Abstract
Background
An observational case series was used to study the virulence characteristics and genotypes of paired Staphylococcus epidermidis isolates cultured from intraocular samples and from periocular environment of patients with postcataract surgery endophthalmitis.
Methods
Eight S. epidermidis isolates were obtained from three patients (2 from patients #1 and 2 and 4 from patient #3) whose vitreous and/or anterior chamber (AC) specimens and preoperative lid/conjunctiva samples were culture positive. Cultures were identified by API-Staph phenotypic identification system and genotypically characterized by Fluorescent Amplified Fragment Length Polymorphism (FAFLP) and checked for their antimicrobial susceptibility. The isolates were tested for biofilm-production and methicillin-resistance (MR) by PCR amplification of icaAB and mecA gene respectively.
Results
Four out of eight S. epidermidis strains showed multiple drug resistance (MDR). All the eight strains were PCR positive for mecA gene whereas seven out of eight strains were positive for icaAB genes. In all three patients FAFLP typing established vitreous isolates of S. epidermidis strains to be indistinguishable from the strains isolated from the patient's conjunctival swabs. However, from patient number three there was one isolate (1030b from lid swab), which appeared to be nonpathogenic and ancestral having minor but significant differences from other three strains from the same patient. This strain also lacked icaAB gene. In silico analysis indicated possible evolution of other strains from this strain in the patient.
Conclusion
Methicillin-resistant biofilm positive S. epidermidis strains colonizing the conjunctiva and eyelid were responsible for postoperative endophthalmitis (POE).
Background
Endophthalmitis is an important ocular disease that is most commonly caused by postoperative and posttraumatic introduction of bacteria into the posterior segment of the eye. In severe infections, visual acuity is greatly diminished or completely lost [1]. Indigenous ocular flora is presumed to be a source of infectious organisms in bacterial postoperative endophthalmitis (POE). Bacteria have been recovered from the anterior chamber (AC) during the cataract wound closure in many cases [2]. A few studies have examined the genetic relationship between organisms isolated from the eyelid/conjunctiva, and organisms recovered from the vitreous of patients with POE [3,4].
Coagulase-negative staphylococci (CoNS) have been reported as the most frequent cause of bacterial POE [5]. Among CoNS, S. epidermidis seems to be predominantly present as normal flora of the lids and conjunctiva and implicated as a source of infection [1,3,4]. Based on the current literature, one may hypothesize that the commensal organisms turn virulent either prior to or once they gain intraocular access. With reference to intraocular infections, the transformation of commensal bacteria into virulent forms has not been studied at molecular level. Our aim was to investigate the genetic characteristics in strains isolated from extraocular samples and intraocular specimens and determine the variations, if any. We analyzed paired S. epidermidis isolates from an individual patient's eyelid/conjunctiva and vitreous/AC specimen by using FAFLP, a recently developed potential molecular strains-typing technique. In addition, we attempted to detect genes such as icaAB, mecA that might enhance virulence among the isolates by virtue of their role in biofilm formation, invasiveness and development of resistance to antibiotics.
Methods
We examined 10 cases of POE after cataract surgery. All cases were referred to L. V. Prasad eye institute for management. Samples were collected from the eyelids and conjunctiva using cotton swabs from patients scheduled to undergo pars plana vitrectomy and intraocular antibiotic injection for the management of endophthalmitis. During surgery AC fluid and/or vitreous biopsy was collected from all cases. While the swabs were inoculated onto blood agar and brain heart infusion broth, the intraocular fluids were processed for bacteria and fungi as described earlier [6]. A total of eight isolates of S. epidermidis from three patients were included in the study (Table 1).
Table 1.
