Abstract
The UL36 open reading frame encoding the tegument protein ICP1/2 represents the largest open reading frame in the genome of herpes simplex virus type 1 (HSV-1). Polypeptides homologous to the HSV-1 UL36 protein are present in all subfamilies of Herpesviridae. We sequenced the UL36 gene of the alphaherpesvirus pseudorabies virus (PrV) and prepared a monospecific polyclonal rabbit antiserum against a bacterial glutathione S-transferase (GST)-UL36 fusion protein for identification of the protein. The antiserum detected a >300-kDa protein in PrV-infected cells and in purified virions. Interestingly, in coprecipitation analyses using radiolabeled infected-cell extracts, the anti-UL36 serum reproducibly coprecipitated the UL37 tegument protein, and antiserum directed against the UL37 protein coprecipitated the UL36 protein. This physical interaction could be verified using yeast two-hybrid analysis which demonstrated that the UL37 protein interacts with a defined region within the amino-terminal part of the UL36 protein. By use of immunogold labeling, capsids which accumulate in the cytoplasm in the absence of the UL37 protein (B. G. Klupp, H. Granzow, E. Mundt, and T. C. Mettenleiter, J. Virol. 75:8927-8936, 2001) as well as wild-type intracytoplasmic and extracellular virions were decorated by the anti-UL36 antiserum, whereas perinuclear primary enveloped virions were not. We postulate that the physical interaction of the UL36 protein, which presumably constitutes the innermost layer of the tegument (Z. Zhou, D. Chen, J. Jakana, F. J. Rixon, and W. Chiu, J. Virol. 73:3210-3218, 1999), with the UL37 protein is an important early step in tegumentation during virion morphogenesis in the cytoplasm.
Infectious herpesvirus particles contain more than 30 virus-encoded proteins which are assembled into the four morphologically differentiable components of the herpesvirus virion: the inner nucleoprotein core containing the double-stranded DNA genome, the icosahedral capsid shell, the tegument located between the capsid and envelope, and the lipid envelope containing viral (glyco)proteins (reviewed in reference 31). The requirements and molecular interactions which result in formation of herpesvirus capsids are well characterized (reviewed in references 16 and 36). However, much less is known about the molecular details of tegumentation or envelopment. It is now generally accepted that herpesvirus particles mature via an envelopment-deenvelopment-reenvelopment pathway (reviewed in reference 28). Intranuclear capsids bud through the inner nuclear membrane, thereby acquiring a primary envelope and, presumably, also a primary tegument (17). This budding process requires the presence of the products of the UL31 and UL34 genes of herpes simplex virus type 1 (HSV-1) (6, 30, 32) and pseudorabies virus (PrV) (14, 20). Primary enveloped virions differ in morphology (15, 17) and biochemical composition (20, 35) from mature virus particles. This can be explained by loss of the primary envelope and at least part of the primary tegument by fusion of the primary envelope with the outer leaflet of the nuclear membrane and by translocation of capsids into the cytoplasm. Final tegumentation then occurs in the cytoplasm, and capsids acquire a final envelope by budding into trans-Golgi vesicles (reviewed in references 13 and 28).
Although the overall pathway of herpesvirus virion maturation appears clear, the molecular details are largely unknown. For example, in HSV-1, more than 15 proteins have been hypothesized or demonstrated to be components of the tegument, and more than 11 virally encoded proteins reside in the virion envelope (reviewed in references 28 and 36). How these complex structures are assembled into a functional virus particle is currently under intense study. Whereas herpesvirus capsids exhibit icosahedral symmetry, it has long been assumed on the basis of electron microscopic evidence that the tegument is a largely unstructured virion component. However, recent cryoelectron microscopic data demonstrated that at least the innermost part of the tegument which is in contact with the capsid displays some symmetry in that contact points are located at and around the vertices of the capsid (38). It has been hypothesized that the interaction between tegument and capsid shell involves the largest protein found in herpesviruses, the product of the HSV-1 UL36-homologous genes (26, 38). Indeed, some time ago it had been proposed that the HSV-1 UL36 protein physically interacts with the major capsid protein (26). Lack of the UL36 protein resulted in accumulation of unenveloped HSV-1 capsids in the cytoplasm (11). An increase in the presence of unenveloped HSV-1 capsids in the cytoplasm was also observed in the absence of the UL37 protein (12), another tegument component (25, 33, 34). We recently showed that in cells infected with a UL37 deletion mutant of PrV, aggregations of capsids exhibiting an ordered arrangement were present in the cytoplasm (21). Capsids appeared not to contact each other directly but via extensions which seemed to emanate from the vertices of the capsids. We hypothesized that in the absence of the UL37 protein, the UL36 gene product is deposited onto the capsid but further tegumentation is blocked. Formation of aggregates may thus be caused by aberrant UL36-UL36 interaction. In this scenario, a possible physical interaction between the UL37 and UL36 proteins would constitute an important early step in tegumentation.
