Abstract
Background
The extracellular matrix has a major effect upon the malignant properties of bladder cancer cells both in vitro in 3-dimensional culture and in vivo. Comparing gene expression of several bladder cancer cells lines grown under permissive and suppressive conditions in 3-dimensional growth on cancer-derived and normal-derived basement membrane gels respectively and on plastic in conventional tissue culture provides a model system for investigating the interaction of malignancy and extracellular matrix. Understanding how the extracellular matrix affects the phenotype of bladder cancer cells may provide important clues to identify new markers or targets for therapy.
Methods
Five bladder cancer cell lines and one immortalized, but non-tumorigenic, urothelial line were grown on Matrigel, a cancer-derived ECM, on SISgel, a normal-derived ECM, and on plastic, where the only ECM is derived from the cells themselves. The transcriptomes were analyzed on an array of 1186 well-annotated cancer derived cDNAs containing most of the major pathways for malignancy. Hypervariable genes expressing more variability across cell lines than a set expressing technical variability were analyzed further. Expression values were clustered, and to identify genes most likely to represent biological factors, statistically over-represented ontologies and transcriptional regulatory elements were identified.
Results
Approximately 400 of the 1186 total genes were expressed 2 SD above background. Approximately 100 genes were hypervariable in cells grown on each ECM, but the pattern was different in each case. A core of 20 were identified as hypervariable under all 3 growth conditions, and 33 were hypervariable on both SISgel and Matrigel, but not on plastic. Clustering of the hypervariable genes showed very different patterns for the same 6 cell types on the different ECM. Even when loss of cell cycle regulation was identified, different genes were involved, depending on the ECM. Under the most permissive conditions of growth where the malignant phenotype was fully expressed, activation of AKT was noted. TGFβ1 signaling played a major role in the response of bladder cancer cells to ECM. Identification of TREs on genes that clustered together suggested some clustering was driven by specific transcription factors.
Conclusion
The extracellular matrix on which cancer cells are grown has a major effect on gene expression. A core of 20 malignancy-related genes were not affected by matrix, and 33 were differentially expressed on 3-dimensional culture as opposed to plastic. Other than these genes, the patterns of expression were very different in cells grown on SISgel than on Matrigel or even plastic, supporting the hypothesis that growth of bladder cancer cells on normal matrix suppresses some malignant functions. Unique underlying regulatory networks were driving gene expression and could be identified by the approach outlined here.
Background
Bladder cancer has the fourth highest incidence in men and seventh in women in the US [1]. Superficial papillary bladder cancers, though they may grow very large, do not invade and generally can be managed by excision. The more aggressive invasive bladder cancers that develop de novo have a five-year survival rate of 5% or less if metastasis has occurred [2,3]. Although 85% of bladder cancers first appear as superficial papillary carcinomas, up to two-thirds of patients experience recurrence, with approximately 15–25% progressing to higher grade tumors [2,3]. Approximately half of the deaths from bladder cancer result from progression. The extracellular matrix plays a crucial role in the development and evolution of bladder cancers [4-6]. The finding that specific genetic alterations frequently associated with bladder cancer are detectable in histologically normal urothelium of patients with bladder cancer [7] suggests that the malignant phenotype can be suppressed in vivo. This observation strongly suggests that understanding how the extracellular matrix modulates the phenotype of bladder cancer cells is highly relevant to understanding the processes of recurrence and progression.
Models for investigating interactions between the extracellular matrix (ECM) and bladder cancer cells using 3-dimensional culture have shown the ECM plays a crucial role in modulating the phenotype of bladder cancer cells [4,5,8-10]. Previous studies in our laboratory show suppression of the malignant phenotype when cells are cultured on a normal ECM [9,10]. On Matrigel, which is prepared from a cancer-modified ECM, bladder cancer cells recapitulate their in vivo phenotype. In contrast, when grown on SISgel, which is prepared from normal basement membrane ECM, the tumorigenic cell lines lose their invasiveness or ability to form papillary structures. Instead, they form either multiple or single layers resembling normal urothelium. By systematically varying both malignancy in the form of the cell line and the ECM on which they are grown, the interaction between these biological variables can be examined at the level of gene transcription. In this study, expression of 1186 well-annotated cancer-associated genes was measured on a cDNA array. The study design involved 6 cell lines varying from non-malignant to highly malignant grown on two ECM preparations and on plastic. Because practical considerations limited the number of replicates to 2, the dataset was inherently "noisy." We therefore developed statistically valid, novel approaches to data analysis that can identify biologically relevant patterns in a noisy dataset. Application of these methods for the given dataset yielded clear distinction in gene expression between different cell lines and/or matrixes.
