively, K3 could exploit TAP/tapasin as a means to target class I molecules for destruction. However, K3 expression does not result in the rapid turnover of TAP/tapasin (Fig. 3 and 5). Therefore, K3 does not appear to interact directly with TAP/tapasin, suggesting that K3 can remain bound to class I while it is associated with TAP/tapasin. Association of K3 with TAP/tapasin is surprising in light of the fact that β2m assembly is a prerequisite for optimal heavy-chain association with TAP/tapasin (42) and K3 shows predominant association with β2m-free heavy chains. Thus, we suspect that the class I/K3 complexes associated with the peptide loading complex represent the few class I molecules that assemble with β2m.
In summary, we show here that K3 is predominantly detected in association with incompletely assembled class I molecules, including ones bound to TAP and other members of the peptide loading complex. These findings raise the possibility that K3 may block interaction of class I heavy chains with certain molecular chaperones to impair their assembly with β2m or that K3 may use molecular chaperones to help target incompletely assembled class I molecules for destruction.
ADDENDUM IN PROOF
J. M. Boname and P. G. Stevenson (Immunity 15:627-636, 2001) recently reported that the K3 protein of γHV68 associates with newly synthesized Db molecules, resulting in ubiquitination of the class I heavy chain.
Acknowledgments
The first three authors contributed equally to this study.
This work was supported by NIH grants AI16789, AI46553, AI42793, AI07163, AI01498, AI45019, HL60090, and CA74730.
We thank Eva-Marie Wormstall for help with production of antibody to K3 protein. Susan Harris helped with computer graphics.
REFERENCES
- 1.Ahn, K., A. Gruhler, B. Galocha, T. R. Jones, E. J. H. J. Wietz, H. L. Ploegh, P. A. Peterson, Y. Yang, and K. Früh. 1997. The ER-luminal domain of the HCMV glycoprotein US6 inhibits peptide translocation by TAP. Immunity 6:613-621. [DOI] [PubMed] [Google Scholar]
- 2.Andersson, M., S. Paabo, T. Nilsson, and P. A. Peterson. 1985. Impaired intracellular transport of class I MHC antigens as a possible means for adenoviruses to evade immune surveillance. Cell 43:215-222. [DOI] [PubMed] [Google Scholar]
- 3.Bai, A., and J. Forman. 1997. The effect of proteasome inhibitor lactacystin on the presentation of TAP-dependent and TAP-independent peptide epitopes by class I molecules. J. Immunol. 159:2139-2146. [PubMed] [Google Scholar]
- 4.Beck, J. C., T. H. Hansen, S. E. Cullen, and D. R. Lee. 1986. Slower processing, weaker β2-m association, and lower surface expression of H-2Ld are influenced by its amino terminus. J. Immunol. 137:916-923. [PubMed] [Google Scholar]
- 5.Burgert, H.-G., and S. Kvist. 1985. An adenovirus type 2 glycoprotein blocks cell surface expression of human histocompatibility class I antigens. Cell 41:987-997. [DOI] [PubMed] [Google Scholar]
- 6.Carreno, B. M., J. C. Solheim, M. Harris, I. Stroynowski, J. M. Connolly, and T. H. Hansen. 1995. TAP associates with a unique class I conformation, whereas calnexin associates with multiple class I forms in mouse and man. J. Immunol. 155:4726-4733. [PubMed] [Google Scholar]