In this report we identify the PrV UL36 protein and demonstrate that it is present in the intracytoplasmic capsid aggregates formed in the absence of UL37 but that it is not detectable in perinuclear primary enveloped virions. Moreover, we show that the UL36 and UL37 proteins do indeed physically interact.
Sequence of the PrV UL36 gene
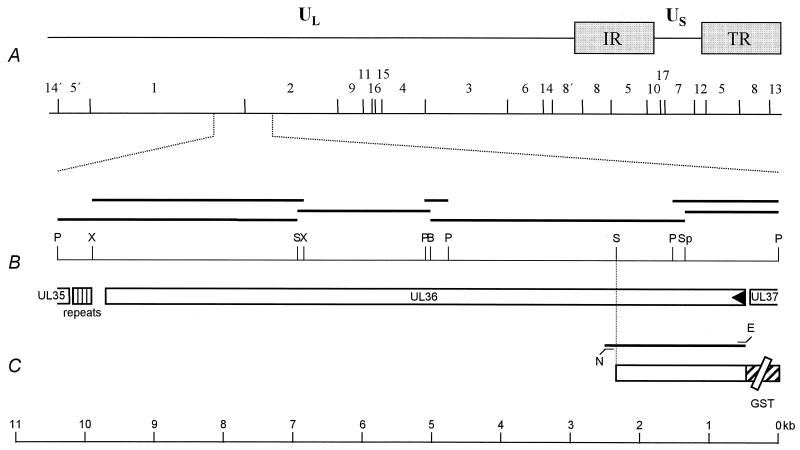
The 5′ part of the UL36 sequence has been deposited in the EMBL database under accession no. AJ318065 (21). To complete the UL36 sequence, the adjacent 3.7-kb SphI/BamHI fragment (Fig. 1) of the PrV-Ka strain (19) was cloned and subjected to nested deletion reaction, and nested deletion clones were sequenced. An overlapping 1.5-kb PstI fragment was sequenced to verify the region spanning the SphI site. The 1.9-kb BamHI/SalI and 3.5-kb SalI/PstI subfragments of BamHI fragment 1 were also cloned, subjected to nested deletion reactions, and sequenced (Fig. 1). To verify the sequence spanning the BamHI site, a 0.3-kb PstI fragment was cloned and sequenced, as well as a 3.0-kb XcmI fragment that encompasses the SalI site (Fig. 1). Sequencing was performed on double-stranded plasmid DNA using the Sequenase-7-deaza-dGTP Sequencing Kit (Amersham Pharmacia Biotech, Freiburg, Germany). Each nucleotide was sequenced at least twice on each DNA strand. The now completed PrV UL36 gene sequence comprises 9,255 bp (nucleotides [nt] 470 to 9724 [GenBank accession no. AJ422133]) with a GC content of 76% encoding a 3,084-amino-acid (aa) protein with a predicted molecular mass of 324 kDa. A putative TATA box is located at nt 427 to 435 and overlaps the poly(A) signal of the UL37 gene. The poly(A) signal of the UL36 open reading frame (ORF) is located at nt 9723 to 9728 and overlaps the stop codon at nt 9722 to 9724. Downstream from the UL36 ORF, there is a GT cluster at nt 9764 to 9771. The PrV UL36 protein is homologous to the corresponding proteins of equine herpesvirus 1 (gene 24) (37), HSV-1 (24), and varicella-zoster virus (gene 22) (10), with 41, 39, and 37% identical amino acids, respectively. Although overall amino acid identity to homologous proteins of beta- (7) and gammaherpesviruses (1) is low, all UL36 homologs except that of Epstein-Barr virus (gene BPLF1) (1) contain leucine zipper motifs which in the PrV homolog are located at aa 779 to 800 and 827 to 848, indicating potential for protein-protein interactions. In the PrV UL36 protein, these leucine zipper motifs are present downstream from the UL37 binding domain (aa 312 to 398; see below). Moreover, all UL36 homologs contain at least one consensus sequence for N-linked glycosylation which is located at aa 1744 to 1746 in the PrV protein. Downstream from the gene is a region of repeated sequences from nt 9905 to 10106. Within this region, there are 21 imperfect 9- or 10-mer repeats of the sequence 5′-GGGGGACTT-3′ or, alternatively, of the sequence 5′-GGGGGGCATT-3′ or 5′-GGGGGGATT-3′. The functional importance of this region is unknown. On the complementary strand, the stop codon of the UL35 ORF is located at nt 10234 to 10232, with a poly(A) signal present at nt 10219 to 10214. For an overall outline of this genomic region, see Fig. 1.
FIG. 1.
