Abstract
A viral reservoir of human immunodeficiency virus type 1 (HIV-1)-infected, resting CD4+ T cells persists despite suppression of plasma viremia by combination antiretroviral therapy. In a longitudinal analysis of three patients treated with a five-drug regimen, both R5 and X4 HIV-1 variants persisted in the cellular reservoir for up to 3 years.
Resting CD4+ T cells have been identified as a reservoir for replication-competent human immunodeficiency virus type 1 (HIV-1) even when suppression of HIV-1 plasma viremia by combinations of three or more antiretroviral drugs is achieved (4, 5, 8, 9, 21, 26, 27). These cells are thought to be latently infected, having been generated by reversal of infected cells from an activated to a resting state. Ongoing viral replication during potent antiretroviral therapy has been demonstrated by episodes of detectable plasma viral RNA (6, 21), episomal cDNA circles (22), viral RNA transcripts (10, 11, 15, 16), and ongoing genetic evolution (12). This residual replication may contribute to the apparent long half-life of the reservoir by reinfection of CD4+ T cells and thereby refeeding of the reservoir (14).
We have previously shown that coreceptor usage of HIV-1 is an important determinant for T-cell tropism during untreated HIV-1 infection. Non-syncytium-inducing (NSI), CCR5-utilizing variants (R5 variants) can be isolated from CCR5+ memory CD4+ T cells, whereas syncytium-inducing (SI), CXCR4-utilizing HIV-1 variants (X4 variants) can be isolated from both memory and naive CXCR4+ CD4+ T cells (3, 24). Since CCR5 is mainly expressed on activated CD4+ T cells (2, 18) and CXCR4 is expressed at higher levels on resting CD4+ T cells, it is conceivable that R5 and X4 HIV-1 variants differ in their capacity to generate a latent infection and to persist during therapy. Although the presence of both R5 and X4 HIV-1 variants in the viral reservoir has previously been reported (19), longitudinal, quantitative data are not available to address this possibility. Here, we performed an in-depth study of the dynamics of R5 and X4 HIV-1 variants in three patients (patients 08, 14, and 15) in whom X4 HIV-1 variants had evolved before initiation of therapy.
All participants were antiretroviral therapy naive when they started a five-drug regimen consisting of zidovudine, lamivudine, abacavir, nevirapine, and indinavir. Low-dose ritonavir was added to this drug regimen to enhance indinavir concentrations in serum, seminal plasma, and cerebrospinal fluid. The drug regimen was changed in case of toxicity but consisted of at least four drugs during the entire study period. At baseline, the patients had low CD4+ T cell numbers (30 to 130 cells/μl), high viral RNA levels in plasma (4.5 to 5.1 log copies/ml), and a high cellular infectious load (137 to 386 tissue culture infectious dose (TCID)/106 CD4+ T cells). After initiation of therapy, CD4+ T cell numbers gradually increased and viral RNA load in plasma declined to levels below the limit of detection (5 copies/ml) (25) (Fig. 1). Throughout follow-up, the plasma viral RNA load in general remained below the limit of detection, although episodes of intermittent detectable viremia were monitored in patients 08 and 14.
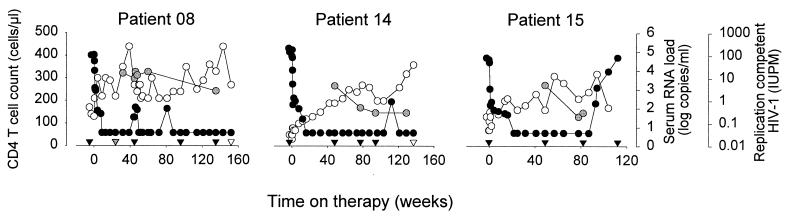
FIG. 1.
