Abstract
African swine fever virus (ASFV) polyprotein pp220, encoded by the CP2475L gene, is an N-myristoylated precursor polypeptide that, after proteolytic processing, gives rise to the major structural proteins p150, p37, p34, and p14. These proteins localize at the core shell, a matrix-like virus domain placed between the DNA-containing nucleoid and the inner envelope. In this study, we have examined the role of polyprotein pp220 in virus morphogenesis by means of an ASFV recombinant, v220i, containing an inducible copy of the CP2475L gene regulated by the Escherichia coli repressor-operator system. Under conditions that repress pp220 expression, the virus yield of v220i was about 2.6 log units lower than that of the parental virus or of the recombinant grown under permissive conditions. Electron microscopy revealed that pp220 repression leads to the assembly of icosahedral particles virtually devoid of the core structure. Analysis of recombinant v220i by immunoelectron microscopy, immunoblotting, and DNA hybridization showed that mutant particles essentially lack, besides the pp220-derived products, a number of major core proteins as well as the viral DNA. On the other hand, transient expression of the CP2475L gene in COS cells showed that polyprotein pp220 assembles into electron-dense membrane-bound coats, whereas a mutant nonmyristoylated version of pp220 does not associate with cellular membranes but forms large cytoplasmic aggregates. Together, these findings indicate that polyprotein pp220 is essential for the core assembly and suggest that its myristoyl moiety may function as a membrane-anchoring signal to bind the developing core shell to the inner viral envelope.
African swine fever virus (ASFV), a large enveloped-DNA-containing virus, is the only member of the new family Asfarviridae (18, 19, 38, 44). ASFV infects soft ticks of the Ornithodoros genus and different species of suids and is responsible for a highly lethal disease of domestic pigs. ASFV is unique among deoxyviruses in that it resembles the poxviruses in its genome structure but is morphologically similar to the iridoviruses (38, 44). The viral genome, a double-stranded DNA molecule of 170 to190 kbp, encodes more than 150 polypeptides, including structural proteins and a variety of enzymes involved in DNA replication and repair, gene transcription, and protein modification. Also, the ASFV genome encodes a number of proteins potentially involved in the modulation of the virus-host interaction (38, 43, 45).
The virus particle contains about 50 proteins (22) and consists of several concentric domains with an overall icosahedral shape and an average diameter of 200 nm (3, 4, 14). The viral core is composed of two domains: a central DNA-containing nucleoid and a surrounding thick protein coat referred to as the core shell. The core is enwrapped by an inner lipid envelope and an icosahedral protein capsid. Extracellular particles possess an additional envelope derived from the plasma membrane (9). ASFV particles are formed within cytoplasmic viral factories (3, 9, 10, 33) from precursor membranous structures (3), which are thought to be cisternal structures derived from the endoplasmic reticulum (ER) (4, 37). Morphological evidence indicates that viral membranes become icosahedral particles by the progressive construction of the outer capsid layer, which is composed mainly of viral protein p72 (3, 24). Biochemical data also suggest that protein p72 is assembled in a time-dependent fashion into large membrane-bound complexes that may correspond to capsid-like structures (17). Concomitantly, the developing particles engulf the core material through a complex and poorly understood process that probably begins with the construction of the core shell underneath the viral envelope (3, 37). Subsequently, the viral DNA and nucleoproteins would be encapsulated and finally condensed into the characteristic electron-dense nucleoid (3, 11). The resulting intracellular mature virions are infectious (6), although their relevance for in vivo infection has not yet been established. A fraction of the intracellular particles reach the plasma membrane, likely by a microtubule-mediated transport (2, 15), and are released by budding (9) to give rise to the infectious extracellular enveloped virions.
One of the most striking features of ASFV gene expression is the synthesis of polyprotein precursors to produce several major structural components. The largest ASFV gene, CP2475L, encodes polyprotein pp220, an N-myristoylated polypeptide that after proteolytic processing gives rise to proteins p150, p37, p34, and p14 (40). These proteins are present in equimolecular amounts within the core shell of the mature virions and account for about one-fourth of their total protein mass (3). On the other hand, the major proteins p35 and p15 are processing products of the higher-molecular-weight precursor pp62, encoded by gene CP530R (42). The two polyproteins are expressed late after infection and processed through an ordered cascade of proteolytic events that take place after the second glycine of the consensus motif Gly-Gly-X (31, 40, 42). We have recently shown that the viral protein pS273R is a cysteine proteinase responsible for polyprotein processing (5). Interestingly, ASFV proteinase localizes within the core region of the assembling virions and within the core shell of the mature particles. Together, these data indicate that polyprotein expression and processing must be relevant for core formation.
This report analyzes the role of polyprotein pp220 in ASFV morphogenesis. We describe the construction and properties of a conditional lethal mutant of ASFV with an inducible copy of gene CP2475L. The results indicate that polyprotein pp220 is crucial for the assembly and envelopment of the core shell and for the subsequent steps of core formation, including DNA encapsidation and nucleoid maturation. We also show that precursor pp220, when expressed alone, assembles into large membrane-bound coats reminiscent of the core shell and that N myristoylation is required for its anchoring to membranes.
MATERIALS AND METHODS
Cells and viruses.
Vero and COS-7 cells were obtained from the American Type Culture Collection and grown in Dulbecco's modified Eagle's medium (DMEM) containing 10 or 5% fetal calf serum (FCS), respectively. The ASFV strain BA71V, adapted to grow in Vero cells, and vGUSREP, a BA71V-derived recombinant that expresses the Escherichia coli lac repressor, have already been described (20, 24). The recombinant vaccinia virus vTF7-3 expressing bacteriophage T7 RNA polymerase (23) was kindly provided by Bernard Moss.
Antibodies.
The monospecific rabbit polyclonal sera against the structural proteins p150, p37/p14, p34, p35, p15, pS273R, pE120R, p72, and p12 and the mouse monoclonal antibody 17K.G12 against the structural protein p17 have been described previously (5, 6, 7, 32, 39, 40, 42). The monoclonal antibody specific for single- and double-stranded DNAs (MAB3032) was purchased from Chemicon International. To prepare antibodies against proteins p10 (34) and pA104R (8), the complete open reading frames (ORFs) K78R and A104R, respectively, were expressed in E. coli and purified using the His-Patch ThioFusion expression system according to the procedure of the manufacturer (Invitrogen BV). Antibodies against the purified soluble recombinant proteins were raised in rabbits.
Plasmid construction.
The intermediate transfer vectors pIND1 and pIND2, designed to allow inducible expression of a target gene after homologous recombination with virus vGUSREP, have been reported previously (6). They contain a cassette formed by the viral inducible promoter p72.I, the lacZ gene under control of the strong late promoter p72 (24), and two multiple cloning sites to allow the cloning of the target gene and the corresponding upstream and downstream flanking sequences.
A synthetic DNA fragment of 1,045 bp, which contains the nucleotide sequence from position −1079 to −55 relative to the translation initiation codon of the CP2745L gene, was obtained by PCR using BA71V genomic DNA as a template and the oligonucleotides 5′-TATGGTACCAATCATATAAGAATAAC and 5′-CGGCGGCCGCGACCAGGACTGCTTCTC, which contain KpnI and NotI restriction sites (underlined) at their respective 5′ ends. Plasmids pIND1.pp220.Fl and pIND2.pp220.Fl were generated by inserting the KpnI- and NotI-digested PCR fragment into the KpnI- and NotI-linearized plasmids pIND1 and pIND2, respectively. An 8-kb XbaI DNA fragment containing the complete CP2745L ORF was excised from pGEM-CP2475L (5) and inserted into the XbaI sites of plasmids pIND1.pp220.Fl and pIND2.pp220.Fl to obtain the final transfer vectors pIND1.pp220 and pIND2.pp220, respectively. These vectors differ in the direction of transcription of the lacZ gene with respect to the inducible promoter.
Generation of recombinant virus v220i.
Recombinant viruses were generated essentially as previously described (36) with minor modifications. Briefly, Vero cells were infected with virus vGUSREP (24) and transfected with plasmid pIND1.pp220 or pIND2.pp220 in the presence of different concentrations of IPTG (isopropyl-β-d-thiogalactopyranoside). At 48 h postinfection (hpi), the cells were harvested and the recombinant viruses were isolated by sequential rounds of plaque purification in the presence of IPTG. The inducer concentration at which virus production was maximal, 0.15 mM, was chosen for the growth and purification of the recombinant virus. Similar results were obtained with the two plasmids used, and one virus clone from the pIND2.pp220-transfected cells was selected for further characterization. The genomic structure of this recombinant virus, named v220i, was confirmed by DNA hybridization analysis.
Plaque assays.
Vero cell monolayers, in six-well plates, were infected with 600 PFU of recombinant v220i or parental BA71V. After 1 h, the inoculum was removed and the cells were overlaid with DMEM containing 0.6% Noble agar and 2% FCS in the presence or absence of 0.15 mM IPTG. Five days later, the medium was removed and the monolayers were stained with 1% crystal violet.
One-step virus growth curves.
