Abstract
The synthesis of the hepadnavirus relaxed circular DNA genome requires two template switches, primer translocation and circularization, during plus-strand DNA synthesis. Repeated sequences serve as donor and acceptor templates for these template switches, with direct repeat 1 (DR1) and DR2 for primer translocation and 5′r and 3′r for circularization. These donor and acceptor sequences are at, or near, the ends of the minus-strand DNA. Analysis of plus-strand DNA synthesis of duck hepatitis B virus (DHBV) has indicated that there are at least three other cis-acting sequences that make contributions during the synthesis of relaxed circular DNA. These sequences, 5E, M, and 3E, are located near the 5′ end, the middle, and the 3′ end of minus-strand DNA, respectively. The mechanism by which these sequences contribute to the synthesis of plus-strand DNA was unclear. Our aim was to better understand the mechanism by which 5E and M act. We localized the DHBV 5E element to a short sequence of approximately 30 nucleotides that is 100 nucleotides 3′ of DR2 on minus-strand DNA. We found that the new 5E mutants were partially defective for primer translocation/utilization at DR2. They were also invariably defective for circularization. In addition, examination of several new DHBV M variants indicated that they too were defective for primer translocation/utilization and circularization. Thus, this analysis indicated that 5E and M play roles in both primer translocation/utilization and circularization. In conjunction with earlier findings that 3E functions in both template switches, our findings indicate that the processes of primer translocation and circularization share a common underlying mechanism.
Hepadnaviruses are a family of DNA viruses whose primary site of replication is the liver (for reviews, see references 3 and 13). Each family member displays a narrow host range. Infections can be either acute or chronic. Liver disease is a common, but not obligatory, consequence of infection. It is thought that the host's immune response to the infection is a major contributor to the development of liver disease. Humans that are chronically infected with hepatitis B virus (HBV) are at an increased risk for the development of primary liver cancer, making HBV one of the leading causes of this malignancy worldwide.
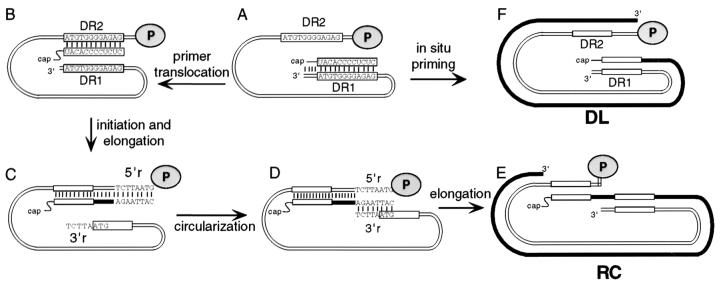
Hepadnaviruses replicate their genomes through reverse transcription of an RNA precursor (16). DNA replication occurs within the viral capsid in the cytoplasm of the infected cell. Only after synthesis of a significant portion of plus-strand DNA are capsids able efficiently to leave the cell as enveloped virions. Capsids with immature genomes inefficiently exit the cell as enveloped virions. Therefore, virus production is dependent on correct and efficient execution of each of the steps of reverse transcription within the infected cell. As with other reverse transcription schemes, template switching is integral to hepadnavirus DNA synthesis (5, 17). Template switching is the process in which the DNA strand undergoing synthesis switches from one template to another template. Sequence identity of the donor and acceptor templates provides the DNA strand undergoing the switch the opportunity to anneal. Complementarity ensures that DNA synthesis resumes at the correct location. The predominate end product of hepadnaviral genome replication is relaxed circular (RC) DNA. Three template switches are required for its synthesis: one during first, or minus-strand, synthesis (17) and two during second, or plus-strand, synthesis (5). The synthesis of the two strands of hepadnaviral DNA is sequential: only upon completion of the synthesis of minus-strand DNA does the synthesis of plus-strand DNA begin (16) (Fig. 1A). The primer for the initiation of plus-strand DNA synthesis is generated upon completion of minus-strand synthesis by the final RNase H cleavage of the initial RNA template by the viral reverse transcriptase (7) (Fig. 1A). This primer, which can be 18 or 19 nucleotides (nt), is derived from the 5′ terminus of the pregenomic RNA template (5). The sequence of the 12 nt at the 3′ end of the plus-strand primer is referred to as direct repeat 1 (DR1) because a second copy of these 12 nt, called DR2, is found at a second location within the genome. For most minus-strand templates, the primer switches positions before plus-strand synthesis begins (5). The primer translocates and presumably base pairs with the DR2 site, which is located within 50 nt of the 5′ end of the minus strand (Fig. 1B). For a minority of the minus-strand templates, the plus-strand primer is not translocated but instead is extended in situ leading to the synthesis of a duplex linear form of the genome (15) (Fig. 1F). Translocated primers initiate plus-strand synthesis from DR2 and are elongated to the 5′ end of the minus-strand template (Fig. 1C). Then the elongating plus-strand switches to use the 3′ end of the minus-strand as template (Fig. 1D). A short terminal redundancy of 7 or 8 nt on the minus strand, called r, serves as the donor and acceptor sequence for this process, called circularization (6, 10). Elongation of plus-strand DNA resumes and ultimately yields the mature relaxed circular DNA genome (Fig. 1E). Although the two template switches, primer translocation and circularization, utilize different donor and acceptor sequences, they share the general commonality of switching from one end of the template to the other end. This generalization begs the question whether these template switches share mechanisms.