Location of the PrV UL36 gene. (A) Diagram of the PrV genome shown above a BamHI restriction fragment map (27). The PrV genome is divided into unique long (UL) and unique short (US) regions by internal and terminal repeats (IR and TR, respectively). (B) Enlarged view of the relevant portion of the genome. The locations of the ORFs are shown, and the transcriptional orientation is indicated by the arrowhead. Fragments used for sequencing are indicated by the black bars. The location of the repeated sequence elements separating the UL36 and UL35 ORFs is also indicated. Only relevant cleavage sites are indicated (B, BamHI; E, EcoRI; N, NotI; P, PstI; S, SalI; Sp, SphI, X, XcmI). (C) Construct used for expression of part of UL36 as a GST-UL36 fusion protein (GST part not drawn to scale). At the bottom, the scale for panels B and C is given in kilobase pairs (kb).
Identification and expression kinetics of the PrV UL36 protein
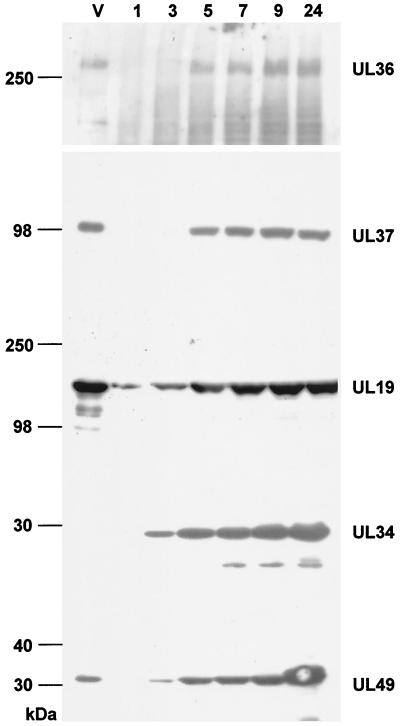
Part of the UL36 ORF was amplified by PCR using primers UL36FOR2 (5′-CACAGAATTCATTTCAGCCATGACGGCCGACG-3′, located at nt 461 to 482 [GenBank accession no. AJ422133], with the UL36 start codon shown in bold type and the EcoRI site introduced for convenient cloning shown in italic type) and UL36REV2 (5′-CACAGCGGCCGCGTGCGCCTGCGCCTCGGC-3′, located at nt 2500 to 2483 [GenBank accession no. AJ422133], with the NotI site introduced for convenient cloning in italics). From this 2.0-kb PCR product, a 1.9-kb EcoRI/SalI fragment encoding aa 1 to 624 of the UL36 protein was subcloned in pGEX-4T-1 (Fig. 1). The ca. 95-kDa GST-UL36 fusion protein was excised after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and used to immunize a rabbit. Generally, immunization was performed five times at 4-week intervals with 100 μg of fusion protein in Freund's incomplete adjuvant. For the first immunization, Freund's complete adjuvant was used and the period between first and second immunization was 6 weeks. Using this monospecific antiserum, a >300-kDa protein was specifically recognized in lysates of PrV-infected cells after SDS-PAGE and Western blotting (Fig. 2). First detectable at 5 h after infection, it increased in amount until 24 h after infection. Expression kinetics was similar to that seen for the UL37 tegument protein (21), whereas other virion constituents, such as the UL34 primary envelope protein (20) and the UL49 tegument protein (4), were already observed at 3 h postinfection. In contrast, the UL19 major capsid protein was detectable as early as 1 h after infection.
FIG. 2.
Identification and kinetics of expression of the PrV UL36 protein. RK13 cells were infected at an MOI of 10 with PrV-Ka and harvested at the time points (hours postinfection) shown above the lanes. Cell lysates were separated by SDS-PAGE on either 5% (for demonstration of UL36 or UL37) or 10% acrylamide gels (for demonstration of UL19, UL34, or UL49). After Western blotting, the separated cell lysates were incubated with polyclonal sera directed against the UL36 protein (diluted 1:105 [this study]), the UL37 protein (diluted 1:105 [21]), the UL19 protein (diluted 1: 2 × 105 [20]), the UL34 protein (diluted 1:105 [20]), or the UL49 protein (diluted 1:105 [4]). Lanes V contained lysates of sucrose gradient-purified PrV-Ka virions. The positions of molecular mass standards (in kilodaltons) are shown to the left of the gel.
The PrV UL36 protein is a component of extracellular virions
To analyze for presence of the UL36 protein in mature extracellular virus particles, virions were purified from the supernatant of infected cells by sucrose-gradient centrifugation (20) and lysed, and viral proteins were separated in SDS-10% PAGE. For separation of high molecular weight proteins, SDS-PAGE with 5% acrylamide in the separating gel was performed. Transfer onto polyvinylidene difluoride membranes (Schleicher & Schüll, Dassel, Germany) was done at 50V for 2 h. As shown in Fig. 2, lanes V, the UL36 protein was readily detectable in extracellular virions as were the UL19 major capsid, and UL37 and UL49 tegument proteins (5, 21). In contrast, the UL34 protein which is a constituent of primary enveloped virions in the perinuclear space but is absent from mature virus particles (20) was not present. In conclusion, the UL36 protein is a constituent of extracellular PrV particles.