Vero cell monolayers, in 24-well plates, were infected with 5 PFU of recombinant v220i or parental BA71V per cell. After a 1-h adsorption, the cells were incubated in DMEM supplemented with 2% FCS. IPTG (0.15 mM) was added immediately after the adsorption period or at 12, 18, or 24 hpi. Infected cells with their culture supernatants were harvested at different times postinfection and sonicated, and titers were determined by plaque assay in the presence of 0.15 mM IPTG.
Immunoprecipitation of polyprotein pp220 from extracts of v220i-infected cells.
Vero cells were mock infected or infected with 10 PFU of BA71V or v220i virus per cell in the absence or in the presence of 0.15 mM IPTG. The cells were pulse-labeled from 12 to 18 hpi with 500 μCi of [35S]methionine-[35S]cysteine (Promix in vitro cell labeling mix; Amersham Pharmacia Biotech) per ml. After labeling, the cells were lysed and immunoprecipitated with anti-pp220/p150 antibodies immobilized on protein A-Sepharose as previously described (6). Total extracts and immunoprecipitated proteins were resolved by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-12% PAGE) and detected by autoradiography. Quantification of protein bands was performed with a GS710 densitometer and Quantity One software (both from Bio-Rad Laboratories).
Purification and analysis of mutant v220i particles.
A simplified version of the procedure described by Carrascosa et al. (12) was used to purify extracellular v220i particles from culture supernatants. Vero cells, cultured in roller bottles (500 cm2), were infected with 5 PFU of recombinant v220i per cell in the presence or absence of 0.15 mM IPTG. At 36 hpi, the culture media were centrifuged at 2,000 rpm for 10 min in a GS3 Sorvall rotor to remove cell debris, and the extracellular virions were concentrated by centrifugation in a GS3 Sorvall rotor at 8,500 rpm for 6 h at 4°C. Virus pellets were washed in phosphate-buffered saline, mixed with Percoll to a final concentration of 45% in phosphate-buffered saline, and subjected to equilibrium gradient sedimentation in a T865 Sorvall rotor at 20,000 rpm for 30 min at 4°C (12). Aliquots of the fractions were analyzed for virus content by negative staining with 2% uranyl acetate (4), for protein content by Western immunoblotting with anti-p150 and anti-p72 antibodies, and for DNA content by dot blot hybridization with an ASFV-specific probe. Fractions 2 to 8 of both gradients, containing virus particles essentially free of contaminant membrane vesicles, were subsequently pooled and gel filtered through a Sephacryl S-1000 (Amersham Pharmacia Biotech) column to remove Percoll particles (12).
For immunoblotting of v220i particles, samples were electrophoresed in SDS-12% polyacrylamide gels, transferred to nitrocellulose, and probed with the indicated antibodies against ASFV structural proteins. Protein detection was performed using peroxidase-conjugated antibodies and the ECL system (Amersham Pharmacia Biotech).
For dot blot hybridization analysis, equivalent amounts of the gradient fractions were diluted in 100 μl of H2O containing 100 ng of sheared salmon sperm DNA per ml and applied to nitrocellulose membrane (Protran [Schleicher & Schuell]; pore size, 0.2 μm) in a vacuum manifold. The DNA was denatured, neutralized, and bound to the membrane according to the manufacturer's protocol. The probe was the BA71V EcoRI fragment C′ labeled with [32P]dCTP by random priming with a NEBlot kit (New England Biolabs). Hybridization was carried out at 65°C as described previously (35). The radioactive DNA signal was visualized by autoradiography.
For electron microscopy of purified v220i particles, virus pellets were fixed with 2% glutaraldehyde in 200 mM HEPES (pH 7.4) for 1 h at room temperature and processed for conventional Epon embedding as described below.
Electron microscopy.
For conventional Epon section analysis, Vero cells were infected with 10 PFU of BA71V or recombinant v220i per cell in the absence or presence of 0.15 mM IPTG and fixed at the indicated times with 2% glutaraldehyde in 200 mM HEPES (pH 7.4) for 1 h at room temperature. Postfixation was carried out with 1% OsO4 and 1.5% K3Fe(CN)6 in H2O at 4°C for 30 min. Samples were dehydrated with acetone and embedded in Epon according to standard procedures.
For freeze-substitution, the v220i-infected Vero cells were fixed at the indicated times after infection for 1 h with 4% formaldehyde and 0.1% glutaraldehyde in 200 mM HEPES (pH 7.2) on ice. The specimens were cryoprotected with 30% glycerol for 30 min, rapidly frozen in liquid propane (−180°C), and stored in liquid nitrogen. Freeze-substitution and embedding in Lowicryl K4M were carried out as described previously (3, 4). Ultrathin sections were collected on nickel grids coated with Formvar and carbon and processed for immunogold labeling of ASFV proteins as described previously (3, 4). For DNA detection, sections were preincubated for 30 min at 37°C with 20 μg of proteinase K per ml in a buffer containing 25 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 10 mM CaCl2. Labeling was performed as described previously (27) using a monoclonal antibody to DNA and a goat anti-mouse antibody coupled to 10-nm-diameter gold particles. Specimens were examined at 80 kV in a Jeol 1010 electron microscope. For quantification of the immunolabeling experiments, four to six electron micrographs of virus factories were analyzed for each condition at a final magnification of ×75,000. Gold particles were counted on cross-sectioned virions.
Site-directed mutagenesis of gene CP2475L.
Plasmid pGEM-CP2475L, containing the CP2475L gene under control of the T7 polymerase promoter, has been described elsewhere (5). Plasmid pGEM-CP2475L-G2A, in which the glycine residue at position 2 of ORF CP2475L was mutated to alanine, was obtained as follows. Site-directed mutagenesis was performed by PCR using plasmid pGEM-CP2475L as a template; the oligonucleotide 5′-GAGATgcatgcATGGCTAACCGTGGATCTTCAACC, which contains a point mutation (underlined) and an SphI restriction site (lowercase), as the upstream primer; and the oligonucleotide 5′-GGTGGAACCGCTCGAGGATCGGGCAG as the downstream primer. The resulting amplified fragment was digested with SphI and EcoRV and cloned into the SphI- and EcoRV-digested plasmid pGEM-CP2475L. The presence of the G2A mutation within the resulting plasmid was confirmed by sequencing.
Transient expression in COS cells.
COS-7 cells were transfected for 1 h at 37°C with 250 ng of DNA of the plasmids pGEM-CP2475L and pGEM-CP2475L-G2A per 105 cells by using Lipofectamine Plus reagent (Life Technologies) and following the manufacturer's indications. The transfected cells were then infected with 5 PFU of the recombinant vaccinia virus vTF7-3 expressing T7 RNA polymerase (22) per cell.
For metabolic labeling experiments, the cells were incubated from 3 to 6 hpi with 500 μCi of [35S]methionine-[35S]cysteine in methionine- and cysteine-free DMEM per ml or with 250 μCi of [3H]myristic acid (Amersham Pharmacia Biotech) in DMEM supplemented with 2% delipidated FCS per ml. After the labeling periods, the cells were lysed and processed for immunoprecipitation with an anti-pp220/p150 antibody.
Cell fractionation was performed at 15 h after infection as previously described (6). The expression of myristoylated and nonmyristoylated polyprotein pp220 was analyzed by Western immunoblotting of equivalent amounts of the different cellular fractions.
For electron microscopy, COS-7 cells were transfected with plasmid pGEM-CP2475L or pGEM-CP2475L-G2A and infected with 5 PFU of vTF7-3 per cell as described above. In some experiments, the cells were incubated in the presence of 40 μg of cytosine arabinoside, an inhibitor of vaccinia virus DNA replication and late protein synthesis, per ml. At 15 hpi, the cells were processed for Epon embedding or freeze-substitution.
RESULTS
Generation of ASFV recombinant v220i.
To study the role of polyprotein pp220 in virus replication, we constructed the ASFV recombinant v220i, in which expression of the polyprotein-encoding gene CP2475L is controlled by the E. coli lac operator-repressor system (Fig. 1A). v220i virus was obtained from vGUSREP, a recombinant virus derived from the BA71V strain that constitutively expresses the E. coli lac repressor (24). To allow the inducible expression of polyprotein pp220, vGUSREP was modified by replacing the original promoter of gene CP2475L by an inducible promoter, p72.I, composed of the strong late viral promoter p72.4 and the operator sequence O1 (24). The genome structure of the resulting v220i virus was confirmed by DNA hybridization (data not shown).
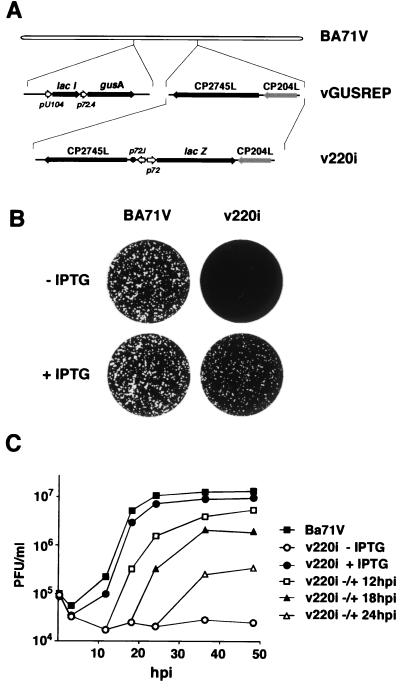
FIG. 1.