Changes in viral load and CD4+ T cell numbers in response to antiretroviral therapy with a five-drug regimen with three patients who developed X4 variants before initiation of therapy. Data from patients 08 and 014 were available from a previous study (van Rij et al., submitted for publication). At approximately 87 weeks of therapy, patient 15 reported noncompliance with therapy and subsequently stopped therapy for 10 weeks. Patient 08 was treated with anti-CD3 and recombinant IL-2 during five consecutive days after 46 weeks on therapy (20). Open circles represent CD4+ T cell numbers, black circles represent viral RNA load in serum, and gray circles represent the cellular infectious load in HLA-DR− CD4+ T cells. Black triangles at the x axis represent time points selected for phenotypic and sequence analyses of viral clones. A gray triangle at the x axis of patient 08 represents a time point from which only viral sequences were analyzed. White triangles reflect time points analyzed for in vivo viral tropism for naive and memory CD4+ T cells.
The proportion of HIV-1-infected HLA-DR− CD4+ T cells was determined by cocultivation of these cells with healthy donor peripheral blood mononuclear cells (PBMC) after overnight activation with phytohemagglutinin (PHA), recombinant human interleukin 2 (IL-2), and irradiated allogeneic PBMC, as previously described (R. P. van Rij et al., submitted for publication). Briefly, after overnight activation, cells were washed and cocultivated with PHA-stimulated healthy donor PBMC under limiting dilution conditions in 96- or 24-well plates. Weekly, half of the cells were transferred to new tissue culture plates containing fresh PHA-stimulated PBMC, and culture supernatant was analyzed for p24 production. Virus could be cultured from HLA-DR− CD4+ T cells at all time points, including the last time point analyzed, which was after 135, 94, and 82 weeks of therapy for patient 08, 14, and 15, respectively. After cessation of therapy for 10 weeks, plasma RNA levels increased to pretreatment levels and the cellular infectious load increased to 74 TCID/106 CD4+ T cells in patient 15.
To study the dynamics of R5 and X4 HIV-1 virus populations during treatment, the syncytium-inducing phenotype in the MT2 T-cell line was determined during isolation of biological virus clones from HLA-DR− CD4+ T cells. MT2 cells express CXCR4 but not CCR5, and infection and formation of syncytia by HIV-1 is a rapid indication of the ability to use CXCR4. When half of the cells from each well were transferred to a new tissue culture plate each week, MT2 cells were added to the remainder of the cultures, and the presence of SI HIV-1 variants was assessed by visual inspection for the presence of syncytia (3, 24).
Since all virus clones obtained at each time point were analyzed, percentages of NSI and SI HIV-1 variants reflect the in vivo distribution of cells infected with these variants. Before the start of therapy, both NSI and SI HIV-1 variants coexisted in all patients (54 to 76% of the virus clones had an SI phenotype [Fig. 2A]). After initiation of therapy, both NSI and SI virus variants persisted in patient 08 up to 152 weeks of therapy (36% SI phenotype). Predominantly SI HIV-1 variants were isolated from patients 14 and 15 (90 and 100% SI phenotype, respectively), but this difference was not significantly different from baseline distributions of NSI and SI variants (P = 0.22 [patient 14] and 0.15 [patient 15] [Fisher exact test]). After cessation of therapy, both NSI and SI HIV-1 variants were obtained from patient 15, despite the inability to culture NSI HIV-1 variants during treatment.
FIG. 2.
Phenotype of biological virus clones obtained before and during treatment with five antiretroviral drugs. (A) Percentage of virus clones obtained at baseline and at several time points during therapy with SI (black bars) and NSI (gray bars) phenotypes in the MT2 T-cell line. The bars labeled ART represent combined data from all time points during treatment. The absolute number of analyzed clones is indicated at the top of the bars. The bar labeled “off therapy” for patient 15 represents virus clones obtained after cessation of therapy for 10 weeks. (B) Coreceptor usage in U87 cells transfected with CD4 and either CCR3, CCR5, or CXCR4. U87 cells were infected with a 500 50% tissue culture infectious dose of each virus clone and cultured for 21 days, and the culture supernatant was harvested weekly for analysis of p24 production. Bar fills indicate coreceptor usage: CCR5 (R5, white bars), CCR5 and CXCR4 (R5X4, light gray), CCR3, CCR5, and CXCR4 (R3R5X4, dark gray) and CXCR4 (black bars). Pretherapy clones (pre) and clones combined from various time points during treatment (ART) are shown. NT, not tested. (C) Dependency of virus clones on CCR5 for infection of primary cells. End-point dilution titration was performed on cells from a healthy donor homozygous for the 32-bp deletion in CCR5 (Δ32) and on cells from at least two healthy blood donors (++). Viral clones from all three patients before and during therapy are shown. Open circles represent NSI clones that are able to use CCR5 and CXCR4 in U87 cells.