Coimmunoprecipitation of UL36 and UL37 proteins
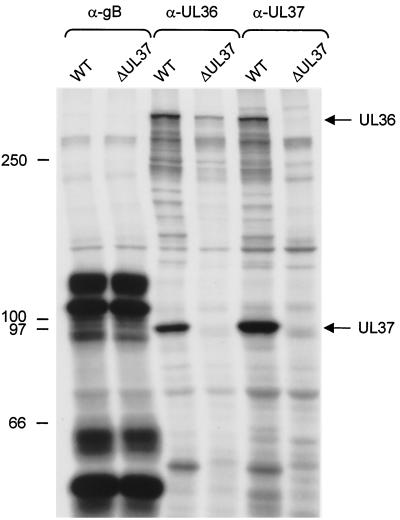
To further characterize the UL36 protein, RK13 cells were infected at a multiplicity of infection (MOI) of 10 with wild-type PrV-Ka (Fig. 3, lanes WT), which had been radiolabeled with [35S] methionine/cysteine (100 μCi/ml) (Tran-S35-Label; ICN, Eschwege, Germany) from 2 to 24 h postinfection, and cleared lysates were precipitated (23) with monoclonal antibody (MAb) against glycoprotein B (diluted 1:20) (29) or monospecific antiserum directed against the UL36 protein (diluted 1:100) or the UL37 protein (diluted 1:100) (21) (Fig. 3). As shown in Fig. 3, the anti-UL36 serum precipitated a protein with a molecular mass of >300 kDa, which corresponds to the size of the UL36 gene product as deduced from the sequence and as judged from Western blotting (see above). Surprisingly, in addition to several minor proteins of different sizes, the anti-UL36 serum also precipitated a prominent protein of ca. 100 kDa. This protein comigrated with the UL37 protein as precipitated by the anti-UL37 serum which, however, also precipitated the >300-kDa protein. Apart from the difference in intensity of the UL36- and UL37-specific signals, both precipitations were nearly identical. That the 100-kDa protein indeed represents the UL37 gene product was verified by its absence in PrV-ΔUL37-infected cells (Fig. 3). Whereas in the absence of the UL37 protein the UL36 gene product was not precipitated by the anti-UL37 serum, the UL36 protein was still precipitated by the anti-UL36 serum in the absence of the UL37 gene product. Thus, reciprocal coprecipitation indicated that the UL36 and UL37 proteins of PrV physically interact to form a complex. As a control, the 110-kDa nonglycosylated precursor as well as the glycosylated uncleaved 120-kDa precursor of gB and the cleaved ca. 55- and 65-kDa subunits were precipitated in equal amounts from cells infected with either virus.
FIG. 3.
Coimmunoprecipitation of UL36 and UL37 proteins. Lysates from metabolically radiolabeled RK13 cells infected with PrV-Ka (wild type [WT]) or PrV-ΔUL37 were precipitated with the UL36-specific antiserum (α-UL36), the UL37-specific antiserum (α-UL37), or a MAb directed against gB (α-gB). Precipitates were analyzed by SDS-5% PAGE under reducing conditions, and labeled protein was visualized by fluorography. The positions of the UL36 and UL37 proteins (to the right of the gel) and molecular mass markers (to the left of the gel) are shown.