(A) Genomic structure of the ASFV recombinant virus v220i. The recombinant virus v220i was obtained from vGUSREP, a BA71V-derived recombinant virus, which contains the Lac repressor-encoding gene lacI inserted into the nonessential thymidine kinase locus. In the v220i virus, the promoter of the polyprotein pp220-encoding gene CP2475L was replaced by an inducible promoter, p72.I, which is composed of a strong late promoter (p72.4) and the operator sequence O1 (•) from the E. coli lac operon. The reporter genes lacZ and gusA, used for selection and purification of the recombinants, are also shown. (B) Plaque phenotype of v220i. Monolayers of Vero cells were infected with parental BA71V or recombinant v220i in the presence (+) or absence (−) of 0.15 mM IPTG. Plaques were visualized with 1% crystal violet 5 days after infection. (C) One-step growth curves of v220i. Vero cells were infected with 5 PFU of v220i per cell in the presence or absence of 0.15 mM IPTG. At the indicated times of infection, the total virus titer of each sample was determined by plaque assay on Vero cells in the presence of the inducer. Parental BA71V infections were also titrated as a control. Recombinant v220i was also grown under restrictive conditions for 12, 18, or 24 h and then induced with IPTG. At different times after induction, the infectious virus was titrated as described above.
Inducer dependence of recombinant v220i.
To test the inducer dependence of recombinant v220i virus, a plaque assay was performed at different concentrations of the inducer ranging from 0 to 1 mM IPTG. The lysis plaque number was maximal at 0.15 mM IPTG (not shown). Figure 1B shows the plaque phenotypes of parental BA71V and recombinant v220i viruses in the presence or absence of 0.15 mM IPTG. Under permissive conditions, the plaque number was similar for both parental and recombinant viruses, although the plaque size was slightly smaller in v220i infections. Omission of IPTG led to a dramatic decrease in plaque formation by v220i virus compared with control BA71V virus.
In a further approach, one-step growth curves for v220i virus were performed in the presence or absence of 0.15 mM IPTG. As shown in Fig. 1C, under permissive conditions the growth curve of recombinant v220i virus was similar to that obtained with parental BA71V virus. In the absence of inducer, the yields of v220i virus did not increase over time, remaining about 2.6 log units below those observed under permissive conditions from 24 to 48 hpi. In the same experiment, we also tested the ability of v220i grown for different times under nonpermissive conditions to produce infectious particles upon IPTG addition. As shown in Fig. 1C, the later the time of IPTG addition, the lower was the final virus titer obtained. When IPTG was added at 12 hpi, an early time for virus assembly (3, 10), the maximal virus yield, reached at 48 hpi, was about 60% of that observed for v220i grown under permissive conditions throughout the infection. When the inducer was added at 18 or 24 hpi, the maximal yields were 20 and 4% of those obtained with the control, respectively. Together, the results of the plaque assays and one-step growth curves indicate that recombinant v220i is an IPTG-dependent conditional lethal mutant.
Inducible expression of polyprotein pp220.
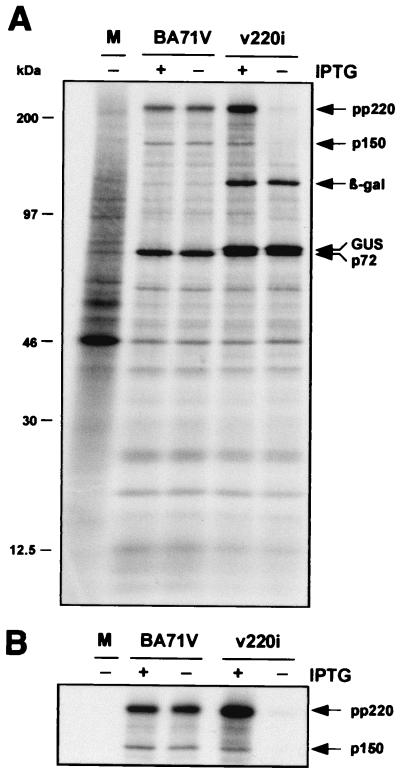
To confirm that the expression of polyprotein pp220 can be regulated by inducer during v220i infections, cells infected with the recombinant under permissive or nonpermissive conditions were labeled with [35S]methionine-[35S]cysteine from 12 to 18 hpi, a period in which late gene expression is in progress. Mock- and BA71V-infected cells labeled under the same conditions were used as negative and positive controls, respectively. As shown in Fig. 2A, similar overall protein profiles were observed when parental BA71V and recombinant v220i infections were compared, with the exception that extra bands corresponding to the reporter β-glucuronidase (GUS) and β-galactosidase gene products were expressed in the cells infected with the inducible recombinant. As expected, during BA71V infections, synthesis and processing of polyprotein pp220 were equivalent in the presence and absence of IPTG. In contrast, during v220i infections, the pp220 expression level was drastically reduced when the inducer was not added. This inhibition became more evident when the extracts were immunoprecipitated with a serum against both the polyprotein pp220 and the mature structural protein p150 (Fig. 2B). Densitometric quantification revealed that under permissive conditions, polyprotein expression was about 150% of that determined for control BA71V infections, whereas under nonpermissive conditions, it was reduced to about 5%. In summary, these results show that the conditional lethal phenotype of recombinant v220i is related to the inducer-dependent expression of the viral polyprotein.
FIG. 2.
Inducible expression of polyprotein pp220. Vero cells were either mock infected (lane M) or infected with parental BA71V or recombinant v220i virus in the presence (+) or absence (−) of IPTG. The cells were pulse-labeled with [35S]methionine-[35S]cysteine from 12 to 18 hpi, lysed, and either analyzed directly by SDS-PAGE (A) or first immunoprecipitated with a serum against polyprotein pp220 and its derived product p150 (B). The positions of the polyprotein pp220, the structural protein p150, the major capsid protein p72, and the reporter β-galactosidase and GUS proteins are indicated. Note that GUS protein migrates slightly slower than capsid protein p72. The electrophoretic mobilities of molecular mass markers are indicated at the left in panel A.
Electron microscopy of v220i-infected cells.
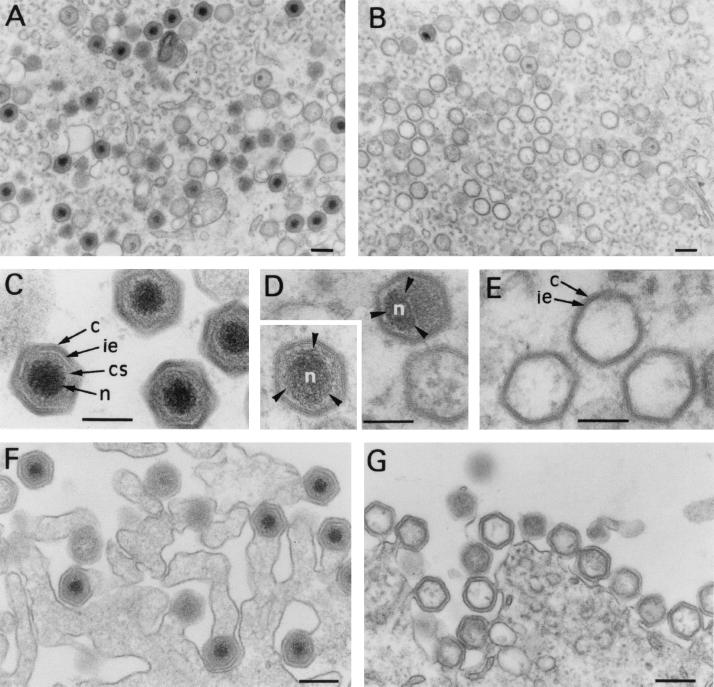
To investigate the effect of pp220 repression on virus morphogenesis, v220i-infected cells maintained for 24 h under permissive or nonpermissive conditions were analyzed by electron microscopy (Fig. 3). In the presence of inducer, the appearance of the cytoplasmic virus factories was similar to that described for parental BA71V virus (3); all stages of virus assembly, including large numbers of mature virions, were detected (Fig. 3A). In contrast, when IPTG was omitted, intracellular mature particles were essentially absent (Fig. 3B). Instead, large quantities of icosahedral virions lacking a proper core were evident at the assembly sites. Compared with the intracellular mature particles (Fig. 3C), most of the defective particles lacked the electron-dense DNA-containing nucleoid as well as the surrounding core shell (Fig. 3B and E). Rather, the central region of the mutant virions appeared to be either occupied by a disorganized and moderately electron-dense content or virtually devoid of core material. A very small fraction of virions contained an electron-dense nucleoid, which, however, was usually altered in size and position (Fig. 3D). Importantly, the repression of pp220 synthesis did not apparently affect the assembly of the inner viral envelope and the outer capsid (Fig. 3E). Moreover, a significant proportion of the defective particles were detected at the plasma membrane during the budding process (Fig. 3G), as occurs with the mature virions produced under permissive conditions (Fig. 3F).