- 7.Coscoy, L., and D. Ganem. 2000. Kaposi's sarcoma-associate herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc. Natl. Acad. Sci. USA 97:8051-8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Degan, E., and D. B. Williams. 1991. Participation of novel 88-kD protein in the biogenesis of murine class I histocompatibility molecules. J. Cell Biol. 112:1099-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fenteany, G., R. F. Standaert, W. S. Lane, S. Choi, E. J. Corey, and S. L. Schreiber. 1995. Inhibition of proteasome activities and subunit-specific modification by lactacystin. Science 268:726-731. [DOI] [PubMed] [Google Scholar]
- 10.Früh, K., K. Ahn, H. Djaballah, P. Sempé, P. M. van Endert, R. Tampé, P. A. Peterson, and Y. Yang. 1995. A viral inhibitor of peptide transporters for antigen presentation. Nature 375:415-418. [DOI] [PubMed] [Google Scholar]
- 11.Gewurz, B. E., R. Gaudet, D. Tortorella, E. W. Wang, H. L. Ploegh, and D. C. Wiley. 2001. Antigen presentation subverted: structure of the human cytomegalovirus protein US2 bound to the class I molecule HLA-A2. Proc. Natl. Acad. Sci. USA 98:6794-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilbert, M. J., S. R. Riddeil, B. Plachter, and P. D. Greenberg. 1996. Cytomegalovirus selectively blocks antigen processing and presentation of its immediate-early gene product. Nature 383:720-722. [DOI] [PubMed] [Google Scholar]
- 13.Hammerling, G. J., U. Hammerling, and H. Lemke. 1979. Isolation of monoclonal antibodies against Ia and H-2 antigens. Immunogenetics 8:433-445. [Google Scholar]
- 14.Harris, M. R., L. Lybarger, Y. Y. L. Yu, N. B. Myers, and T. H. Hansen. 2001. Association of ERp57 with mouse MHC class I molecules is tapasin dependent and mimics that of calreticulin and not calnexin. J. Immunol. 166:6686-6692. [DOI] [PubMed] [Google Scholar]
- 15.Harris, M. R., L. Lybarger, N. B. Myers, C. Hilbert, J. C. Solheim, T. H. Hansen, and Y. Y. L. Yu. 2001. Interactions of HLA-B27 with the peptide loading complex as revealed by heavy chain mutations. Int. Immunol. 13:1275-1282. [DOI] [PubMed] [Google Scholar]
- 16.Hebert, D. H., B. Foellmer, and A. Helenius. 1995. Calnexin and calreticulin promote folding, delay oligomerization and suppress degradation of influenza hemagglutinin in microsomes. EMBO J. 12:2961-2968. [PMC free article] [PubMed] [Google Scholar]
- 17.Hengel, H., W. Brune, and U. H. Koszinowski. 1998. Immune evasion by cytomegalovirus survival strategies of a highly adapted opportunist. Trends Microbiol. 6:190-197. [DOI] [PubMed] [Google Scholar]
- 18.Hill, A., P. Jugovic, I. York, G. Russ, J. Bennink, J. Yewdell, H. Ploegh, and D. Johnson. 1995. Herpes simplex virus turns off the TAP to evade host immunity. Nature 375:411-415. [DOI] [PubMed] [Google Scholar]
- 19.Ishido, S., C. Wang, B.-S. Lee, G. B. Cohen, and J. U. Jung. 2000. Downregulation of major histocompatibility complex class I molecules by Kaposi's sarcoma-associated herpesvirus K3 and K5 proteins. J. Virol. 74:5300-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones, T. R., E. J. H. J. Wiertz, L. Sun, K. N. Fish, J. A. Nelson, and H. L. Ploegh. 1996. Human cytomegalovirus US3 impairs transport and maturation of the major histocompatibility complex class I heavy chains. Proc. Natl. Acad. Sci. USA 93:11327-11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones, T. R., and L. Sun. 1997. Human cytomegalovirus US2 destabilizes major histocompatibility complex class I heavy chains. J. Virol. 71:2970-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levitskaya, J., A. Sharipo, A. Leonchiks, A. Ciechanover, and M. G. Masucci. 1997. Inhibition of ubiquitin/proteasome-dependent protein degradation by the Gly-Ala repeat domain of the Epstein-Barr virus nuclear antigen 1. Proc. Natl. Acad. Sci. USA 94:12616-12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lie, W.