Clinical characteristics of the patients having POE with details of strains and their origin. All patients underwent phacoemulsification with intraocular lens (IOL) implantation.
| Patient No. | Age/Sex | Predisposing factors/Systemic disease | Site of isolation | Strain ID | Species | Antibiotypea | mecA/icaAB | Visual Acuity |
| 1 | 43/M | Eye lid swelling | AC fluid | L778a/03 | S.epidermidis | Oxa, G | +/+ | 20/60 |
| Eye lid | L778b/03 | S.epidermidis | Oxa, G | +/+ | ||||
| 2 | 60/M | Hypertension | Vitreous | L849a/03 | S.epidermidis | Oxa | +/+ | 20/25 |
| Eye lid | L849b/03 | S.epidermidis | Oxa | +/+ | ||||
| 3 | 58/F | Diabetic retinopathy | Vitreous | L1030a/03 | S.epidermidis | Ak, Ca, Cip, G, Oxa | +/+ | 20/40 |
| Eye lid | L1030b/03 | S.epidermidis | Ak, Ca, Cip, G, Oxa | +/- | ||||
| AC fluid | L1030c/03 | S.epidermidis | Ak, Ca, Cip, G, Oxa | +/+ | ||||
| Conjunctiva | L1030d/03 | S.epidermidis | Ak, Ca, Cip, G, Oxa | +/+ |
a Antibiotic resistance; Oxa – Oxacillin; G – Gentamicin; Ak – Amikacin; Ca – Ceftazidime; Cip – Ciprofloxacin.
The organisms were identified by using API Staph identification system (bioMerieux, France). Sensitivity to several antibiotics namely amikacin, ceftazidime, ciprofloxacin, gentamicin, and oxacillin was determined by Kirby Bauer disk diffusion method. Genomic DNA was isolated from all the eight strains according to methods described earlier [7]. PCR primers and conditions for amplification of icaA and icaB genes were as described by Frebourg et al [8]. Strains were checked for the presence of mecA by PCR that corresponds to unique penicillin-binding protein (PBP2a), directly associated with methicillin resistance (MR). All the strains were subjected to FAFLP by using EcoRI+0 and MseI+A primer combinations for selective PCR. FAFLP experiment and analysis (PE Biosystems, UK) were done as per the manufacturer's instructions. FAFLP electropherograms were analyzed using Genescan 3.7 and Genotyper 3.7 softwares (PE Biosystems) as described earlier [9]. The percentage similarities/differences between the patterns generated by different strains were calculated using the Dice correlation coefficient. A dendrogram representing the similarity coefficients was constructed using neighbor joining method [9].
Results
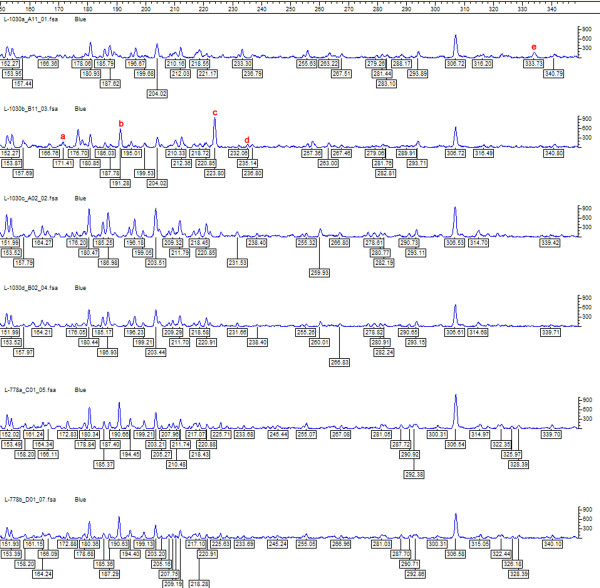
Table 1 shows the clinical characteristics of all three cases included in the study. Paired strains obtained from each of these patients showed identical antibiotic sensitivity profile. Six out of eight strains were resistant to more than one antibiotic tested. MR was demonstrated in all the eight strains with the amplification of mecA. Seven out of eight strains showed icaAB gene amplification specific for biofilm production. FAFLP profiles revealed that intraocular S. epidermidis were indistinguishable from the conjunctival/lid isolates in all three patients (having < 1 % genetic distance among the paired strains)(figure 1). Three of the four strains from the third patient were identical and clonally originating, while the fourth strain (1030b) had minor differences from the rest in having four polymorphic fragments (figure 2). This strain was also icaAB negative. 1030b strain showed closer similarity with 1030a (< 3% genetic distance) and clustered with it in dendrogram (figure 1). Thus, we believe that in the third patient, POE was caused by strains that were clonally evolving and having minor genetic variations.