FIG. 3.
Electron microscopy of v220i-infected cells. Ultrathin Epon sections of v220i-infected cells incubated for 24 h in the presence (A, C, and F) or in the absence (B, D, E, and G) of IPTG are shown. While under permissive conditions (A), the assembly sites contain large amounts of mature virions, under restrictive conditions (B), they contain essentially icosahedral core-less particles. Compared with the intracellular mature virions (C), most of the defective particles (E) lack the core shell (cs) and the electron-dense nucleoid (n) but contain the inner envelope (ie) and the capsid (c). A minor population of mutant v220i particles (D) contain the nucleoid, but its size and position frequently appear to be altered. Defective v220i particles are released by budding at the plasma membrane (G), as occurs with the mature particles produced under permissive conditions (F). Bars, 200 nm (A, B, F, and G) and 100 nm (C, D, and E).
Together, these observations indicate that polyprotein pp220 is essential for the assembly of the core but not for the assembly of the external domains into icosahedral particles.
Immunoelectron microscopy of intracellular defective v220i particles.
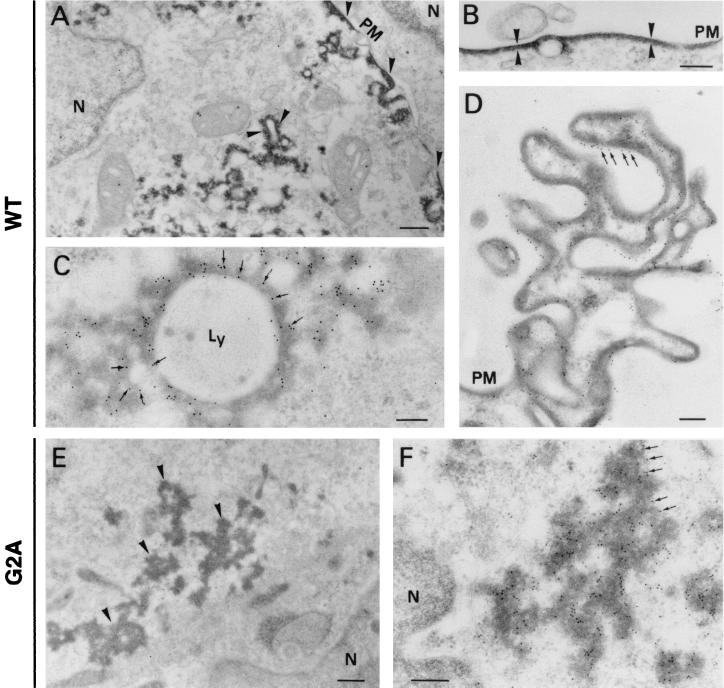
To explore in more detail the consequences of pp220 repression for virus formation, we compared the composition of the defective v220i particles with that of the mature virions by immunoelectron microscopy. Cells infected with recombinant v220i under permissive or restrictive conditions were processed at 24 hpi by freeze-substitution followed by Lowicryl embedding. Ultrathin sections from both samples were then incubated with a repertoire of antibodies against major structural proteins followed by protein A-gold. Table 1 shows a quantification of the labeling density obtained for each antibody on mature icosahedral particles generated in the presence of IPTG and on defective virions (without a proper core) produced in the absence of inducer. Examples of the labeling patterns for some of these proteins are illustrated in Fig. 4. Antibodies against peripheral proteins, like the capsid proteins p72 (24) (Fig. 4A and B) and pE120R (6) or the transmembrane polypeptide p17 (41), labeled mature and defective core-less particles equally. In contrast, the labeling of proteins localized at the core shell, like the pp220-derived protein p150 (3) (Fig. 4C and D) or the pp62-derived protein p35 (42; our unpublished results), was practically absent in the defective v220i particles. Similarly, the detection of nucleoid markers, such as the DNA-binding proteins pA104R (8) (Fig. 4E and F) and p10 (34; our unpublished results), within defective core-less particles was considerably reduced. The exception was a very minor population of nucleoid-containing particles that probably result from the residual expression of the CP2475L gene under restrictive conditions.
TABLE 1.
Quantification of structural virus proteins within v220i particles by immunoelectron microscopya
| Protein | Gold particles per virion (mean ± SEM)b
|
Ratio (+/−) | |
|---|---|---|---|
| Mature virions (+ IPTG) | Defective virions (− IPTG) | ||
| p72 (capsid) | 1.06 ± 0.11 | 0.96 ± 0.09 | 1.1 |
| pE120R (capsid) | 1.92 ± 0.22 | 2.13 ± 0.15 | 0.9 |
| p17 (inner envelope) | 1.13 ± 0.18 | 1.10 ± 0.10 | 1.0 |
| p150 (core shell) | 1.73 ± 0.16 | 0.09 ± 0.03 | 19.2 |
| p35 (core shell) | 0.41 ± 0.07 | 0.08 ± 0.02 | 5.1 |
| p10 (nucleoid) | 0.92 ± 0.11 | 0.04 ± 0.01 | 23.0 |
| pA104R (nucleoid) | 1.08 ± 0.13 | 0.24 ± 0.05 | 4.5 |
Immunogold labeling was as described in the legend to Fig. 4.
Quantification was performed for closed icosahedral virions at the assembly sites. For each protein, n ranged from 50 to 100 mature virions and from 100 to 200 defective virions.
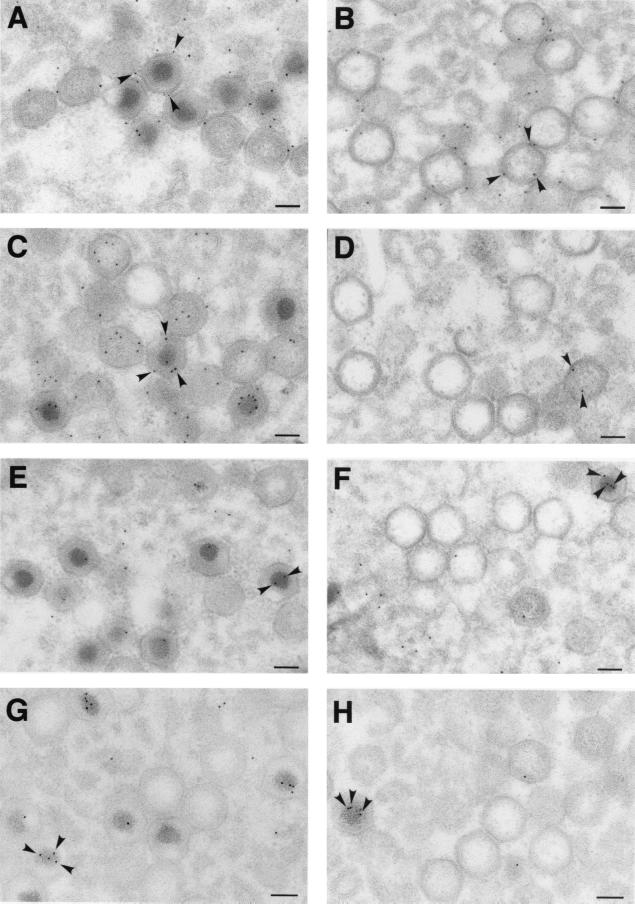
FIG. 4.
Immunoelectron microscopy of defective v220i particles. Vero cells were infected with recombinant v220i in the presence (A, C, E, and G) or absence (B, D, F, and H) of IPTG. At 24 hpi, the cells were fixed and processed by freeze-substitution. Ultrathin Lowicryl sections were incubated with antibodies against the capsid protein p72 (A and B), the core shell protein p150 (C and D), and the nucleoid protein pA104R (E and F), followed by incubation with protein A-gold (10 nm diameter). Sections were also labeled with a monoclonal anti-DNA antibody followed by a goat anti-mouse antibody conjugated to 10-nm-diameter gold particles (G and H). The arrowheads indicate representative labeling of the capsids (A and B), the core shells (C and D), and the nucleoids (E, F, G, and H) of particles at the virus factories. Note that p72 labeling is detected to similar extents on virions produced under permissive and nonpermissive conditions, while the labeling of core proteins and DNA is significantly weaker on the defective v220i particles. The images in panels D, F, and H have been selected to show one example of positively labeled virions (arrowheads). Bars, 100 nm.
To ascertain if mutant v220i particles were able to encapsidate the viral genome, an anti-DNA monoclonal antibody was also tested. As shown in Fig. 4G, under permissive conditions the DNA labeling on virions was essentially confined to those containing an electron-dense nucleoid; however, only 44% of the examined mature virions (n = 169) contained gold particles. The fact that not all of the dense mature nucleoids were labeled is probably due to the limited accessibility of the highly packaged viral genome to the anti-DNA antibodies. Under restrictive conditions, the labeling of the defective particles (Fig. 4H) was significantly weaker (about 4 times lower [11% of the virions were labeled; n = 351), thus suggesting that recombinant v220i is a DNA packaging mutant.