FIG. 1.
Model for the synthesis of DHBV plus-strand DNA. Synthesis of plus-strand DNA begins after completion of synthesis of minus-strand DNA. (A) Full-length minus-strand DNA immediately prior to the initiation of synthesis of plus-strand DNA. Thin parallel lines represent minus-strand DNA. Gray oval (labeled P), viral P protein, which is covalently attached to the 5′ terminus of minus-strand DNA. The sequences of DR1 and DR2 are indicated within rectangles. Upon completion of synthesis of minus-strand DNA the final RNase H cleavage generates the plus-strand primer which is derived from the 5′ end of pregenomic RNA. The 3′ end of the primer contains the DR sequence. (B) Primer translocation. At least some portion of the 3′ end of the primer leaves the DR1 site and base pairs with the DR2 site. (C) Initiation of plus-strand DNA synthesis at DR2 and elongation to the 5′ end of minus-strand DNA. The minus-strand template is terminally redundant for 7 or 8 nt. 5′r and 3′r are the names of the terminal redundancies. (D) Circularization. The nascent plus-strand switches templates via complementarity between the 3′ end of the nascent plus-strand and minus-strands. (E) Elongation and completion of plus-strand DNA synthesis yields RC DNA. (F) In situ priming of plus-strand DNA synthesis generates DL DNA.
Studies of plus-strand synthesis for duck hepatitis B virus (DHBV) and heron hepatitis B virus (HHBV) have revealed a complex array of cis-acting sequence requirements (4, 11, 12, 15). In addition to the repeated donor and acceptor sequences, at least three other cis-acting sequences, M, 5E, and 3E, are required for plus-strand DNA synthesis (4). M is located near the middle of the minus strand while 5E and 3E are located near the 5′ and 3′ ends of the template, respectively (Fig. 2A). The mechanism of action of these sequences is not understood. The existence and location of these sequences suggests a complexity to the mechanism of template switching in addition to base pairing between the nascent strand and the acceptor template. To gain insights into the mechanism of the two plus-strand template switches, we have characterized variants of (DHBV) that contain mutations within these cis-acting sequences.
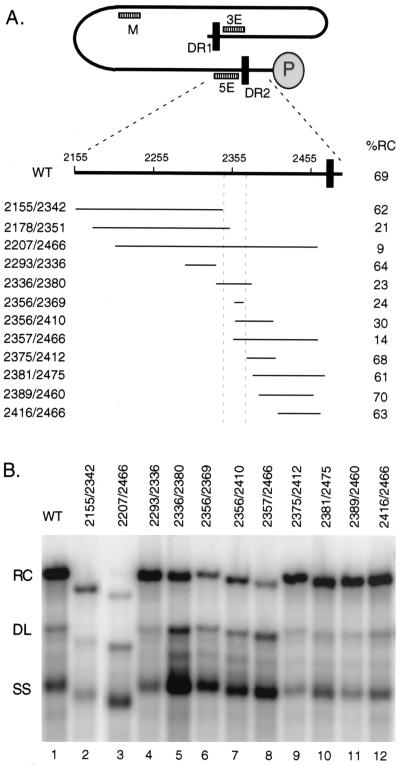
FIG. 2.
Determining 5′ and 3′ boundaries of 5E element. (A) Location of deletions relative to minus-strand DNA. In the top part of the panel, the black line represents full-length minus-strand DNA with DR1 and DR2 indicated with black rectangles. The gray oval labeled “P” represents the P protein which is covalently linked to the 5′ terminus of minus-strand DNA. General locations of 3E, M, and 5E cis-acting sequences are indicated. In the bottom part of the panel is an expanded view of the 5′ end of minus-strand DNA from nt 2155 to 2500. Thin lines represent the nucleotides removed in individual deletion variants. On the left is the name of each variant. The two numbers represent the first and last nucleotides of each deletion. On the right, the percentage of RC DNA synthesized by the wild type and each variant is shown. (B) Southern blot of deletion variants. Lane 1,wild-type (WT) comparison virus; lanes 2 through 12, individual variants as indicated. The positions of RC, DL, and SS DNA are indicated. The blot was hybridized with a genomic-length, minus-strand-specific probe.