Figure 1.
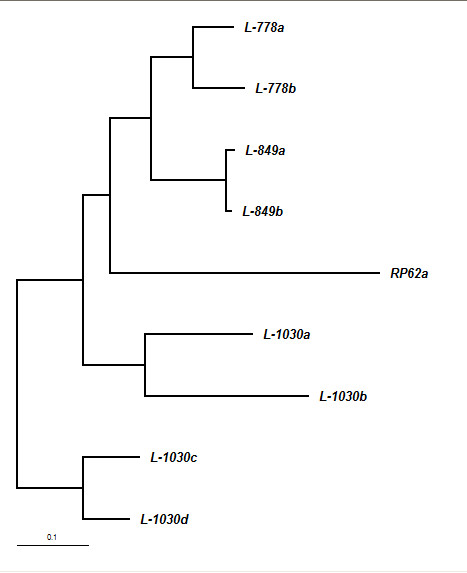
Neighbor joining tree showing similarity levels deduced from the genotyper data derived from FAFLP profiles of all the eight strains isolated from three patients along with the reference strain RP62a.
Figure 2.
S. epidermidis strain specific FAFLP electropherograms depicting the range between 150–350 bp for MseI+A selectivity tested, showing number and fragment sizes as well as the peak heights. The genotyper plots of paired isolates from patient 1 and patient 3. FAFLP patterns in order (top to bottom) are L-1030a, L-1030b, L-1030c, L-1030d, L-778a, and L-778b. Polymorphic fragment in L-1030a designated as 'e' in the figure (333 bp); polymorphic fragments in L-1030b designated as a, b, c, d (171, 191, 223, 235 bp).
Discussion
S. epidermidis present in the patient's periocular flora plays a significant role in causing POE [1,2]. This has been proven by a few studies using pulse field gel electrophoresis (PFGE) technique [2-4]. Application of advanced genotyping tools for typing of bacteria can give useful information regarding species distribution in periocular surfaces as well as help in understanding transmission dynamics and sources of infection, besides being useful in determining strain diversity with precision [4,7]. FAFLP typing in the present investigation established vitreous isolates of S. epidermidis strains were indistinguishable from the strains isolated from the patient's conjunctival swabs in all the 3 cases. These findings underscore the significant role of periocular flora in causation of POE. In this study, we examined the two important virulence factors of S epidermidis namely the biofilm production and multidrug resistance (MDR) of the strains associated with POE [8]. Biofilm- formation (BF) is one of the factors associated with increased virulence wherein icaAB genes were detected significantly more in infecting strains rather than in commensals [8]. MDR strains are known to cause greater intraocular inflammation than the susceptible strains in animal models [10] implying that these strains are more virulent. MR S epidermidis (MRSE) causing POE is difficult to treat. In recent years, the susceptibility of S. epidermidis has changed dramatically wherein nearly half of the strains are MDR. In particular, MR is frequent among S. epidermidis strains on a global scale [10] and the findings were similar in this study. In the present study, all the strains were MR, seven out of eight strains were positive for BF which is striking and substantiates earlier findings [8,10]. Presence of local/systemic risk factors and prophylactic antibiotic may enhance the role of MDR Coagulase negative Staphylococci in causing POE. One of our patients (no. 3) was diabetic for past 10 years, which is a possible risk factor. Information about prior antibiotic usage in our patients was not available. icaAB gene was detected in seven strains in this study indicating biofilm positivity. Strain 1030b appear to be ancestral with the absence of icaAB. 1030b generated four polymorphic bands (171, 191, 223, 235 bp) in FAFLP that were lacking in other three strains obtained from the same patient (figure 2) and this was the only icaAB negative strain. This finding suggests that biofilm-producing clones of resistant S. epidermidis were clonally evolving with changes occurring in their genomes. These clones appear to have genetic alteration that possibly indicates enhanced ability to cause POE. Strain 1030b from the lid of patient 3 is most likely nonpathogenic and ancient while other three strains from the same patient might have evolved from 1030b into more virulent types.