Coreceptor usage of the obtained virus clones was determined by cell-free infection of U87 indicator cell lines, as previously described (24) (Fig. 2B). NSI HIV-1 clones from patient 08 were in general CCR5 restricted (R5), whereas a minority of NSI virus clones were in addition able to infect U87 cells via CXCR4 (R5X4). SI HIV-1 clones could use both CCR5 and CXCR4 (patients 08 and 15), or CCR3, CCR5, and CXCR4 (patient 08), or were restricted to CXCR4 usage (patient 14). No selection for a specific coreceptor usage during therapy was observed.
The dependency on CCR5 for infection of primary cells was determined by the ability to infect PHA-PBMC from a healthy blood donor, lacking CCR5 expression due to a homozygous genotype for the 32-bp deletion in the CCR5 gene (CCR5 Δ32). An absolute association of SI phenotype and the ability to infect CCR5 Δ32 PBMC was observed (Fig. 2C). NSI variants, either R5 or R5X4 in U87 cells, were unable to infect CCR5 Δ32 PBMC, reflecting an absolute dependency on CCR5 for productive infection of primary CD4+ T cells.
The longitudinal isolation of biological R5 and X4 virus clones provided the opportunity to monitor genetic evolution within the R5 and within the X4 virus populations. A 333-bp region spanning the third variable region of gp120 was sequenced (24), and phylogenetic analyses were performed using the Phylip package (version 3.5c and 3.6) (7). As expected, a bootstrap-supported clustering of intrapatient sequences was observed in neighbor-joining phylogenetic trees (data not shown). As previously described (23, 24), R5 and X4 HIV-1 sequences clustered apart within the cluster of patient-specific sequences, supported by bootstrap values ranging from 87 to 100 (data not shown). In none of the patients was evidence obtained for ongoing evolution during antiretroviral treatment. Sequences obtained during therapy clustered with baseline sequences in all patients. From patient 08, sequences that were even identical to baseline sequences, of either the R5 or X4 phenotype, were isolated up to 135 weeks of treatment (data not shown). Using an approach described by Gunthard et al. (12), we were unable to observe an ongoing evolutionary divergence from a deduced most recent common ancestral sequence. R5 and X4 HIV-1 sequences obtained after cessation of therapy for patient 15 were similar or even identical to sequences obtained before therapy.
We have previously shown that R5 HIV-1 variants are isolated mainly from memory CD4+ T cells, whereas X4 HIV-1 variants are equally distributed over naive and memory CD4+ T cells (3, 24). To establish a potential role of naive CD4+ T cells as a cellular reservoir for X4 HIV-1 variants under therapy, we studied whether naive cells were infected in vivo in patients 08 and 14. Patient PBMC were sorted in naive CD27+ CD45RO− and memory CD45RO+ CD4+ T cells and cocultivated with healthy donor PHA-PBMC. Memory CD4+ T cells were infected by either R5 (patient 08) or X4 (patient 14) HIV-1 variants (Table 1). Productive infection of naive CD4+ T cells by X4 HIV-1 variants was observed for patient 08.
TABLE 1.
In vivo tropism for naive and memory CD4+ T cellsa
| Patient no. | No. of wks on therapy | Results for subset
|
|||||
|---|---|---|---|---|---|---|---|
| CD27+ CD45RO− naive
|
CD45RO+ memory
|
||||||
| Infectious load (TCID/106 cells) | % (No.) with SI phenotypeb | Infectious load (TCID/106 cells) | % (No.) with SI phenotypeb | ||||
| 008 | 152 | 0.3 | 100 (1) | 0.7 | 0 (2) | ||
| 014 | 137 | <0.5 (DL)c | NAd | 0.2 | 100 (1) | ||
Patient PBMC isolated from freshly drawn blood were sorted on a MOFLO cell sorter (Cytomation Inc., Ft. Collins, Co.) into naive(CD27+ CD45RO−) and memory (CD45RO+ CD4+) T cells(1,13). The cellular infectious load in these T-cell subsets was determined by cocultivation with healthy donor PHA-stimulated PBMC under limiting dilution conditions.