We have more precisely mapped the location of the 5E sequence. In addition to localizing 5E to a 30-nt segment, we found that 5E contributes to both primer translocation/utilization and circularization. Characterization of several new M variants revealed the same phenotype, a partial defect in primer translocation/utilization, and a partial defect in circularization. These findings are consistent with the notion that the mechanisms of both plus-strand template switches have a common underlying feature. Last, we found that region M can function at an ectopic position in the genome.
MATERIALS AND METHODS
Molecular clones and nomenclature.
All molecular clones, except for one, 2178/2351, were derived from DHBV strain 3 (14). Variant 2178/2351 was derived from DHBV strain 16 and was made by Calvert and Summers (1). The DHBV3 plasmids contained 1.5 copies of the genome, thus being competent for the transcription of pregenomic RNA. Standard procedures were used to make these clones and details describing their construction will be provided upon request. All variant plasmids were sequenced over the relevant regions to establish the exact boundaries of the deleted sequences and to check that no unwanted mutations were present. The name of each variant incorporates the nucleotide positions of the beginning and end of the deletion. For example, variant 2336/2380 has nt 2336 through 2380 deleted.
Cell cultures, transfections, and isolation of viral DNA.
The cell line LMH (2) was used in all transfections. Culturing of LMH cells and their transfection was performed as described previously (8, 9). Viral DNA was extracted from cytoplasmic capsids described by Calvert and Summers (1).
Southern blot analysis.
Southern blotting of viral DNA was performed as previously described (4). Southern blotting of wild-type viral DNA isolated from intracellular capsids reveals three major forms. Each of these DNA forms contains a full-length minus strand (4). Two forms, RC and duplex linear (DL), have a full-length plus-strand initiating from DR2 or DR1, respectively. The third DNA form, called SS, is a full-length minus-strand DNA that is primarily, if not completely, single stranded. For wild-type virus, the three forms are found in characteristic proportions (e.g., see Tables 1 and 3). These proportions reflect the overall efficiency of the individual steps of plus-strand DNA synthesis. The data in Tables 1 and 3 were derived by measuring the levels of RC, DL, and SS DNA for a virus and expressing each of the three DNA forms as a percentage of the total. A deficiency in primer translocation or circularization will lead to a decrease in the proportion of RC DNA and an increase in DL DNA, SS DNA, or both.
TABLE 1.
5E analysis—proportions of replicative intermediates measured by Southern blotting
| DNA form | Mean % (SD) of indicated DNA form in virus varianta
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | 2155/2342 | 2178/2351 | 2207/2466 | 2293/2336 | 2336/2380 | 2356/2369 | 2356/2410 | 2357/2466 | 2375/2412 | 2381/2475 | 2389/2460 | 2416/2466 | |
| RC | 69 (5) | 62 (6) | 21 (5) | 9 (4) | 64 (6) | 23 (6) | 24 (5) | 30 (5) | 14 (3) | 68 (10) | 61 (6) | 70 (4) | 63 (8) |
| DL | 9 (2) | 9 (2) | 16 (4) | 24 (4) | 9 (1) | 21 (5) | 18 (3) | 15 (1) | 28 (5) | 10 (4) | 12 (2) | 8 (1) | 12 (3) |
| SS | 22 (4) | 29 (6) | 63 (9) | 67 (8) | 27 (6) | 56 (9) | 58 (6) | 55 (4) | 58 (7) | 22 (7) | 27 (7) | 22 (7) | 25 (6) |
Values are based on the following numbers of measurements made on intracellular viral DNA isolated from independent transfections: 29 for the wild type (WT), 17 for 2155/2342, 6 for 2293/2336, 15 for 2178/2351, 14 for 2207/2466, 5 for 2336/2380, 16 for 2357/2466, 4 for 2356/2410, 14 for 2356/2369, 16 for 2375/2412, 4 for 2381/2475, 7 for 2389/2460, and 18 for 2416/2466.
TABLE 3.
M analysis—proportions of replicative intermediates measured by Southern blotting
| DNA form | Mean % (SD) of indicated DNA form in virus varianta
|
||||
|---|---|---|---|---|---|
| WT | 724/832 | 721/799 | 799/833 | 798/842 | |
| RC | 69 (5) | 0 | 0 | 14 (1) | 14 (2) |
| DL | 9 (2) | 13 (3) | 14 (3) | 30 (3) | 38 (4) |
| SS | 22 (4) | 87 (3) | 86 (3) | 56 (3) | 48 (3) |
Values are base on the following numbers of measurements made on intracellular viral DNA isolated from independent transfections: 29 for wild type (WT), 9 for 724/832, 4 for 721/799, 5 for 799/833, and 6 for 798/842.