Yeast two-hybrid analysis
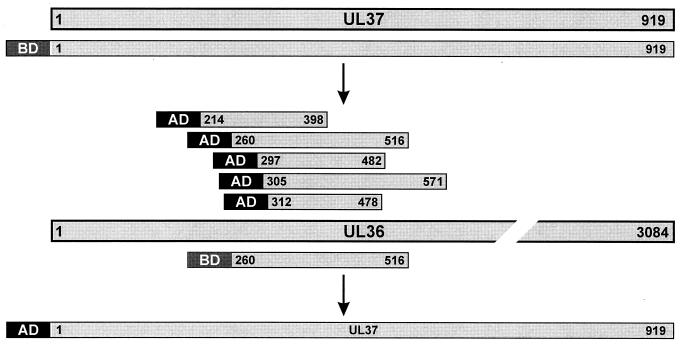
Interaction of the UL37 gene product with other PrV proteins was also assayed using yeast two-hybrid analysis as described recently (14, 18). Briefly, a random expression library of PrV proteins fused to a transcription activating peptide (AD) was constructed after ultrasonic treatment of virion DNA and insertion of fragments of between 500 and 1,000 bp into vector pB42AD (Clontech, Heidelberg, Germany). As bait, the complete UL37 protein was fused to the LexA protein which contains a sequence-specific DNA binding domain (BD [Fig. 4]). For that purpose, the UL37 ORF preceded by 24 bp of originally noncoding sequences was PCR amplified with primers UL37FOR2 and UL37REV (21). The resulting 2,820-bp EcoRI/KpnI fragment was inserted into pUC19 and recloned as a 2,829-bp EcoRI/BamHI fragment into vector pLexA (Clontech). Yeast cells were transformed with the reporter gene plasmid p8op-lacZ, the UL37 bait construct, and the PrV expression library. In transformed yeast cells, interactions between bait and library proteins specifically induced expression of the leu2 and lacZ marker genes, resulting in blue-stained yeast colonies on leucine-free plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). Seven strongly positive yeast clones were further analyzed by selective recloning of the individual library plasmids in Escherichia coli strain KC8 (Clontech) and DNA sequencing of the viral inserts with vector-specific primers. Remarkably, all seven prey plasmids contained parts of the UL36 gene fused in frame to the AD-encoding sequence. Three of these plasmids were identical, but the others possessed different UL36 inserts which overlapped, sharing codons 312 to 398 of the UL36 ORF (Fig. 4). Two-hybrid interactions could be reproduced after cotransfection of yeast cells with the UL37 bait construct and the individual UL36 library plasmids. However, no marker gene expression was detectable when either one of the plasmids was substituted by control vector without insert.
FIG. 4.
Summary diagram of the results of the yeast two-hybrid interaction analyses. The proteins are drawn as bars, and amino acids of the UL36 and UL37 proteins contained within the respective DNA binding (BD) or transcription activating (AD) fusion constructs are indicated above and below the complete protein. Note that the UL36 protein is not drawn to scale.
For reciprocal testing, the viral insert of a prey plasmid containing codons 260 to 515 of the UL36 gene was excised as a 776-bp EcoRI fragment and recloned into the bait vector pLexA. The PCR-amplified UL37 ORF was inserted into EcoRI-and-XhoI-digested vector pB42AD after blunt ending of noncompatible fragment ends with Klenow polymerase. After coexpression of the BD-UL36 and AD-UL37 fusion proteins in yeast cells, blue colonies were again detected on agar plates containing X-Gal (Fig. 4), whereas all vector controls were negative. These results confirm that the UL37 and UL36 proteins of PrV are able to interact directly without the requirement for other viral gene products. The studies further allowed mapping of the responsible interacting domain of the UL36 protein to a stretch of 87 aa.
UL36 protein is added to capsids in the absence of UL37
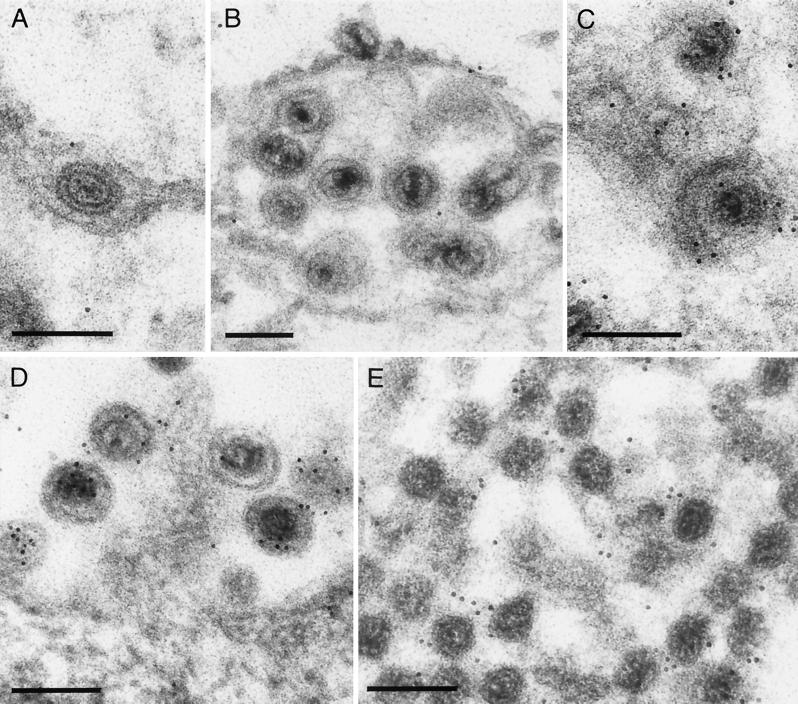
We recently demonstrated that in the absence of the UL37 protein, capsids accumulated in the cytoplasm of infected cells in an orderly arrangement (21). Although not directly contacting each other, they appeared to form contacts via extensions from the vertex region which were hypothesized to consist of or contain the UL36 protein. To test whether the UL36 protein is indeed present on capsids in the absence of the UL37 gene product, cells infected with PrV-ΔUL37 were analyzed by immunoelectron microscopy (20) using the anti-UL36 serum and gold-tagged secondary antibodies. As shown in Fig. 5E, the cytoplasmic aggregates of PrV-ΔUL37 capsids reacted with the anti-UL36 serum, proving that these capsids carry the UL36 protein. The antiserum also labeled intracytoplasmic (Fig. 5C) and extracellular (Fig. 5D) wild-type virus particles, whereas wild-type virions in the perinuclear space (Fig. 5A) were not labeled. For better demonstration of perinuclear virions, cells infected with a virus mutant lacking the US3 protein which had been demonstrated to reproducibly accumulate primary enveloped virions in the perinuclear space were also analyzed (Fig. 5B) (22).