Percentage of clones with SI phenotype in the designated T-cell subsets and, between parentheses, absolute number of obtained virus clones are indicated.
DL, lower detection limit of the assay.
NA, not applicable.
In this longitudinal analysis, we aimed to observe putative differences among R5 and X4 variants in genetic evolution and persistence in the reservoir of infected HLA-DR− CD4+ T cells under treatment. We observed that both R5 and X4 HIV-1 variants can persist under prolonged aggressive therapy with a five-drug regimen. We were unable to observe any evidence for ongoing replication among either R5 or X4 variants by sequence analysis, although intermittent viremia as observed in two patients is suggestive of ongoing replication. We previously demonstrated a differential tropism of R5 and X4 HIV-1 variants for naive and memory CD4+ T cells in the natural course of infection (24). Due to the abundant expression of CXCR4 on naive CD4+ T cells, the frequency of infected naive CD4+ T cells in patients infected with X4 variants is up to 2 logs higher than in patients with R5 variants only (3). Given their longer half-life compared to memory CD4+ T cells (17), naive CD4+ T cells may represent a preferred site for viral persistence, especially in patients infected with X4 HIV-1 variants. In agreement, we were able to isolate X4 virus from naive cells after 3 years of therapy, confirming a previous report of productive infection of naive CD4+ T cells under treatment (19). Finally, our data indicate that prolonged therapy with the currently available antiretroviral drugs does not result in clearance of either R5 or the more pathogenic X4 HIV-1 variants from the viral reservoir but that prolonged suppression of both variants in plasma can be achieved. Further insights into mechanisms of viral persistence in these cells are crucial for understanding and directly targeting cellular reservoirs of HIV-1.
Nucleotide sequence accession number.
Sequences were deposited in GenBank under accession numbers AF355630 to AF355679 (patient 08); AF355726 to AF355748 (patient 14); and AF355680 to AF355725 (patient 15).
Acknowledgments
We are greatly indebted to the patients who volunteered for this study. Recombinant human IL-2 was kindly provided by Chiron (Chiron Benelux BV, Amsterdam, The Netherlands). U87 cell lines were obtained from HongKui Deng and Dan Littman, through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. We thank Berend Hooibrink for FACS sorting and Steven Jansen for patient care.
This study was financially supported by the Dutch AIDS fund (grant 1305).
REFERENCES
- 1.Baars, P. A., M. M. Maurice, M. Rep, B. Hooibrink, and R. A. W. Van Lier. 1995. Heterogeneity of the circulating human CD4+ T-cell population: further evidence that the CD4+CD45RA −D27 −T-cell subset contains specialized primed cells. J. Immunol. 154:17-25. [PubMed] [Google Scholar]
- 2.Blaak, H., L. J. Ran, R. Rientsma, and H. Schuitemaker. 2000. Susceptibility of in vitro stimulated PBMC to infection with NSI HIV-1 is associated with levels of CCR5 expression and β-chemokine production. Virology 267:237-246. [DOI] [PubMed] [Google Scholar]