Primer extension analysis.
A detailed description of the primer extension analysis, its underlying rationale, and calculation of values derived from this analysis can be found in the work of Loeb and Tian (11). Briefly, the primer extension reaction used Vent Exo− thermostable DNA polymerase and 10 reaction cycles. Each sample was analyzed in three primer extension reactions. Inclusion of an internal standard in each sample was necessary for comparison of the level of viral DNA from different reactions. The first primer extension reaction was used to calculate the value C, which was the level of minus-strand DNA in a sample. In the analysis of the 5E variants, the reaction to derive the value C employed a primer that annealed to minus-strand DNA 112 nt from its 5′ end, while the analysis of the M variants used a primer 48 nt from the 5′ end of minus-strand DNA. From the second primer reaction the value B(DR2), which was the level of plus-strand DNA that had initiated from DR2 and had extended at least to the 5′ end of the minus-strand template, was derived. This reaction employed a primer derived from the first 18 nt of minus-strand DNA. The third reaction gave rise to two values, A(DR2) and A(DR1). The value A(DR2) represented the level of plus-strand DNA initiating from DR2 that had successfully circularized and elongated an additional 85 nt. The A(DR1) values represented the level of plus-strand initiation from DR1 (in situ priming). This primer annealed to plus-strand DNA 76 nt from DR1 and 85 nt from the circularization point. From these four primary values a series of ratios was calculated that reflected the efficiency of various events during the synthesis of plus-strand DNA.
RESULTS
Aims and strategies.
Previously, a scanning of the DHBV genome revealed the presence of three cis-acting sequences, named 5E, M, and 3E, that were required for synthesis of RC DNA (4) (general locations relative to minus-strand DNA are indicated in Fig. 2A). All three sequences were found to act during the synthesis of plus-strand DNA, but the mechanism(s) by which they made their contributions was not clear. The aim of this study was to understand better the mechanism(s) by which these sequences contributed to the synthesis of plus-strand DNA. To map more precisely the locations of the 5E and M sequences, we constructed and analyzed new variant viruses that contained small deletions. Because 3E had already been localized (4) to the 11 nt between DR1 and the RNA packaging signal, epsilon, we did not pursue further mapping of 3E. Because 5E and M lie within the P gene, our deletions would likely disrupt P protein function. Therefore, all variants were made in a genetic background that was null for the P protein by virtue of a frameshift mutation near the 5′ end of the P gene (4). Therefore, the wild-type comparison in our studies contained the same P-null mutation. Thus, the molecular clones of the variants were capable of expressing pregenomic RNAs that were templates for reverse transcription and mRNAs for C protein translation. To enable DNA replication it was necessary to provide wild-type P protein which was expressed from a different plasmid and due to a cis defect in encapsidation did not serve as a template for DNA synthesis (4). Plasmid DNAs were cotransfected into LMH cells and after 3 days viral DNA was extracted from cytoplasmic capsids. Viral DNA was analyzed by Southern blotting and primer extension to evaluate the ability of the variants to support RC DNA synthesis.
Mutagenesis of 5E reveals that in addition to primer translocation/utilization, circularization was also affected.
The previous analysis indicated the presence of a cis-acting sequence, named 5E, within 200 nt of the 5′ end of the minus-strand DNA that is required for the synthesis of RC DNA for DHBV (4). Based on the endpoints of three deletion mutants, 5E was localized to the sequence between nt 2343 and 2466, although its precise boundaries were not determined. (For reference, DR2 is between nt 2477 and 2488 and the 5′ end of minus-strand DNA is at nt 2537.) Primer extension analysis of a mutant that had nt 2207 to 2466 deleted (called 2207/2466) indicated a significant reduction in the level of plus-strand DNA primed from DR2. The magnitude of this reduction prohibited the evaluation of the subsequent circularization step. In addition, mutant 2207/2466 had only a two- to threefold increase in the level of plus-strand DNA that was primed in situ from DR1. Thus, disruption of 5E function had two demonstrable effects, a significant lack of utilization of primers normally destined for use at DR2 and a modest increase in in situ priming.