FIG. 5.
Immunoelectron micrograph. RK13 cells were infected with wild-type PrV-Ka (A, C, and D), PrV-ΔUS3 (B), or PrV-ΔUL37 (E) at an MOI of 1 and analyzed 16 h after infection by immunolabeling with the anti-UL36 serum and gold-tagged secondary anti-rabbit antibodies. Label was not detected in primary enveloped virions in the perinuclear space (A) which accumulate in the US3 deletion mutant (B). In contrast, intracytoplasmic (C) and extracellular wild-type virions (D) were labeled with the anti-UL36 serum, as were capsid aggregates in the cytoplasm which form in the absence of the UL37 protein (E) (21). Bars, 200 nm.
The molecular details of how herpesviruses assemble the more than 15 proteins which are present in the tegument of mature virions are largely unknown (28). Moreover, only a few proteins which have been unequivocally identified as integral parts of the tegument are conserved throughout the herpesvirus family and, thus, are thought to play fundamental roles in the tegumentation process. We show here that two conserved tegument proteins, the products of the UL36- and UL37-homologous genes, physically interact with each other. Although not highly conserved in terms of amino acid identities of the gene products, UL36- and UL37-homologous genes and proteins are present in the Alpha- (10, 24, 37), Beta- (7), and Gammaherpesvirinae (1) and may be central for the tegumentation process. This is highlighted by the striking growth defects in the absence of these proteins. A UL36 deletion mutant of HSV-1 is unable to replicate on noncomplementing cells, and in the absence of the UL36 protein, apparently naked capsids accumulate in the cytoplasm (11). Similarly, a UL37-deficient HSV-1 mutant is impaired in secondary envelopment (12). In a UL37 deletion mutant of PrV, titers on noncomplementing cells are reduced ca. 1,000-fold from that of wild-type virus, and ultrastructurally, it was observed that capsids accumulated in the cytoplasm in an ordered arrangement. These capsids lack the typical electron-dense tegument (21). However, we show here that our UL36-specific antiserum decorated the aggregated capsids, which demonstrates that in the absence of the UL37 protein the UL36 gene product is associated with intracytoplasmic capsids. This correlates well with recent data on the structure of the HSV-1 tegument as obtained by cryoelectron microscopy (38). It was shown that the innermost layer of the otherwise largely unstructured tegument exhibits icosahedral symmetry, since it interacted specifically with the vertices of the capsid shell. It was hypothesized that the protein that produces this innermost layer of the tegument is the UL36 protein (38), and indeed there is evidence that the UL36 protein is able to interact with the major capsid protein (26).
Although in the absence of either the HSV-1 UL36 and UL37 protein or the PrV UL37 protein, capsids accumulated in the cytoplasm, the phenotypes are different. In the absence of the PrV UL37 protein, capsids accumulated in an orderly arrangement and, while not contacting each other directly, appeared to make contact via extensions emanating from the vertex regions. Since these clusters reacted with our anti-UL36 serum, we hypothesize that the extensions are formed by the UL36 protein. Moreover, we postulate that these contacts are normally blocked by interaction with the UL37 protein. Thus, interaction with the UL37 protein appears to be an important step in tegumentation after deposition of the UL36 protein onto capsids. A similar physical contact has been proposed to occur between the homologous proteins of human cytomegalovirus, the high-molecular-weight protein (HMWP) which is homologous to the UL36 gene product, and the HMWP-binding protein which is homologous to UL37 (M. E. Harmon and W. Gibson, Proc. Am. Soc. Virol., abstr. W35-4, p. 144, 1996). Thus, interaction between these two tegument components may be a conserved feature in tegumentation of herpesviruses. It should be noted, however, that in HCMV the basic phosphoprotein (pUL32) which has no homologs in the Alpha- or Gammaherpesvirinae has been shown to bind to capsids in vitro (3), and cryoelectron microscopy demonstrated differences in the HCMV tegument structure from that of HSV-1 (8). Therefore, there may be additional tegument-capsid interactions, at least in Betaherpesvirinae. Since corresponding HCMV deletion mutants have not yet been isolated, the functional importance of the observed interactions is unclear.