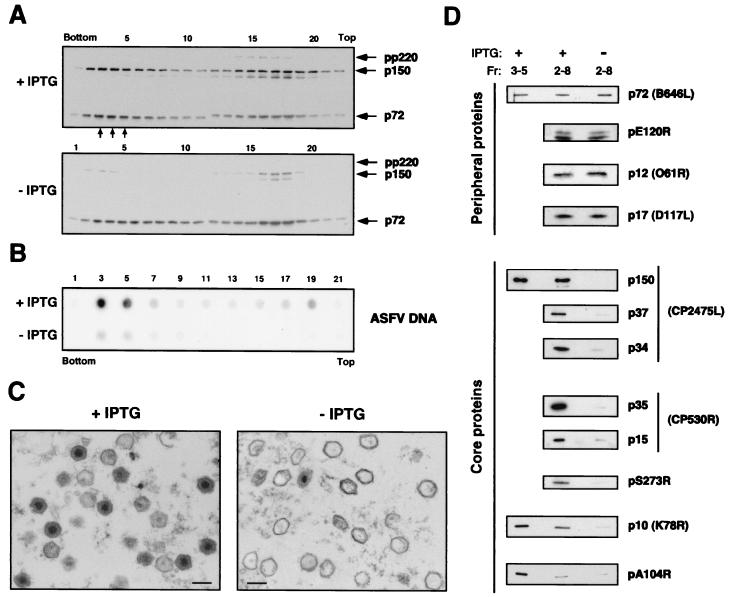
Purification and analysis of extracellular v220i defective particles.
To further characterize the mutant v220i particles, we attempted to purify them from the supernatants of infected cells. Extracellular virions collected from the culture media of permissive and restrictive v220i infections were sedimented into an isopycnic Percoll gradient as previously established (12). Samples were previously adjusted to the same content of capsid protein p72 in order to analyze equivalent amounts of virus particles. Under permissive conditions, a cloudy band corresponding to the sedimentation peak described for infectious ASFV particles (12) was visible close to the bottom of the gradient. No distinctive band was detected in the virus samples obtained from nonpermissive infections. The gradients were then fractionated from the bottom, and each fraction was evaluated for virus content by negative staining (not shown) and by Western immunoblotting with antibodies against protein p72 and the pp220-derived protein p150 (Fig. 5A). In both samples, virus particles were found mainly in two broad zones: one close to the top of the gradient, which contained significant amounts of contaminant membrane vesicles, and the other close to the bottom, which was essentially free of membrane vesicles. A similar virus distribution has been described for parental BA71V virus (12). As shown in Fig. 5A, proteins p72 and p150 were also detected predominantly in the broad peaks of both gradients, whereas unprocessed pp220 precursor was detected only in the top peak containing contaminant membranes. As expected, proteins pp220 and p150 were only weakly detected in the sample obtained under restrictive conditions. Next, we analyzed the gradient fractions for viral DNA content by dot blot hybridization with a specific ASFV DNA probe. As shown in Fig. 5B, under permissive conditions, the viral DNA was detected essentially in fractions 3 to 5, which correspond to the band of infectious virus particles. Under restrictive conditions, DNA hybridization was also maximal around fractions 3 to 5, but the intensity of the signal, as judged by densitometric quantitation, was significantly (5 to 6 times) lower than that of the positive control. This finding supports the conclusion that recombinant v220i is a DNA-deficient mutant.
FIG. 5.
Purification and analysis of extracellular defective v220i particles. (A) Extracellular virions collected from clarified culture supernatants of permissive (+ IPTG) and restrictive (− IPTG) v220i infections were adjusted to the same p72 content and subjected to equilibrium sedimentation in a Percoll gradient. Aliquots of the gradient fractions were analyzed by Western immunoblotting with antibodies against protein p72 and pp220-derived protein p150. The small arrows below fractions 3 to 5 of the + IPTG gradient indicate the position where the infectious ASFV particles band. The positions of proteins pp220, p150, and p72 are indicated. (B) DNA content of v220i particles. Alternate fractions of the Percoll gradients were analyzed by dot blot DNA hybridization with an ASFV probe. (C) Electron microscopy of purified v220i particles obtained in the presence or absence of IPTG. In both samples, fractions 2 to 8 of the Percoll gradients were pooled, gel filtrated through a Sephacryl S-1000 column, and processed by conventional Epon embedding. The material between virus particles mostly corresponds to clusters of Percoll particles. Bars, 200 nm. (D) Western immunoblotting of purified v220i particles with a spectrum of antibodies against ASFV structural proteins. Purified virus particles obtained under permissive (+) and nonpermissive (−) conditions were adjusted to the same p72 content and analyzed by SDS-PAGE and immunoblotting with antibodies to the indicated viral proteins. The mutant v220i particles (−) were collected from fractions 2 to 8 of the Percoll gradients, while the control virions (+) were recovered from fractions 2 to 8 or from fractions 3 to 5. Semipurified particles obtained from clarified culture media were used to analyze the proteinase pS273R.
Since no major differences were observed in the sedimentation profiles of the virions obtained under permissive and nonpermissive conditions, fractions 2 to 8 of both gradients, containing particles essentially free of contaminant vesicles, were pooled for further comparative analyses. Virus samples were gel filtered through a Sephacryl S-1000 column in order to remove Percoll particles and then processed for conventional electron microscopy. As shown in Fig. 5C, whereas the purified extracellular particles obtained under permissive conditions contained predominantly mature dense cores, the particles purified from restrictive v220i infections were mostly electron-lucent icosahedral particles. A fraction of the mutant virions, however, appeared to be partially collapsed, probably due to the purification process or the subsequent processing for electron microscopy.
Next, we analyzed the protein composition of the purified defective v220i particles by Western immunoblotting with a spectrum of antibodies against major ASFV structural proteins. Figure 5D shows the results of a comparison between the mutant and control particles pooled from fractions 2 to 8 of the Percoll gradients. Some ASFV proteins in samples of control mature particles obtained under permissive conditions and collected only from the position where the infectious virus particles band (fractions 3 to 5 of the Percoll gradient) were also analyzed. When the virus samples were adjusted to the same p72 content, other peripheral major proteins, such as the capsid protein pE120R and the transmembrane polypeptides p17 and p12 (1, 6, 13, 24, 41), were detected equally in control and mutant particles. In contrast, the analysis of major core components revealed marked differences between the preparations. Thus, the presence of the mature products derived from polyproteins pp220 (p150, p37, and p34) and pp62 (p35 and p15), as well as the proteinase pS273R, all located at the core shell (3, 5; unpublished data), drastically decreased in the defective v220i particles. Similarly, the levels of the DNA-binding proteins p10 and pA104R (8, 34) were significantly reduced. The differences in these two proteins were even more pronounced when the defective v220i particles were compared with the control particles recovered from the band of infectious particles. This observation probably reflects the fact that the infectious virus band contains essentially mature virions (12), whereas the control sample obtained from fractions 2 to 8 of the Percoll gradients contains a higher proportion of immature virions and therefore a lower content of nucleoid proteins.
In conclusion, the analysis of purified extracellular mutant v220i particles, together with the data obtained from immunoelectron microscopy of intracellular defective virions, indicates that pp220 repression blocks the incorporation of other major components of the core shell and the nucleoid, including the viral genome, into the virus particles.
Effect of pp220 postinduction on virus assembly.