To determine more precisely the location of 5E, several new deletion mutants were analyzed. The structures of these deletion mutants are depicted in Fig. 2A. Results of Southern blotting of these mutants localized 5E to the sequence between nt 2342 and 2374 (Fig. 2B and Table 1). This determination placed 5E approximately 100 nt from DR2. The nucleotides between 5E and DR2 could be deleted with little or no impact on RC DNA synthesis (Fig. 2B, lanes 9 to 12, and Table 1). The Southern blotting analysis of the 5E mutants showed a reduction in the proportion of RC DNA, with modest increases in the proportion of DL DNA and larger increases in the proportion of SS DNA. The increase in the proportion of DL DNA in Southern blot analyses was due to increased in situ priming. Two possibilities for the increase in SS DNA seemed likely: (i) there is a defect in plus-strand primer translocation/utilization that would lead to an increase in full-length minus-strand DNA that did not contain plus-strand DNA or (ii) there is a defect in the template switch that circularizes the genome, which would lead to an increase in an SS DNA intermediate that contained a 50-nt segment of plus-strand DNA. Analyses based on primer extension can determine whether a mutant virus suffers a defect in primer translocation/utilization, circularization, or both (4, 11). This analysis is additionally informative because the magnitude of each type of defect can be determined. To this end, several of the 5E mutants were analyzed by primer extension as described previously (11).
The results from the primer extension analysis of the new 5E variants, summarized in Table 2, indicated two points. First, the primer extension results, in general, agreed with the Southern blotting results (compare Tables 1 and 2). A decrease in the proportion of RC DNA seen by Southern blotting was paralleled with a decrease in the level of plus-strand DNA primed from DR2 and circularized as measured by primer extension. The increase in DL DNA measured in Southern blotting was similar to the increase in in situ priming measured by primer extension. Secondly, the primer extension results indicated that the new 5E variants were partially defective for primer translocation/utilization. For these mutants, a reduced but detectable amount of priming from DR2 was seen (Table 2). In addition, of the plus strands that did initiate from DR2, a reduced fraction, relative to that for the wild type, was competent for circularization (Table 2). These results indicated that the cis-acting sequence 5E contributed to the process of primer translocation/utilization and to the process of circularization. This finding raised the question of whether the cis-acting sequence M, which had previously been shown to contribute to primer translocation/utilization (4), also contributed to circularization.
TABLE 2.
5E analysis—efficiency of plus-strand DNA synthesis as determined by primer extension
| Plus-strand eventb | Steps (as shown in Fig. 1) | Mean % efficiency (SD) of indicated event in virus varianta
|
|||
|---|---|---|---|---|---|
| WT | 2178/2351 | 2356/2369 | 2356/2410 | ||
| Priming from DR2c | A to C | 71 (7) | 30 (9) | 39 (4) | 42 (4) |
| Priming from DR2 and circularizationd | A to E | 55 (8) | 11 (2) | 12 (1) | 12 (1) |
| Circularizatione | C to E | 77 | 37 | 31 | 29 |
| In situ primingf | F | 5 (1) | 12 (5) | 10 (2) | 7 (1) |
| Primer utilization (total)g | (A to C) + F | 76 | 42 | 49 | 49 |
Values are based on the following numbers of measurements made with intracellular viral DNA isolated from independent transfections: wild type (WT), 8; 2178/2351, 3; 2356/2369, 6; 2356/2410, 4.
See Materials and Methods for explanation of the measured values.
B(DR2)/C value.
A(DR2)/C value.
A(DR2)/B(DR2) value.
A(DR1)/C value.
Priming from DR2 plus in situ priming value.
The cis-acting sequence M contributes to primer translocation/utilization and circularization.
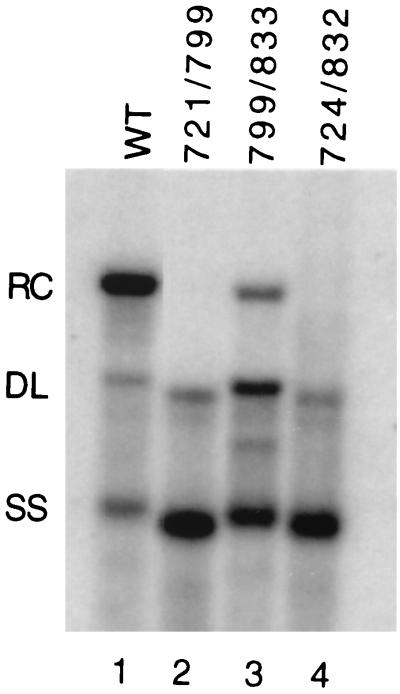
The previous analysis showed that a cis-acting sequence located within the middle of the minus-strand, named M, contributes to the synthesis of plus-strand DNA (4, 12). A mutant virus, with nt 724 to 832 deleted (called 724/832) was found to be significantly deficient in primer translocation/utilization, while a virus with a deletion of nt 832 to 916 appeared normal in its ability to synthesize RC DNA (4). In an attempt to localize the active nucleotides within region M, we characterized two new variants, 721/799 and 799/833, which bisect the original 724/832 deletion. We found that both new variants had mutant phenotypes as judged by Southern blotting (Fig. 3 and Table 3). The phenotype of 721/799 was very similar to the originally described 724/832 variant (Fig. 3, lanes 2 and 4). RC DNA levels were below the limits of detection, and the proportion of DL DNA was 1.5-fold greater than in the wild type, while SS DNA accumulated to high proportions (Table 3). The phenotype of the 799/833 variant was different than that of the 721/799 variant in two respects. It made a higher proportion of DL DNA and it was only partially inhibited for RC DNA synthesis (Table 3).