- 3.Blaak, H., A. B. Van 't Wout, M. Brouwer, B. Hooibrink, E. Hovenkamp, and H. Schuitemaker. 2000. In vivo HIV-1 infection of CD45RA+CD4+ T cells is established primarily by syncytium-inducing variants and correlates with the rate of CD4+ T cell decline. Proc. Natl. Acad. Sci. USA 97:1269-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chun, T.-W., D. Engel, S. B. Mizell, L. A. Ehler, and A. S. Fauci. 1998. Induction of HIV-1 replication in latently infected CD4+ T cells using a combination of cytokines. J. Exp. Med. 188:83-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chun, T.-W., L. Stuyver, S. B. Mizzel, L. A. Ehler, J. M. Mican, M. Baseler, A. Lloyd, M. Nowak, and A. S. Fauci. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 94:13193-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dornadula, G., H. Zhang, B. VanUitert, J. Stern, L. Livornese, M. J. Ingerman, J. Witek, R. J. Kedanis, J. Natkin, J. DeSimone, and R. J. Pomerantz. 1999. Residual HIV-1 RNA in blood plasma of patients taking suppressive highly active antiretroviral therapy. JAMA 282:1627-1632. [DOI] [PubMed] [Google Scholar]
- 7.Felsenstein, J. 1989. PHYLIP-Phylogeny Inference Package. Cladistics 5:164-166. [Google Scholar]
- 8.Finzi, D., J. Blankson, J. D. Siliciano, J. B. Margolick, K. Chadwick, T. Pierson, K. Smith, J. Lisziewicz, F. Lori, C. Flexner, T. C. Quinn, R. E. Chaisson, E. Rosenberg, B. Walker, S. Gange, J. Gallant, and R. F. Siliciano. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512-517. [DOI] [PubMed] [Google Scholar]
- 9.Finzi, D., M. Hermankova, T. Pierson, L. Carruth, C. Buck, R. E. Chaisson, T. C. Quinn, K. Chadwick, J. Margolick, R. Brookmeyer, J. E. Gallant, M. Markowitz, D. D. Ho, D. D. Richman, and R. F. Siliciano. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295-1300. [DOI] [PubMed] [Google Scholar]
- 10.Fischer, M., H. F. Gunthard, M. Opravil, B. Joos, W. Huber, L. R. Bisset, P. Ott, J. Böni, R. Weber, and R. W. Cone. 2000. Residual HIV-RNA levels persist for up to 2.5 years in peripheral blood mononuclear cells of patients on potent antiretroviral therapy. AIDS Res. Hum. Retrovir. 16:1135-1140. [DOI] [PubMed] [Google Scholar]
- 11.Furtado, M. R., D. S. Callaway, J. P. Phair, K. J. Kunstman, J. L. Stanton, C. A. Macken, A. S. Perelson, and S. M. Wolinsky. 1999. Persistence of HIV-1 transcription in peripheral-blood mononuclear cells in patients receiving potent antiretroviral therapy. N. Engl. J. Med. 340:1614-1622. [DOI] [PubMed] [Google Scholar]
- 12.Gunthard, H. F., S. D. W. Frost, A. J. Leigh-Brown, C. C. Ignacio, K. Kee, A. S. Perelson, C. A. Spina, D. V. Havlir, M. Hezareh, D. J. Looney, D. D. Richman, and J. K. Wong. 1999. Evolution of envelope sequences of human immunodeficiency virus type 1 in cellular reservoirs in the setting of potent antiretroviral therapy. J. Virol. 73:9404-9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamann, D., P. A. Baars, M. H. G. Rep, B. Hooibrink, S. R. Kerkhof-Garde, M. R. Klein, and R. A. W. Van Lier. 1997. Phenotypic and functional separation of memory and effector human CD8+ T cells. J. Exp. Med. 186:1407-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho, D. D., and L. Zhang. 2000. HIV-1 rebound after anti-retroviral therapy. Nat. Med. 6:736-737. [DOI] [PubMed] [Google Scholar]
- 15.Imamichi, H., K. A. Crandall, V. Natarajan, M. K. Jiang, R. L. Dewar, S. Berg, A. Gaddam, M. Bosche, J. A. Metcalf, R. T. Davey, Jr., and C. H. Lane. 2001. Human immunodeficiency virus type 1 quasi species that rebound after discontinuation of highly active antiretroviral therapy are similar to the viral quasi species present before initiation of therapy. J. Infect. Dis. 183:36-50. [DOI] [PubMed] [Google Scholar]