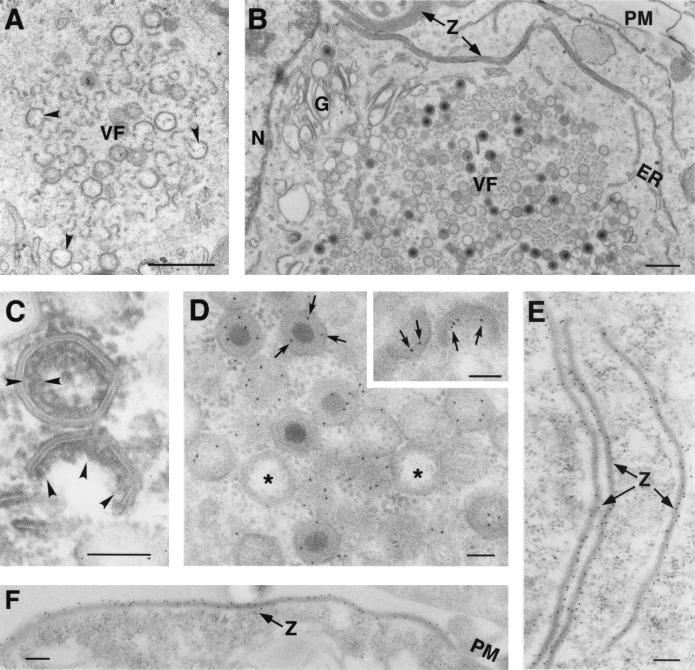
Our next goal was to study the effect of IPTG addition on virus assembly in v220i-infected cells previously maintained under restrictive conditions. Figure 6 shows an electron microscopy analysis of v220i-infected cells maintained during 12 h in the absence of IPTG or incubated with the inducer at 12 hpi for an additional 12-h period. In the first case (Fig. 6A), the assembly areas were relatively small and contained mainly precursor membranous structures as well as icosahedral empty particles, with a significant proportion of them being open. After the induction with IPTG (Fig. 6B), the viral factories increased notably in size and, concomitantly, large amounts of both mature and immature icosahedral particles were observed. The proportion of mature virions was, however, clearly lower than that observed in v220i infections maintained during 24 h under permissive conditions (Fig. 3A), which is consistent with the strong reduction (fivefold) observed in the infectious virus yield (Fig. 1C). Concerning the immature virions, a close inspection revealed that they often appeared to contain a distinctive matrix-like domain underneath the viral envelope (Fig. 6C). Immunoelectron microscopy with an antibody against the polyprotein pp220 confirmed that both mature and developing particles were strongly labeled at the core shell (Fig. 6D). Moreover, specific pp220 labeling was clearly detected in open icosahedral virions in which the core shell appeared to be partially assembled (Fig. 6D, inset). In contrast, no gold particles were found within apparently closed, core-less virions, which were likely formed during the initial 12-h period under restrictive conditions (Fig. 6D). When IPTG was added at later times of infection (from 18 or 24 hpi onwards), the proportion of mature virions decreased and, concomitantly, the number of core-less particles increased (not shown). These findings suggest that core formation occurs when the developing particles are still open. We therefore interpret the partial restoration of infectivity observed after IPTG addition (Fig. 1C) to be a consequence of the de novo assembly of mature virions from precursor membranes, although we cannot completely rule out that it might be due to a further maturation of the preformed icosahedral core-less particles.
FIG. 6.
Effect of pp220 induction on ASFV assembly. Electron microscopy of v220i-infected cells maintained for 12 h in the absence of IPTG (A) or induced from 12 to 24 hpi (B, C, D, E, and F). The samples were processed by conventional Epon embedding (A, B, and C) or by freeze-substitution followed by immunogold labeling with an antibody against pp220 and p150 (D, E, and F). Images in panels A to D correspond to virus factories, whereas those in panels E and F correspond to adjacent areas. Open particles are indicated by arrowheads in panel A. The arrowheads in panel C indicate the core shell of assembling particles. The arrows in panel D and in the insert indicate gold particles (10 nm) at the core shell of a mature virion and two open developing particles, respectively. The asterisks in panel D indicate core-less virions. Panels E and F show zipper-like structures associated with membranes from the ER or from the plasma membrane, respectively. N, nucleus; G, Golgi complex; PM, plasma membrane; VF, virus factories; Z, zipper-like structures. Bars, 500 nm (A and B) and 100 nm (C, D, E, and F).
The examination of v220i-infected cells in which the inducer was added from 12 to 24 hpi provided another interesting observation. The areas contiguous to the assembly sites revealed a high proportion of aberrant viral forms referred to as zipper-like structures (Fig. 6B), which are also found in normal infections in a minor proportion (3). These abnormal arrangements consist of a long and thick (about 30-nm) protein layer, similar to the viral core shell, bound to adjacent membranes of the ER and, less frequently, of the lysosomes and the plasma membrane. As shown in Fig. 6E and F, these structures were intensely labeled by antibodies against polyprotein pp220. No similar forms were found in v220i-infected cells maintained for 12 or 24 h under restrictive conditions. In our view, the accumulation of these pp220-containing aberrant structures could be due, at least in part, to the inability of the core-less particles to recruit the newly synthesized polyprotein.
Transient expression of pp220 in COS cells.
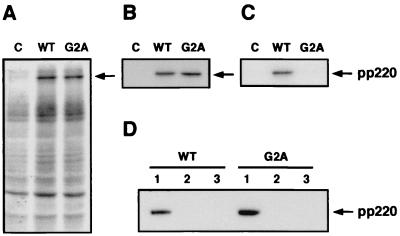
It has previously been reported that polyprotein pp220 is N myristoylated (40). Since myristoylation may serve as a membrane anchoring signal, we studied whether precursor pp220 associates with membranes through its myristic moiety. Transient-expression experiments were performed with COS-7 cells transfected with a plasmid containing the CP2475L gene under control of the T7 bacteriophage promoter and subsequently infected with a recombinant vaccinia virus expressing the T7 RNA polymerase (23). A mutant nonmyristoylable version of polyprotein pp220, in which the N-terminal Gly residue was replaced by Ala, was also expressed. As a negative control, cells were transfected with the empty plasmid and infected with the recombinant vaccinia virus. The transfected cells were labeled with [35S]methionine-[35S]cysteine or with [3H]myristic acid from 3 to 6 hpi, and the extracts were analyzed by SDS-PAGE or immunoprecipitated with an antibody to pp220. Both forms of the ASFV polyprotein were expressed in similar amounts (Fig. 7A) and were efficiently immunoprecipitated from the 35S-labeled extracts (Fig. 7B), but only the wild-type polyprotein was detected after immunoprecipitation of cell extracts labeled with [3H]myristic acid (Fig. 7C). In a second experiment, the transfected cells were homogenized and fractionated into a low-speed (500 × g) sediment and a postnuclear supernatant, which was, in turn, fractionated into a high-speed (100,000 × g) sediment and a supernatant cytoplasmic fraction. Surprisingly, both the myristoylated and the nonmyristoylated forms of pp220 were detected almost exclusively in the low-speed sediment. Since the fractionation experiment may not distinguish between sedimentation due to membrane binding and that due to protein aggregation, we performed electron microscopy of the transfected cells (Fig. 8). Inspection of cells expressing the wild type polyprotein revealed a great accumulation of electron-dense structures associated with the cytosolic sides of a variety of cellular membranes. These pp220-induced membrane-bound coats covered large extensions of the plasma membrane (Fig. 8A, B, and D) or virtually enwrapped intracellular cisternae (Fig. 8A and C), preferentially from the endosomal-lysosomal system. Interestingly, many images (Fig. 8B) of the cross-sectioned coats exhibited a regular thickness (about 30 nm) similar to that of the aberrant zipper-like viral structures (Fig. 6B, E, and F). Equivalent pp220-induced structures were observed in the presence of cytosine arabinoside, a drug that inhibits DNA replication and consequently the morphogenesis of vaccinia virus (not shown). To confirm that these structures were composed of polyprotein pp220, Lowicryl sections of freeze-substituted cells were incubated with an anti-pp220 antibody followed by protein A-gold complexes. As shown in Fig. 8C and D, the electron-dense membrane coats were intensely labeled. On the other hand, the transfected cells expressing the nonmyristoylable polyprotein showed an accumulation of large electron-dense aggregates at the cytoplasm that did not apparently associate with membranes (Fig. 8E). In fact, an extensive examination of the membrane profiles of the cell surfaces did not reveal any structure comparable to those described above. As shown in Fig. 8F, immunoelectron microscopy with an anti-pp220 antibody showed strong labeling on the cytoplasmic aggregates. Thus, the results of the fractionation experiment can be explained by the formation of dense membrane-bound coats by the wild-type polyprotein and of dense cytoplasmic aggregates by the mutant pp220.
FIG. 7.
Transient expression of polyprotein pp220 using the vaccinia virus-T7 RNA polymerase system. COS-7 cells were transfected with plasmid pGEM-T7 containing the wild-type (WT) CP2475L gene or a mutagenized copy (G2A) in which the N-terminal glycine codon was replaced by an alanine codon. pGEM-T7-transfected cells were used as a negative control (lanes C). After transfection, the cells were infected with vTF7-3, a vaccinia virus recombinant expressing T7 RNA polymerase, and labeled with [35S]methionine-[35S]cysteine or with [3H]myristic acid from 3 to 6 hpi. (A) SDS-PAGE of extracts of cells labeled with [35S]methionine-[35S]cysteine. (B) Immunoprecipitation of the 35S-labeled lysates with an anti-pp220 antibody. (C) Immunoprecipitation of the 3H-labeled extracts with an anti-pp220 antibody. (D) Cells transfected with the wild-type and mutagenized versions of the CP2475L gene were fractionated into a low-speed (500 × g) sediment (lanes 1) and a postnuclear supernatant, which was in turn fractionated into a soluble cytoplasmic fraction (lanes 2) and a high-speed (100,000 × g) sediment (lanes 3). The position of polyprotein pp220 is indicated.
FIG. 8.