-R., N. B. Myers, J. M. Connolly, J. Gorka, D. R. Lee, and T. H. Hansen. 1991. The specific binding of peptide ligand to Ld class I major histocompatibility complex molecules determines their antigen structure. J. Exp. Med. 173:449-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lybarger, L., Y. Y. L. Yu, T. Chun, C.-R. Wang, A. G. Grandea III, L. Van Kaer, and T. H. Hansen. 2001. Tapasin enhances peptide-induced expression of H2-M3 molecules, but is not required for the retention of open conformers. J. Immunol. 167:2097-2105. [DOI] [PubMed] [Google Scholar]
- 25.Machold, R. P., E. J. H, J. Wiertz, T. R. Jones, and H. L. Ploegh. 1997. The HCMV gene products US11 and US2 differ in their ability to attack allelic forms of murine major histocompatibility complex (MHC) class I heavy chains. J. Exp. Med. 185:363-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maryanski, J. L., P. Pala, G. Gorradin, B. R. Jordan, and J. C. Cerottini. 1986. H-2 restricted cytolytic T cells specific for HLA can recognize a synthetic HLA peptide. Nature 324:578-579. [DOI] [PubMed] [Google Scholar]
- 27.Miller, D. M., and D. D. Sedmak. 1999. Viral effects on antigen processing. Curr. Opin. Immunol. 11:94-99. [DOI] [PubMed] [Google Scholar]
- 28.Myers, N. B., M. R. Harris, J. M. Connolly, L. Lybarger, Y. Y. L. Yu, and T. H. Hansen. 2000. Kb, Kd, and Ld molecules share common tapasin dependencies as determined using a novel epitope tag. J. Immunol. 165:5656-5665. [DOI] [PubMed] [Google Scholar]
- 29.Nossner, E., and P. Parham. 1995. Species-specific differences in chaperone interaction of human and mouse MHC class I molecules. J. Exp. Med. 181:327-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oi, V. T., P. P. Jones, J. W. Goding, and L. A. Herzenberg. 1978. Properties of monoclonal antibodies to mouse Ig allotypes, H-2 and Ia antigen. Curr. Top. Microbiol. Immunol. 81:115-129. [DOI] [PubMed] [Google Scholar]
- 31.Oldridge, J., and M. Marsh. 1998. Nef—an adaptor adaptor? Trends Cell Biol. 8:302-305. [DOI] [PubMed] [Google Scholar]
- 32.Ortman, B., M. J. Androlewicz, and P. Cresswell. 1994. MHC class I/β2-microglobulin complexes associate with TAP transporters before peptide binding. Nature 368:864-867. [DOI] [PubMed] [Google Scholar]
- 33.Ozato, K., N. Mayer, and D. H. Sachs. 1980. Hybridoma cell lines secreting monoclonal antibodies to mouse H-2 and Ia antigens. J. Immunol. 124:533-540. [PubMed] [Google Scholar]
- 34.Pamer, E., and P. Cresswell. 1998. Mechanisms of MHC class I-restricted antigen processing. Annu. Rev. Immunol 16:323-358. [DOI] [PubMed] [Google Scholar]
- 35.Piguet, V., L. Wan, C. Borel, A. Mangasarian, N. Demaurex, G. Thomas, and D. Trono. 2000. HIV-1 Nef protein binds to the cellular protein PACS-1 to downregulate class I major histocompatibility complexes. Nat. Cell Biol. 2:163-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ploegh, H. L. 1998. Viral strategies of immune evasion. Science 280:248-253. [DOI] [PubMed] [Google Scholar]
- 37.Pretell, J., R. S. Greenfield, and S. S. Tevethia. 1979. Biology of simian virus 40 (SV40) transplantation antigen (TrAg). V. In vitro demonstration of SV40 TrAg in SV40 infected nonpermissive mouse cells by the lymphocyte mediated cytotoxicity assay. Virology 97:32-41. [DOI] [PubMed] [Google Scholar]