Predictive insilico AFLP methods with EcoRI+0 and MseI+A selectivity used on sequenced genomes of two S. epidermidis strains that are in the public domain and their extrapolation and comparison with our data showed interesting results. 1030b strain showed differential amplification of four genomic regions whereas 1030a, 1030c, 1030d lacked amplification of these regions which might be due to the mutation {insertion and deletion (Indels)} in EcoRI and MseI restriction site sequences. These corresponding polymorphisms are mapped to ORFs such as SE0884, SE0177, SE2068, SE0841 in ATCC 12228 and SE0775, SE2397, SE 2081, SE1995 in RP62a strain. Modification in at least two of the above ORFs namely SE0775 (fbe gene) and SE2081 putatively coding for fibronectin/fibrinogen binding protein and an adhesion protein respectively, seems to have definite role in increasing virulence. One recent study has showed that Fbe is a major factor involved in adherence of S. epidermidis to fibrinogen [11]. Antibodies against Fbe could block adherence of the bacteria to fibrinogen-coated surfaces. SE2081 is putative adhesion lipoprotein that may also have role in adhesion. Mutations in these two ORFs in the genomes of 1030a, 1030c, and 1030d might have enhanced their virulence favoring their survival under stressed condition.
Conclusion
Refined molecular typing tool like FAFLP potentially presents as a highly sensitive rapid technique for strain typing. Methicillin-resistant biofilm positive S. epidermidis strains colonizing the conjunctiva and eyelid may be responsible for POE. The versatile results of our study show that FAFLP can detect subtle genetic variations in the colonizing strains that may explain their possible role in altered virulence.
List of abbreviations used
AC: Anterior chamber
POE: Postoperative endophthalmitis
CoNS: Coagulase negative staphylococci
FAFLP: Fluorescence amplified fragment length polymorphisms
MDR: Multiple Drug Resistance
MR: Methicillin resistance
BF: Biofilm formation
MRSE: Methicillin Resistant Staphylococcus epidermidis
PFGE: Pulse Field Gel Electrophoresis
IOL: Intraocular lens
Competing interests
The author(s) declare that they have no competing interests
Authors' contributions
PK: Conception and design, performed FAFLP experiments, analysis and interpretation, writing the article, provision of materials, patients, or resources, obtaining funding.
AD: Data collection, provision of materials, patients, or resources, literature search
NA: Analysis and interpretation, technical, or logistic support
AP: Data collection, provision of materials, patients, or resources
TD: Critical revision of the article, administrative, technical, or logistic support
SEH: Critical revision of the article, final approval of the article
SS: Conception and design, analysis and interpretation, writing the article, provision of materials, patients, or resources
All authors have read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Acknowledgments
Acknowledgements
PK thanks the Department of Science and Technology (DST), Government of India for a Young Scientist award and project grant (SR/FTP/LS-A-49/2001).
Contributor Information
Prashanth Kenchappa, Email: prashant@cdfd.org.in.
Aparna Duggirala, Email: aparna_duggirala@rediffmail.com.
Niyaz Ahmed, Email: niyaz@cdfd.org.in.
Avinash Pathengay, Email: avinash@lvpei.org.
Taraprasad Das, Email: tpd@lvpei.org.
Seyed E Hasnain, Email: hasnain@cdfd.org.in.
Savitri Sharma, Email: savitri@lvpei.org.