- 16.Kuster, H., M. Opravil, P. Ott, E. Schlaepfer, M. Fischer, H. F. Gunthard, R. Lüthy, R. Weber, and R. W. Cone. 2000. Treatment-induced decline of human immunodeficiency virus-1 p24 and HIV-1 RNA in lymphoid tissue of patients with early human immunodeficiency virus-1 infection. Am. J. Pathol. 156:1973-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCune, J. M., M. B. Hanley, D. Cesar, R. Halvorsen, R. Hoh, D. Schmidt, E. Wieder, S. Deeks, S. Siler, R. Neese, and M. Hellerstein. 2000. Factors influencing T cell turnover in HIV-1-seropositive patients. J. Clin. Investig. 105:R1-R8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ostrowski, M. A., S. J. Justement, A. Cantanzaro, C. A. Hallahan, L. A. Ehler, S. B. Mizell, P. N. Kumar, J. Mican, T.-W. Chun, and A. S. Fauci. 1998. Expression of chemokine receptors CXCR4 and CCR5 in HIV-1-infected and uninfected individuals. J. Immunol. 161:3195-3201. [PubMed] [Google Scholar]
- 19.Pierson, T., J. Hoffman, J. Blankson, D. Finzi, K. Chadwick, J. B. Margolick, C. Buck, J. D. Siliciano, R. W. Doms, and R. F. Siliciano. 2000. Characterisation of chemokine receptor utilization of viruses in the latent reservoir for human immunodeficiency virus type 1. J. Virol. 74:7824-7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prins, J. M., S. Jurriaans, R. M. E. van Praag, H. Blaak, R. P. van Rij, P. T. A. Schellekens, I. J. M. Ten Berge, S.-L. Yong, C. H. Fox, M. T. L. Roos, F. De Wolf, J. Goudsmit, H. Schuitemaker, and J. M. A. Lange. 1999. Immuno-activation with anti-CD3 and recombinant human IL-2 in HIV-1 patients on potent antiretroviral therapy. AIDS 13:2405-2410. [DOI] [PubMed] [Google Scholar]
- 21.Ramratnam, B., J. E. Mittler, L. Zhang, D. Boden, A. Hurley, F. Fang, C. A. Macken, A. S. Perelson, M. Markowitz, and D. D. Ho. 2000. The decay of the latent reservoir of replication-competent HIV-1 is inversely correlated with the extent of residual viral replication during prolonged anti-retroviral therapy. Nat. Med. 6:82-85. [DOI] [PubMed] [Google Scholar]
- 22.Sharkey, M. E., I. Teo, T. Greenough, N. Sharova, K. Luzuriaga, J. L. Sullivan, R. P. Bucy, L. G. Kostrikis, A. Haase, C. Veryard, R. E. Davaro, S. Cheeseman, J. S. Daly, C. Bova, R. T. Ellison, B. Mady, K. Kew Lai, G. Moyle, M. Nelson, B. Gazzard, S. Shaunak, and M. Stevenson. 2000. Persistence of episomal HIV-1 infection intermediates in patients on highly active anti-retroviral therapy. Nat. Med. 6:76-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van 't Wout, A. B., H. Blaak, L. J. Ran, M. Brouwer, C. Kuiken, and H. Schuitemaker. 1998. Evolution of syncytium-inducing and non-syncytium-inducing biological virus clones in relation to replication kinetics during the course of HIV-1 infection. J. Virol. 72:5099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Rij, R. P., H. Blaak, J. A. Visser, M. Brouwer, R. Rientsma, S. Broersen, A. M. De Roda Husman, and H. Schuitemaker. 2000. Differential coreceptor expression allows for independent evolution of non-syncytium-inducing and syncytium-inducing HIV-1. J. Clin. Investig. 160:1039-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weverling, G. J., J. M. A. Lange, S. Jurriaans, J. M. Prins, V. Lukashov, D. W. Notermans, M. T. L. Roos, H. Schuitemaker, R. M. W. Hoetelmans, S. A. Danner, J. Goudsmit, and F. De Wolf. 1998. Alternative multidrug regimen provides improved suppression of HIV-1 replication over triple therapy. AIDS 12:F117-F122. [DOI] [PubMed] [Google Scholar]
- 26.Wong, J. K., M. Hezareh, H. F. Günthard, D. Havlir, C. C. Ignacio, C. A. Spina, and D. D. Richman. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291-1295. [DOI] [PubMed] [Google Scholar]
- 27.Zhang, L., B. Ramratnam, K. Tenner-Racz, Y. He, M. Vesanen, S. Lewin, A. Talal, P. Racz, A. S. Perelson, B. T. Korber, M. Markowitz, and D. D. Ho. 1999. Quantifying residual HIV-1 replication in patients receiving combination antiretroviral therapy. N. Engl. J. Med. 340:1605-1613. [DOI] [PubMed] [Google Scholar]