FIG. 3.
Southern blotting of region M variants. Lane 1, wild-type (WT) comparison; lane 2, deletion variant with nt 721 to 799 removed; lane 3, deletion variant with nt 799 to 833 removed; lane 4, deletion variant with nt 724 to 832 removed. The positions of RC, DL, and SS DNA are indicated. The blot was hybridized with a genomic-length, minus-strand-specific probe.
Next, we performed the primer extension analysis on the 721/799 and 799/833 variants to evaluate whether they were partially defective for primer utilization and circularization, as are the 5E variants. A summary of this analysis is presented in Table 4. (Four of the five values for wild-type virus in Table 4 are lower than the wild-type values reported in Table 2. These four values were all normalized to the level of minus-strand DNA. Different primers were used to measure the level of minus-strand DNA in Tables 2 and 4. The primer used in the analysis presented in Table 4 was 48 nt from the minus-strand 5′ end [primer 2489+], while the primer used in Table 2 was 112 nt away [primer 2425+]. We think that the difference in the wild-type values in Tables 2 and 4 is due to the use of different primers in the measurement of minus-strand DNA.) The level of plus-strand priming from DR2 for the 721/799 virus was significantly reduced relative to that of the wild-type virus (Table 4). This was accompanied by a small increase in in situ priming (Table 4). Clearly, the 721/799 virus had a significant defect in primer utilization. In addition, for the 721/799 variant the plus strands initiating from DR2 were not circularizing with the same efficiency as the wild type (Table 4). As with the 5E variant viruses, 721/799 suffered a partial defect in both primer translocation/utilization and circularization, resulting in a significant reduction in RC DNA synthesis. The analysis of the 799/833 virus indicated similar deficiencies, except that the magnitude of the effect differed from that of 721/799. 799/833 had a circularization defect that was similar in magnitude to that of 721/799. Relative to the wild type, it supported reduced levels of priming from DR2. But 799/833 had a fourfold increase, relative to the wild type, in in situ priming (Table 4). Thus, for 799/833, total primer utilization was only slightly less than that of the wild type.
TABLE 4.
M analysis—efficiency of plus-strand DNA synthesis as determined by primer extension
| Plus-strand eventb | Steps(as shown in Fig. 1) | Mean % efficiency (SD) of indicated event in virus varianta
|
||
|---|---|---|---|---|
| WT | 721/799 | 799/833 | ||
| Priming from DR2c | A to C | 44 (12) | 8 (1) | 19 (5) |
| Priming from DR2 and circularizationd | A to E | 39 (9) | 2 (1) | 6 (1) |
| Circularizatione | C to E | 87 | 25 | 32 |
| In situ primingf | F | 4 (2) | 8 (2) | 16 (3) |
| Primer utilization (total)g | (A to C) + F | 48 | 16 | 35 |
Values are based on the following numbers of measurements made with intracellular viral DNA isolated from independent transfections: wild type (WT), 6; 721/799, 4; 799/833, 4.
See Materials and Methods for explanation of the measured values.
B(DR2)/C value.
A(DR2)/C value.
A(DR2)/B(DR2) value.
A(DR1)/C value.
Priming from DR2 plus in situ priming value.
Region M can function at an ectopic location.