- 38.Reddehase, M. J., J. B. Rothbard, and U. H. Koszinowski. 1989. A pentapeptide as minimal antigenic determinant for MHC class I-restricted T lymphocytes. Nature 337:651-653. [DOI] [PubMed] [Google Scholar]
- 39.Schwartz, O., V. Maréchal, S. La Gall, F. Lemonnier, and J.-M. Heard. 1996. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat. Med. 2:338-342. [DOI] [PubMed] [Google Scholar]
- 40.Smith, J. D., N. B. Myers, J. Gorka, and T. H. Hansen. 1992. Disparate interaction of peptide ligand with nascent versus mature class I major histocompatibility complex molecules: comparisons of peptide binding to alternative forms of Ld in cell lysates and at the cell surface. J. Exp. Med. 175:191-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, J. D., N. B. Myers, J. Gorka, and T. H. Hansen. 1993. Model for the in vivo assembly of nascent Ld class I molecules and for the expression of unfolded Ld molecules at the cell surface. J. Exp. Med. 178:2035-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Solheim, J. C., M. R. Harris, C. S. Kindle, and T. H. Hansen. 1997. Prominence of β2-microglobulin, class I heavy chain conformation, and tapasin in the interactions of class I heavy chain with calreticulin and TAP. J. Immunol. 158:2236-2241. [PubMed] [Google Scholar]
- 43.Spiliotis, E. T., H. Manley, M. Osorio, C. Zuniga, and M. Edidin. 2001. Selective export of MHC class I molecules from the ER after their dissociation from TAP. Immunity 13:841-851. [DOI] [PubMed] [Google Scholar]
- 44.Stevenson, P. G., S. Efstathiou, P. C. Doherty, and P. J. Lehner. 2000. Inhibition of MHC class I-restricted antigen presentation by γ2-herpesviruses. Proc. Natl. Acad. Sci. USA 97:8455-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Virgin, H. W., IV, P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wiertz, E. J. H. J., T. R. Jones, L. Sun, M. Bogyo, H. J. Geuze, and H. L. Ploegh. 1996. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84:769-779. [DOI] [PubMed] [Google Scholar]
- 47.Wiertz, E. J. H. J., D. Tortorella, M. Bogyo, J. Yu, W. Mothes, T. R. Jones, T. A. Rapoport, and H. L. Ploegh. 1996. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 384:432-438. [DOI] [PubMed] [Google Scholar]
- 48.Williams, D. B., S. J. Swiedler, and G. W. Hart. 1985. Intracellular transport of membrane glycoproteins: two closely related histocompatibility antigens differ in their rates of transit to the cell surface. J. Cell Biol. 101:725-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.York, I. A., C. Roop, D. W. Andrews, S. R. Riddell, F. L. Graham, and D. C. Johnson. 1994. A cytosolic herpes simplex virus protein inhibits antigen presentation to CD8+ T lymphocytes. Cell 77:525-535. [DOI] [PubMed] [Google Scholar]
- 50.Yu, Y. Y. L., N. B. Myers, C. M. Hilbert, M. R. Harris, G. K. Balendiran, and T. H. Hansen. 1999. Definition and transfer of a serological epitope specific for peptide empty forms of MHC class I. Int. Immunol. 11:1897-1905. [DOI] [PubMed] [Google Scholar]
- 51.Ziegler, H., R. Thäle, P. Lucin, W. Murani, T. Flohr, H. Hengel, H. Farrell, W. Rawlinson, and U. H. Koszinowski. 1997. A mouse cytomegalovirus glycoprotein retains MHC class I complexes in the ERGIC/cis-Golgi compartments. Immunity 6:57-66. [DOI] [PubMed] [Google Scholar]
- 52.Ziegler, H., W. Muranyi, H.-G. Burgert, E. Kremmer, and U. H. Koszinowski. 2000. The luminal part of the murine cytomegalovirus glycoprotein gp40 catalyzes the retention of MHC class I molecules. EMBO J. 19:870-881. [DOI] [PMC free article] [PubMed] [Google Scholar]