Subcellular localization of myristoylated and nonmyristoylated pp220 in transfected COS-7 cells. Transfected COS-7 cells expressing myristoylable (wild-type [WT]) (A, B, C, and D) and nonmyristoylable (G2A) (E and F) polyprotein pp220 were processed by conventional Epon embedding (A and B) or by freeze-substitution and Lowicryl embedding (C, D, E, and F). The Lowicryl sections were incubated with an antibody against polyprotein pp220 followed by protein A-gold (10 nm diameter). Note that the myristoylated polyprotein assembles into electron-dense membrane coats (arrowheads in panels A and B), whereas the nonmyristoylated polyprotein forms large cytoplasmic aggregates (arrowheads in panel E). The arrowheads in panel B indicate the regular thickness (about 30 nm) of a dense coat bound to the plasma membrane. The arrows in panels C and D indicate pp220 labeling at dense coats bound to lysosomes and the plasma membrane, whereas the arrows in panel F indicate pp220 labeling at dense cytoplasmic aggregates. N, nucleus; PM, plasma membrane; Ly, lysosomes. Bars, 500 nm (A and E) and 200 nm (B, C, D, and F).
In conclusion, these results show that polyprotein pp220 associates with lipid membranes and that this binding is dependent on its N myristoylation. Also, the formation of the membrane-bound coats by pp220 suggests a capability of self-assembly into a core shell-like domain.
DISCUSSION
The characterization of an ASFV recombinant, v220i, which inducibly expresses the gene CP2475L, and the analysis of the role of N myristoylation in the membrane binding of polyprotein pp220 have provided evidence on the pivotal role of the polyprotein in virus morphogenesis.
Analysis of recombinant v220i led to two important findings: (i) the polyprotein pp220 is essential for the core assembly, and (ii) the formation of icosahedral particles is not dependent on core maturation. Under nonpermissive conditions, v220i was essentially unable to form lysis plaques, and its infectious virus yield was about 2.6 log units lower than that obtained in control infections. At the electron microscopic level, repression of pp220 synthesis caused a dramatic effect on virus morphogenesis. Intracellular mature virions were rarely found within the virus factories, whereas large quantities of icosahedral particles essentially devoid of the characteristic electron-dense nucleoid and the surrounding core shell were observed. Interestingly, the assembly of the outermost viral domains, i.e., the inner envelope, the capsid, and the outer envelope, was not apparently affected under restrictive conditions. Indeed, the defective icosahedral particles were efficiently released from the plasma membrane, as occurs with the mature virions in normal infections. This mutant phenotype is consistent with the localization of the pp220-derived proteins p150, p37, p34, and p14 at the core shell, a matrix-like domain placed between the internal DNA-containing nucleoid and the inner viral envelope (3). According to our working model for ASFV morphogenesis, based mainly on the ultrastructural analysis of viral intermediates from parental BA71V infections, the core assembly takes place, at least in its initial stages, within the developing icosahedral particles while they are still open (3). The first stage of this sequential maturational process would be the progressive construction of the core shell underneath the inner lipid envelope. The subsequent uptake of the DNA together with the viral nucleoproteins would lead to the formation of the nucleoid, whose final condensation might take place once the virus particles are already closed (3, 11). From this model, it seems conceivable that a blockage in the assembly of the core shell will have major effects on the subsequent steps of core maturation, as occurs with recombinant v220i. In support of this view is the fact that the defective v220i particles lack not only the pp220-derived proteins but also other examined major core proteins from the core shell (p35 and p15) and the nucleoid (p10 and pA104R). Moreover, the mutant v220i virions were shown to be DNA-deficient particles. This finding, together with the absence of preformed nucleoids or highly dense DNA-containing structures within both normal (3) and mutant v220i (this report) viral factories, is also consistent with the idea that DNA condensation and nucleoid maturation occur within the developing particles and depend on the previous assembly of the core shell. It therefore seems likely that the core-less v220i particles produced under nonpermissive conditions are defective virions irreversibly arrested in their assembly rather than maturation intermediates.
Collectively, our findings show that polyprotein pp220 is directly or indirectly involved in the incorporation of other major core components, including the viral genome, into the virus particles. An interesting question that remains to be addressed is the specific role of polyprotein processing in core assembly. It is tempting to speculate that proteolytic processing might be an essential mechanism for the patio-temporal control of the interactions between the core proteins. Thus, only properly processed pp220 precursors would enter the assembly pathway and lead to the subsequent incorporation of other core components like the nucleoid proteins and the viral genome. In relation to this, it should be noted that other complex DNA-containing viruses, such as the adenoviruses and the poxviruses, use proteolytic processing at cleavage sequences similar or identical to those of ASFV for the maturation of their core precursors. In the case of vaccinia virus, conditional lethal mutations of several genes were shown to affect both the cleavage of the precursor proteins and the assembly of the core (16, 21, 27, 28, 29). A remarkable example is the temperature-sensitive mutant ts16, which exhibits a blockage in the maturation of the core structure and in the proteolytic processing of the core precursors P4a, P4b, and L4 but not in DNA uptake (21, 28). Interestingly, the ts16 mutation maps to the I7 gene (21), which encodes a core protein having some similarity to the recently identified SUMO-1-specific proteases as well as to the proteinases of adenovirus and ASFV (30). It will be of interest to know whether the repression of ASFV proteinase has consequences for core assembly similar to those reported for vaccinia virus mutant ts16 or to those described here for ASFV recombinant v220i.
Besides proteolytic processing, the other major modification of polyprotein pp220 is the covalent addition of myristic acid to its N-terminal glycine residue (40). Using a COS cell expression system, we found that polyprotein pp220 assembles into large membrane-bound coats somewhat reminiscent of the core shell of the mature virus particles and to the aberrant zipper-like viral structures (3, 4, 24). In contrast, a mutant nonmyristoylated version of the polyprotein did not associate with membranes but formed large cytoplasmic aggregates. These data strongly suggest that N myristoylation functions as a membrane-anchoring signal and that polyprotein pp220 possesses an intrinsic capability of self-assembly into a higher-order structure. Both properties could be exploited during ASFV morphogenesis for the assembly of the viral core. Polyprotein pp220 could bind to the inner viral envelope via the myristoyl moiety and multimerize underneath the lipid layer to construct the protein scaffold of the developing core shell. As described above, the proteolytic processing of precursor pp220 might trigger the interactions required for the subsequent incorporation of additional core components. In relation to this, the possible role of polyprotein pp220 as a matrix-like linker protein is similar to that described for the matrix proteins of other enveloped viruses (25). For example, some retroviral Gag polyproteins use N myristoylation for targeting to the plasma membrane and are able to multimerize to produce virus-like particles (26). Interestingly, while polyprotein pp220 preferentially associates with ER-derived viral membranes in ASFV infections (3, 4), it is bound predominantly to the plasma membrane and endosome-lysosome membranes when expressed in COS cells. This observation suggests that, as occurs for other myristoylated proteins, proper membrane targeting of polyprotein pp220 involves other factors besides its acylation.
In summary, this report shows that polyprotein pp220 is essential for the assembly and envelopment of the core shell and for the subsequent steps of core maturation, including DNA encapsidation and nucleoid condensation, but that it is not required for the construction of the external viral domains. The generation of icosahedral virus-like particles by using core-deficient mutants like the one described here may facilitate the identification and characterization of the proteins that compose the viral envelopes and the capsid and may also provide a useful tool for the investigation of effective vaccines against ASFV infection.
Acknowledgments
We thank J. Salas and A. Alejo for critical reading of the manuscript. We also thank M. Guerra, M. Rejas, and M. J. Bustos for technical assistance.
This study was supported by grants from the Dirección General de Investigación Científica y Técnica (BMC 2000-1485), the European Community (FAIR-CT97-3441), and the Ministerio de Educación y Cultura (AGF98-1352-CE) and by an institutional grant from Fundación Ramón Areces. G. Andrés was supported by a fellowship from Comunidad Autónoma de Madrid.