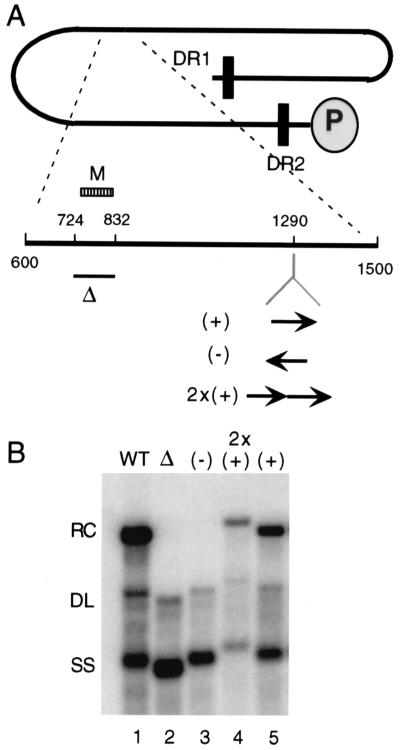
When the sequence between nt 724 and 832 is deleted from DHBV, very little RC DNA is made (4) (Fig. 4B, lane 2). To determine whether M can function at a position in the genome other than its natural location, we reintroduced M, within the context of the original 724/832 deletion, into the unique KpnI site at nt 1290 (Fig. 4A). When the fragment containing nt 724 to 832 was reintroduced in an opposite orientation, the Southern blot profile of the resultant virus was very similar to that for the original 724/832 deletion, that is, the synthesis of very little, if any, RC DNA and an increase in the proportion of SS DNA (Fig. 4B, lane 3). But when this fragment was inserted in its natural orientation at nt 1290, a substantial restoration in RC DNA synthesis was achieved (Fig. 4B, lane 5). Introduction of two copies of the sequence did not result in a further increase in the proportion of RC DNA (Fig. 4B, lane 4). We did not perform a parallel analysis on the 5E sequence.
FIG. 4.
Region M element can function at an ectopic site in the genome. (A) Diagram of full-length minus-strand DNA with expanded view of nt 600 to 1500. The position of the 724/832 deletion is indicated with a bar (Δ). The deleted fragment was reintroduced at the KpnI site at nt 1290. (+), virus with a single copy of the deleted fragment inserted into the KpnI site in the sense orientation; (−), virus with a single copy of the deleted fragment inserted into the KpnI site in the opposite orientation; 2x(+), virus with tandem copies of the deleted fragment inserted into the KpnI site in the sense orientation. (B) Southern blot of variants. The positions of RC, DL, and SS DNA are indicated. The blot was hybridized with a genomic-length, minus-strand-specific probe.
DISCUSSION
We present evidence that the DHBV 5E cis-acting element is located within a short sequence of approximately 30 nt that is 100 nt 3′ of DR2 on minus-strand DNA. More significantly, we found that viruses with 5E mutations are partially defective for plus-strand primer translocation/utilization and for circularization. In a similar fashion, we found that mutations within region M are partially defective for primer translocation/utilization and for circularization. Lastly, we found that region M sequences can function at an ectopic site in the DHBV genome in an orientation-dependent manner.
Our studies indicate that 5E and M make contributions to two processes, primer translocation/utilization and circularization, during plus-strand DNA synthesis. By affecting primer utilization, primers that normally are destined for translocation to DR2 are not used efficiently at DR2 or DR1. This leads to the accumulation of an SS DNA intermediate that is not associated with plus-strand DNA. But the defect in primer translocation/utilization for our 5E and M mutant viruses is only partial. Only some of the capsids in cells expressing these mutants are deficient for this step. Other capsids successfully carry out primer translocation. A subset of these capsids is then defective for the next template switch, circularization. The net sum of these partial defects in primer translocation and circularization is the observed decrease in the levels of RC DNA. Temporally, primer translocation and circularization occur at different times, yet our analysis indicates that 5E and M make contributions to both template switches. We think that these results indicate that the mechanisms of primer translocation and circularization share a common component. The exact nature of this component and the overall mechanism is not clear from the present data set. A possibility is that 5E and M contribute to plus-strand DNA synthesis by juxtaposing the ends of the minus-strand DNA, which is where the donor and acceptor sites for the two template switches are located.
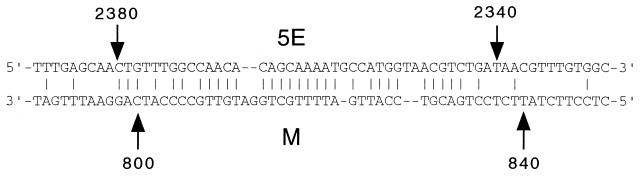
Our mapping studies have localized 5E to the vicinity of nt 2340 to 2380. Inspection of the nucleotide sequence of DHBV3 within these 5E boundaries and the M boundaries revealed the potential for a partial duplex to form between these sequences (Fig. 5). The sequences of heron hepatitis B virus (GenBank accession number M22056) and Ross Goose hepatitis B virus (GenBank accession number M95589) also have the potential to form similar but not identical patterns of base pairs (data not shown). Whether 5E and M function by forming an imperfect duplex remains to be determined.
FIG. 5.