The UL36-homologous genes are the largest ORFs present in herpesviruses. The size of the resulting ca. 2,000- to 3,000-aa proteins (2,241-aa protein for human cytomegalovirus [7]; 3,421-aa protein for equine herpesvirus 1 [37]) would permit the formation of large extensions from the capsid shell. These extensions may be responsible for keeping the bulk of the tegument at a distance from the capsid shell, as observed by the presence of a clear halo between the capsid and the tegument in mature virions and in aggregates of tegumented capsids that form when envelopment is blocked (4, 5). They may also carry multiple interacting domains besides those which bind UL37 protein, the major capsid protein, or as postulated above, other UL36 proteins. In this context it is important to note that a ts mutation in the HSV-1 UL36 protein has been shown to result in a defect in release of viral DNA from incoming capsids (2). Thus, the UL36 protein may serve important functions during entry and egress. Moreover, the HSV-1 UL36 protein was suggested to bind the genomic packaging sequence and play a role in DNA packaging (9). In addition, the absence of the HSV-1 UL37 protein, besides impairing secondary envelopment, also led to a decrease in the efficiency of primary envelopment (12). Whether these two phenotypes are correlated is unclear at present. However, the absence of the PrV UL37 protein clearly interferes with intracytoplasmic tegumentation of capsids during egress (21), indicating that the PrV UL36-UL37 interaction is relevant for cytoplasmic stages of virion morphogenesis. Although virion formation is heavily impaired in the absence of the UL37 protein, a low-level production of infectious extracellular virus particles still occurs (21). So far it is unclear whether these virions contain the UL36 protein and which tegument proteins may be responsible for recruiting the UL36-decorated capsids into virus particles in the absence of UL37.
Besides confirming results from coprecipitation analyses that the UL36 and UL37 proteins of PrV physically interact, our yeast two-hybrid studies also outlined the interacting domain on the UL36 protein. When the complete UL37 protein was used as bait, five different expression clones of UL36 were selected. They specified UL36-related proteins from aa 214 to 571, with a common overlap from aa 312 to 398. Thus, the region between aa 312 and 398 of the 3,084-aa PrV UL36 protein comprises the UL37-interacting domain. This domain is located amino terminal from two putative leucine zipper motifs which may effect dimerization of the UL36 protein.
In summary, we have shown that the PrV UL36 and UL37 tegument proteins physically interact and that, in the absence of the UL37 protein, the UL36 protein is still detectable on intracytoplasmic capsids. However, the absence of the PrV (21) or HSV-1 UL37 protein (12), as well as the absence of the HSV-1 UL36 protein (11), impaired or blocked further tegumentation and envelopment. Therefore, we propose that tegumentation of intracytoplasmic capsids starts with interaction of the UL36 protein with the capsid shell, followed by interaction of the UL37 protein with UL36. By this mechanism, an ordered addition of the innermost layers of the tegument to nascent virions may be achieved. Further protein-protein interactions with nonconserved tegument components may then account for the complete assembly of a functional tegument. Finally, interaction between tegument proteins and carboxy-terminal portions of viral glycoproteins present in trans-Golgi vesicles is likely to drive final envelopment (4, 5).
Nucleotide sequence accession number. The sequence obtained has been deposited in GenBank under accession no. AJ422133.
Acknowledgments
Part of this work was supported by the Deutsche Forschungsgemeinschaft (DFG Me 854/5-1).
We thank Nadine Müller, Uta Hartwig, Charlotte Ehrlich, and Petra Meyer for expert technical assistance.