REFERENCES
- 1.Alcami, A., A. Angulo, C. Lopez-Otín, M. Muñoz, J. M. Freije, A. L. Carrascosa, and E. Viñuela. 1992. Amino acid sequence and structural properties of protein p12, an African swine fever virus attachment protein. J. Virol. 66:3860-3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alves de Matos, A. P., and Z. G. Carvalho. 1993. African swine fever virus interaction with microtubules. Biol. Cell 78:229-234. [DOI] [PubMed] [Google Scholar]
- 3.Andrés, G., C. Simón-Mateo, and E. Viñuela. 1997. Assembly of African swine fever virus: role of polyprotein pp220. J. Virol. 71:2331-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrés, G., R. García-Escudero, C. Simón-Mateo, and E. Viñuela. 1998. African swine fever virus is enveloped by a two-membraned collapsed cisterna derived from the endoplasmic reticulum. J. Virol. 72:8988-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrés, G., A. Alejo, C. Simón-Mateo, and M. L. Salas. 2001. African swine fever virus protease: a new viral member of the SUMO-1-specific protease family. J. Biol. Chem. 276:780-787. [DOI] [PubMed] [Google Scholar]
- 6.Andrés, G., R. García-Escudero, E. Viñuela, M. L. Salas, and J. M. Rodriguez. 2001. African swine fever virus structural protein pE120R is essential for virus transport from the assembly sites to the plasma membrane but not for infectivity. J. Virol. 75:6758-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Angulo, A., E. Viñuela, and A. Alcamí. 1993. Inhibition of African swine fever virus binding and infectivity by purified recombinant virus attachment protein p12. J. Virol. 67:5463-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borca, M. V., P. M. Irusta, G. F. Kutish, C. Carrillo, C. L. Afonso, T. Burrage, and D. L. Rock. 1996. A structural DNA-binding protein of African swine fever virus with similarity to bacterial histone-like proteins. Arch. Virol. 141:301-313. [DOI] [PubMed] [Google Scholar]
- 9.Breese, S. S., Jr., and C. J. DeBoer. 1966. Electron microscope observation of African swine fever virus in tissue culture cells. Virology 28:420-428. [DOI] [PubMed] [Google Scholar]
- 10.Brookes, S. M., L. K. Dixon, and R. M. E. Parkhouse. 1996. Assembly of African swine fever virus: quantitative ultrastructural analysis in vitro and in vivo. Virology 224:84-92. [DOI] [PubMed] [Google Scholar]
- 11.Brookes, S. M., A. D. Hyatt, T. Wise, and R. M. E. Parkhouse. 1998. Intracellular virus DNA distribution and the acquisition of the nucleoprotein core during African swine fever virus particle assembly: ultrastructural in situ hybridisation and DNase-gold labelling. Virology 249:175-188. [DOI] [PubMed] [Google Scholar]
- 12.Carrascosa, A. L., M. del Val, J. F. Santarén, and E. Viñuela. 1985. Purification and properties of African swine fever virus. J. Virol. 54:337-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carrascosa, A. L., I. Sastre, and E. Viñuela. 1991. African swine fever virus attachment protein. J. Virol. 65:2283-2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carrascosa, J. L., J. M. Carazo, A. L. Carrascosa, N. García, A. Santisteban, and E. Viñuela. 1984. General morphology and capsid fine structure of African swine fever virus particles. Virology 132:160-172. [DOI] [PubMed] [Google Scholar]
- 15.Carvalho, Z. G., A. P. Alves de Matos, and C. Rodrigues-Pousada. 1988. Association of African swine fever virus with the cytoskeleton. Virus Res. 11:175-192. [DOI] [PubMed] [Google Scholar]
- 16.Cassetti, M. C., M. Merchlinsky, E. J. Wolffe, A. S. Weisberg, and B. Moss. 1998. DNA packaging mutant: repression of the vaccinia virus A32 gene results in noninfectious, DNA-deficient, spherical, enveloped particles. J. Virol. 72:5769-5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cobbold, C., and T. Wileman. 1998. The major structural protein of African swine fever virus, p73, is packaged into large structures, indicative of viral capsid or matrix precursors, on the endoplasmic reticulum. J. Virol. 72:5215-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costa, J. V. 1990. African swine fever virus, p. 247-270. In G. Darai (ed.), Molecular biology of iridoviruses. Kluwer Academic Publishers, Boston, Mass.
- 19.Dixon, L. K., J. V. Costa, J. M. Escribano, D. L. Rock, E. Viñuela, and P. J. Wilkinson. 2000. The Asfarviridae, p. 159-165. In M. H. V. Van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carsten, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Seventh report of the International Committee for the Taxonomy of Viruses. Academic Press, New York, N.Y.
- 20.Enjuanes, L., A. L. Carrascosa, M. A. Moreno, and E. Viñuela. 1976. Titration of African swine fever virus. J. Gen. Virol. 32:471-477. [DOI] [PubMed] [Google Scholar]
- 21.Ericsson, M., S. Cudmore, S. Shuman, R. C. Condit, G. Griffiths, and J. Krijnse Locker. 1995. Characterization of ts16, a temperature-sensitive mutant of vaccinia virus. J. Virol. 69:7072-7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esteves, A., M. I. Marques, and J. V. Costa. 1986. Two-dimensional analysis of African swine fever virus proteins and proteins induced in infected cells. Virology 152:192-206. [DOI] [PubMed] [Google Scholar]
- 23.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.García-Escudero, R., G. Andrés, F. Almazán, and E. Viñuela. 1998. Inducible gene expression from African swine fever virus recombinants: analysis of the major capsid protein p72. J. Virol. 72:3185-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garoff, H., R. Hewson, and D. J. Opstelten. 1998. Virus maturation by budding. Microbiol. Mol. Biol. Rev. 62:1171-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gheysen, D., E. Jacobs, F. de Foresta, C. Thiriart, M. Francotte, D. Thines, and M. de Wilde. 1989. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 59:103-112. [DOI] [PubMed] [Google Scholar]
- 27.Heljasvaara, R., D. Rodriguez, C. Risco, J. L. Carrascosa, M. Esteban, and J. R. Rodriguez. 2001. The major core protein P4a (A10L gene) of vaccinia virus is essential for correct assembly of viral DNA into the nucleoprotein complex to form immature viral particles. J. Virol. 75:5778-5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kane, E. M., and S. Shuman. 1993. Vaccinia virus morphogenesis is blocked by a temperature-sensitive mutation in the I7 gene that encodes a virion component. J. Virol. 67:2689-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klemperer, N., J. Ward, E. Evans, and P. Traktman. 1997. The vaccinia virus I1 protein is essential for the assembly of mature virions. J. Virol. 71:9285-9294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, S. J., and M. Hochstrasser. 1999. A new protease required for cell-cycle progression in yeast. Nature 398:246-251. [DOI] [PubMed] [Google Scholar]
- 31.López-Otín, C., C. Simón-Mateo, L. Martinez, and E. Viñuela. 1989. Gly-Gly-X, a novel consensus sequence for the proteolytic processing of viral and cellular proteins. J. Biol. Chem. 264:9107-9110. [PubMed] [Google Scholar]
- 32.Martínez-Pomares, L., C. Simón-Mateo, C. López-Otín, and E. Viñuela. 1997. Characterization of the African swine fever virus structural protein p14.5: a DNA binding protein. Virology 229:201-211. [DOI] [PubMed] [Google Scholar]
- 33.Moura Nunes, J. F., J. D. Vigario, and A. M. Terrinha. 1975. Ultrastructural study of African swine fever virus replication in cultures of swine bone marrow cells. Arch. Virol. 49:59-66. [DOI] [PubMed] [Google Scholar]
- 34.Muñoz, M., J. M. P. Freije, M. L. Salas, E. Viñuela, and C. López-Otín. 1993. Structure and expression in Escherichia coli of the gene coding for protein p10 of African swine fever virus. Arch. Virol. 130:93-107. [DOI] [PubMed] [Google Scholar]
- 35.Parry, H. D., and L. Alphey. 1995. The utilization of cloned DNAs to study gene organization and expression, p. 143-191. In D. M. Glover and B. D. Hames (ed.), DNA cloning 1: core techniques. Oxford University Press, New York, N.Y.
- 36.Rodríguez, J. M., F. Almazán, E. Viñuela, and J. F. Rodríguez. 1992. Genetic manipulation of African swine fever virus: construction of recombinant viruses expressing the β-galactosidase gene. Virology 188:239-250. [DOI] [PubMed] [Google Scholar]
- 37.Roullier, I., S. M. Brookes, A. D. Hyatt, M. Windsor, and T. Wileman. 1998. African swine fever virus is wrapped by the endoplasmic reticulum. J. Virol. 72:2373-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salas, J., M. L. Salas, and E. Viñuela. 1999. African swine fever virus: a missing link between poxviruses and iridoviruses?, p. 467-480. In E. Domingo, R. G. Webster, and J. J. Holland (ed.), Origin and evolution of viruses. Academic Press, London, United Kingdom.
- 39.Sanz, A., B. García-Barreno, M. L. Nogal, E. Viñuela, and L. Enjuanes. 1985. Monoclonal antibodies specific for African swine fever virus proteins. J. Virol. 54:199-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simón-Mateo, C., G. Andrés, and E. Viñuela. 1993. Polyprotein processing in African swine fever virus: a novel gene expression strategy for a DNA virus. EMBO J. 12:2977-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simon-Mateo, C., J. M. Freije, G. Andres, C. Lopez-Otin, and E. Vinuela. 1995. Mapping and sequence of the gene encoding protein p17, a major African swine fever virus structural protein. Virology 206:1140-1144. [DOI] [PubMed] [Google Scholar]
- 42.Simón-Mateo, C., G. Andrés, F. Almazán, and E. Viñuela. 1997. Proteolytic processing in African swine fever virus: evidence for a new structural polyprotein, pp62. J. Virol. 71:5799-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tulman, E. R., and D. L. Rock. 2001. Novel virulence and host range genes of African swine fever virus. Curr. Opin. Microbiol. 4:456-461. [DOI] [PubMed] [Google Scholar]
- 44.Viñuela, E. 1987. Molecular biology of African swine fever virus, p. 31-49. In Y. Becker (ed.), African swine fever. Nijhoff, Boston, Mass.
- 45.Yáñez, R. J., J. M. Rodríguez, M. L. Nogal, L. Yuste, C. Enriquez, J. F. Rodríguez, and E. Viñuela. 1995. Analysis of the complete nucleotide sequence of African swine fever virus. Virology 208:249-278. [DOI] [PubMed] [Google Scholar]