Predicted base pairing pattern between minus-strand sequences of 5E and M of DHBV3. Nucleotide coordinates are indicated.
It is worth noting that mutations within 5E or M that lead to a failure to initiate plus-strand DNA synthesis at DR2 do not result in a concomitant increase in priming from DR1, indicating that lack of priming from DR2 does not automatically result in priming from DR1. Either the mechanism of regulation of in situ priming is distinct from the mechanism of action of 5E and M or the mechanism by which 5E and M positively contributes to priming from DR2 and circularization also positively influences the efficiency of in situ priming.
In our initial characterization of regions 5E and M, we analyzed two mutants with either nt 724 to 832 (region M) or nt 2207 to 2466 (region 5E) deleted (4). We reported that both of these mutants suffered large defects in primer utilization such that evaluating their ability to perform circularization was difficult. In our present analysis we studied these mutants again (data not shown). Our ability to carry out our quantitative primer extension analysis (11) has allowed us to conclude now that the 724/832 and 2207/2266 mutants are also partially defective for both primer translocation/utilization and circularization.
Acknowledgments
We thank Ru Tian for excellent technical assistance and Bill Sugden for critical review of the manuscript.
This work was supported by NIH grants R29 GM50263, P01 CA22443, P30 CA07175, and T32 CA09135 and ACS grant JFRA-651.
REFERENCES
- 1.Calvert, J., and J. Summers. 1994. Two regions of an avian hepadnavirus RNA pregenome are required in cis for encapsidation. J. Virol. 68:2084-2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Condreay, L. D., C. E. Aldrich, L. Coates, W. S. Mason, and T. T. Wu. 1990. Efficient duck hepatitis B virus production by an avian liver tumor cell line. J. Virol. 64:3249-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ganem, D. 1996. Hepadnaviridae and their replication, p. 2703-2737. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 4.Havert, M. B., and D. D. Loeb. 1997. cis-acting sequences in addition to donor and acceptor sites are required for template switching during synthesis of plus-strand DNA for duck hepatitis B virus. J. Virol. 71:5336-5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lien, J.-M., C. E. Aldrich, and W. S. Mason. 1986. Evidence that a capped oligoribonucleotide is the primer for duck hepatitis B virus plus-strand DNA synthesis. J. Virol. 57:229-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lien, J. M., D. J. Petcu, C. E. Aldrich, and W. S. Mason. 1987. Initiation and termination of duck hepatitis B virus DNA synthesis during virus maturation. J. Virol. 61:3832-3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loeb, D. D., R. C. Hirsch, and D. Ganem. 1991. Sequence-independent RNA cleavages generate the primers for plus strand DNA synthesis in hepatitis B viruses; implication for other reverse transcribing elements. EMBO J. 10:3533-3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loeb, D. D., and R. Tian. 1995. Transfer of the minus strand DNA during hepadnavirus replication is not invariable but prefers a specific location. J. Virol. 69:6886-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loeb, D. D., R. Tian, and K. J. Gulya. 1996. Mutations within DR2 independently reduce the amount of both minus- and plus-strand DNA synthesized during hepatitis B virus replication. J. Virol. 70:8684-8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loeb, D. D., K. J. Gulya, and R. Tian. 1997. Sequence identity of the terminal redundancies on the minus-strand DNA template are necessary but not sufficient for the template switch during hepadnaviral plus-strand DNA synthesis. J. Virol. 71:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loeb, D. D., and R. Tian. 2001. Mutations that increase in situ priming also decrease circularization for duck hepatitis B virus. J. Virol. 75:6492-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mueller-Hill, K., and D. D. Loeb. 1996. Previously unsuspected cis-acting sequences for DNA replication revealed by characterization of a chimeric heron/duck hepatitis B virus. J. Virol. 70:8310-8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seeger, C., and W. S. Mason. 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64:51-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sprengel, R., C. Kuhn, H. Will, and H. Schaller. 1985. Comparative sequence analysis of duck and human hepatitis B virus genomes. J. Med. Virol. 15:323-333. [DOI] [PubMed] [Google Scholar]
- 15.Staprans, S., D. D. Loeb, and D. Ganem. 1991. Mutations affecting hepadnavirus plus-strand DNA synthesis dissociate primer cleavage from translocation and reveal the origin of linear viral DNA. J. Virol. 65:1255-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Summers, J., and W. S. Mason. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29:403-415. [DOI] [PubMed] [Google Scholar]
- 17.Wang, G.-H., and C. Seeger. 1993. Novel mechanism for reverse transcription in hepatitis B viruses. J. Virol. 67:6507-6512. [DOI] [PMC free article] [PubMed] [Google Scholar]