REFERENCES
- 1.Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. F. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, P. Tuffnell, and B. G. Barrell. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature (London) 310:207-211. [DOI] [PubMed] [Google Scholar]
- 2.Batterson, W., D. Furlong, and B. Roizman. 1983. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J. Virol. 45:397-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baxter, M., and W. Gibson. 2001. Cytomegalovirus basic phosphoprotein (pUL32) binds to capsids in vitro through its amino one-third. J. Virol. 75:6865-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brack, A. R., B. G. Klupp, H. Granzow, R. Tirabassi, L. W. Enquist, and T. C. Mettenleiter. 2000. Role of the cytoplasmic tail of pseudorabies virus glycoprotein E in virion formation. J. Virol. 74:4004-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brack, A. R., J. M. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J. Virol. 73:5364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, Y. E., C. Van Sant, P. W. Krug, A. E. Sears, and B. Roizman. 1997. The null mutant of the UL31 gene of herpes simplex virus 1: construction and phenotype in infected cells. J. Virol. 71:8307-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchinson, T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barrell. 1990. Analysis of the protein coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 8.Chen, D. H., H. Jiang, M. Lee, F. Liu, and Z. H. Zhou. 1999. Three-dimensional visualization of tegument/capsid interactions in the intact human cytomegalovirus. Virology 260:10-16. [DOI] [PubMed] [Google Scholar]
- 9.Chou, J., and B. Roizman. 1989. Characterization of DNA sequence-common and sequence-specific proteins binding to cis-acting sites for cleavage of the terminal a sequence of the herpes simplex virus 1 genome. J. Virol. 63:1059-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1759-1816. [DOI] [PubMed] [Google Scholar]
- 11.Desai, P. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desai, P., G. Sexton, J. McCaffery, and S. Person. 2001. A null mutation in the gene encoding the UL37 polypeptide of herpes simplex virus type 1 abrogates virus maturation. J. Virol. 75:10259-10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enquist, L. W., P. J. Husak, B. W. Banfield, and G. A. Smith. 1999. Infection and spread of alphaherpesviruses in the nervous system. Adv. Virus Res. 51:237-347. [DOI] [PubMed] [Google Scholar]
- 14.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host cell nucleus and represent components of primary enveloped but not of mature virions. J. Virol. 76:364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gershon, A. A., D. L. Sherman, Z. Zhu, C. A. Gabel, R. T. Ambron, and M. D. Gershon. 1994. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J. Virol. 68:6372-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibson, W. 1996. Structure and assembly of the virion. Intervirology 39:389-400. [DOI] [PubMed] [Google Scholar]
- 17.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 75:3675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gyuris, J., E. Golemis, H. Chertkov, and R. Brent. 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75:791-803. [DOI] [PubMed] [Google Scholar]
- 19.Kaplan, A. S., and A. Vatter. 1959. A comparison of herpes simplex and pseudorabies virus. Virology 7:394-407. [DOI] [PubMed] [Google Scholar]
- 20.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klupp, B. G., H. Granzow, E. Mundt, and T. C. Mettenleiter. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 75:8927-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2001. Effect of pseudorabies virus US3 protein on nuclear membrane localization of the UL34 protein and virus egress from the nucleus. J. Gen. Virol. 82:2363-2371. [DOI] [PubMed] [Google Scholar]
- 23.Lukács, N., H.-J. Thiel, T. C. Mettenleiter, and H.-J. Rziha. 1985. Demonstration of three major species of pseudorabies virus glycoproteins and identification of a disulfide-linked glycoprotein complex. J. Virol. 53:166-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the unique long region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 25.McLauchlan, J., K. Liefkens, and N. D. Stow. 1994. The herpes simplex virus type 1 UL37 gene product is a component of virus particles. J. Gen. Virol. 75:2047-2052. [DOI] [PubMed] [Google Scholar]
- 26.McNabb, D., and R. J. Courtney. 1992. Characterization of the large tegument protein (ICP1/2) of herpes simplex virus type 1. Virology 190:221-232. [DOI] [PubMed] [Google Scholar]
- 27.Mettenleiter, T. C. 2000. Aujeszky's disease (pseudorabies) virus: the virus and molecular pathogenesis-state of the art, June 1999. Vet. Res. 31:99-115. [DOI] [PubMed] [Google Scholar]
- 28.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nixdorf, R., B. G. Klupp, and T. C. Mettenleiter. 2001. Role of the cytoplasmic tails of pseudorabies virus glycoproteins B, E and M in intracellular localization and virion incorporation. J. Gen. Virol. 82:215-226. [DOI] [PubMed] [Google Scholar]
- 30.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roizman, B. 1996. Herpesviridae, p. 2221-2231. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 32.Roller, R., Y. Zhou, R. Schnetzer, J. Ferguson, and D. DeSalvo. 2000. Herpes simplex virus type 1 UL34 gene product is required for viral envelopment. J. Virol. 74:117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmitz, J. B., A. Albright, P. Kinchington, and F. Jenkins. 1995. The UL37 protein of herpes simplex virus type 1 is associated with the tegument of purified virions. Virology 206:1055-1066. [DOI] [PubMed] [Google Scholar]
- 34.Shelton, L. G., M. Pensiero, and F. Jenkins. 1990. Identification and characterization of the herpes simplex virus type 1 protein encoded by the UL37 open reading frame. J. Virol. 64:6101-6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skepper, J., A. Whiteley, H. Browne, and A. Minson. 2001. Herpes simplex virus nucleocapsids mature to progeny virions by an envelopment-deenvelopment-reenvelopment pathway. J. Virol. 75:5697-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stevens, A., and P. G. Spear. 1997. Herpesvirus capsid assembly and envelopment. In W. Chiu, R. Burnett, and R. Garcea (ed.) Structural biology of viruses, p. 312-351. Oxford University Press, New York, N.Y.
- 37.Telford, E., M. S. Watson, K. McBride, and A. J. Davison. 1992. The DNA sequence of equine herpesvirus-1. Virology 189:304-316. [DOI] [PubMed] [Google Scholar]
- 38.Zhou, Z., D. Chen, J. Jakana, F. J. Rixon, and W. Chiu. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 73:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]