References
- Kresloff MS, Castellario AA, Zarbin MA. Endophthalmitis. Surv Ophthalmol. 1998;43:193–224. doi: 10.1016/S0039-6257(98)00036-8. [DOI] [PubMed] [Google Scholar]
- Leong JK, Shah R, McCluskey PJ, Benn RA, Taylor RF. Bacterial contamination of the anterior chamber during phacoemulsification cataract surgery. J Cataract Refract Surg. 2002;28:826–33. doi: 10.1016/S0886-3350(01)01160-9. [DOI] [PubMed] [Google Scholar]
- Speaker MG, Milch FA, Shah MK, Eisner W, Kreiswirth BN. Role of external bacterial flora in the pathogenesis of acute postoperative endophthalmitis. Ophthalmology. 1991;98:639–49. doi: 10.1016/s0161-6420(91)32239-5. [DOI] [PubMed] [Google Scholar]
- Bannerman TL, Rhoden DL, McAllister SK, Miller JM, Wilson LA. The source of coagulase-negative staphylococci in the Endophthalmitis Vitrectomy Study. A comparison of eyelid and intraocular isolates using pulsed-field gel electrophoresis. Arch Ophthalmol. 1997;115:357–61. doi: 10.1001/archopht.1997.01100150359008. [DOI] [PubMed] [Google Scholar]
- Han DP, Wisniewski SR, Wilson LA, Barza M, Vine AK, Doft BH, Kelsey SF. Spectrum and susceptibilities of microbiologic isolates in the endophthalmitis vitrectomy study; the endophthalmitis vitrectomy study group. Am J Ophthalmol. 1996;122:1–17. doi: 10.1016/s0002-9394(14)71959-2. [DOI] [PubMed] [Google Scholar]
- Das T, Choudhary K, Sharma S, Jalali S, Nuthethi R, Endophthalmitis Research Group Clinical profile and outcome in Bacillus endophthalmitis. Ophthalmology. 2001;108:1819–1825. doi: 10.1016/S0161-6420(01)00762-X. [DOI] [PubMed] [Google Scholar]
- Sloos JH, Janssen P, van Boven CP, Dijkshoorn L. AFLP typing of Staphylococcus epidermidis in multiple sequential blood cultures. Res Microbiol. 1998;149:221–8. doi: 10.1016/S0923-2508(98)80082-X. [DOI] [PubMed] [Google Scholar]
- Frebourg NB, Lefebvre S, Baert S, Jean-Franç Lemeland O. PCR-Based assay for discrimination between invasive and contaminating Staphylococcus epidermidis strains. J Clin Microbiol. 2000;38:877–880. doi: 10.1128/jcm.38.2.877-880.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed N, Bal A, Khan AA, Alam M, Kagal A, Arjunwadkar V, Rajput A, Majeed AA, Rahman SA, Banerjee S, Joshi S, Bharadwaj R. Whole genome fingerprinting and genotyping of multiple drug resistant (MDR) isolates of Pseudomonas aeruginosa from endophthalmitis patients in India. Infect Genet Evol. 2002;1:237–242. doi: 10.1016/S1567-1348(02)00030-8. [DOI] [PubMed] [Google Scholar]
- Mino de Kaspar H, Shriver EM, Nguyen EV, Egbert PR, Singh K, Blumenkranz MS, Ta CN. Risk factors for antibiotic-resistant conjunctival bacterial flora in patients undergoing intraocular surgery. Graefes Arch Clin Exp Ophthalmol. 2003;41:730–3. doi: 10.1007/s00417-003-0742-5. [DOI] [PubMed] [Google Scholar]
- Pei L, Flock JI. Lack of fbe, the gene for a fibrinogen-binding protein from Staphylococcus epidermidis, reduces its adherence to fibrinogen coated surfaces. Microb Pathog. 2001;31:185–93. doi: 10.1006/mpat.2001.0462. [DOI] [PubMed] [Google Scholar]