Methods
Cell culture
SV-HUC-1, TCCSUP, RT4 and J82 cells were obtained from the American Type Culture Collection, Bethesda, MD, which provided information allowing the cells to be ranked by malignancy of the tumor of origin. The 253 J and 253 JB-V cells were provided by Dr. Colin Dinney [11]. The former is derived from a metastatic tumor, while the latter is a highly metastatic variant cloned in Dr. Dinney's laboratory after 5 passages of 253 J cells in the bladder walls of nude mice. Although invasive and metastatic, the tumor morphology is papillary. The ranking according to malignancy from lowest to highest is: SV-HUC-1 (non-malignant but immortalized), RT4 (low grade), 253 J (high grade) 253 JB-V (high grade), J82 (high grade), TCCSUP (high grade). Excepting the non-malignant SV-HUC-1 cells and possibly the TCCSUP cells (see below), all the cancer cell lines are transitional cell carcinomas (TCC). Details of cell culture on Matrigel and SISgel have been reported previously [9,10]. Culture on both matrices plus plastic in which the proliferation rate is approximately the same in all 3 has been described previously [12].
Array protocol
RNA was isolated from the cells growing in 3 dimensional culture using the RNeasy kit (Qiagen) by adding 300 μl lysis buffer to the culture well and pipeting up and down to lyse the cells and dissolve the gel. The RNA was isolated from the lysate using a QIAshredder spin column to complete homogenization followed by proteinase K digestion, washing, DNase 1 treatment and elution from RNeasy spin column. Yield of total RNA and the purity were assessed by spectrophotometry. The yield was approximately 2.4 – 6 μg RNA per culture in 30 μl. First strand synthesis was carried out at 42°C for 1 h with reagents supplied by Clontech as part of the SMART II kit, except that Superscript II (Invitrogen) reverse transcriptase was substituted. The SMART II oligonucleotide was included to capture full length cDNA at the 5'end and to permit a proportional 2,000 – 5,000 fold amplification [13]. Labeled probe was produced from an aliquot of the amplified probe with Klenow fragment and 32P-labeled nucleotide. After probe purification, 15 × 106 cpm of probe was hybridized in duplicate to the Clontech Human Cancer 1.2 array on nylon following the protocol provided by the company. The Clontech Human Cancer 1.2 array is a focused array consisting of 1186 well-annotated cancer-associated genes. The results were visualized by exposing a phosphoimager screen overnight and then reading with the Packard Cyclone phosphoimager and Optiquant software.
Data normalization
Data were normalized as described in detail elsewhere [14]. In brief, the normalization method relies on a number of low expression genes to provide an estimate of non-specific binding. This information is then used to perform a Z transformation on the data. Once normalized, the data are "unbiased" through robust linear regression to allow comparisons across different arrays. Fitted data are then used to find a homoscedastic group of gene variances that will be used as an internal standard of measurement noise [15]. Expressed genes (expression levels 2 standard deviations above the mean of the background) were noted, anti-log transformed, and used for subsequent analysis. This filtering step minimizes false positives, though at the cost of the low-expression genes, which are measured with low precision anyway.
Hypervariable gene identification
The logic of this step is that the variability within a dataset consists of both experimental error and variability introduced by the biological variables of interest. Moreover, while genes that do not vary significantly may be representing ongoing physiologic processes, they are unlikely to be correlated with malignancy or the effect of ECM since they do not change with either of those variables. Dozmorov, et al. [14] presented a method to identify the subset of genes that most likely represent experimental variability. Those genes with statistically significant variability greater than this subset represent the hypervariable genes that are most likely to represent the action of biological variables. Thus, in this study any gene that showed variable expression across cell lines exceeding the estimate of technical variability would be considered hypervariable. In order to reduce noise in the dataset and to insure adequate statistical power, the list was filtered to remove genes that were not expressed 2 SD above background in at least 4 of 6 cell lines for growth on each ECM condition. This filtering reduced the 1186 genes to roughly 400 for each matrix, with about 100 being identified as hypervariable.
Correlational clustering
The expression values of the hypervariable genes were clustered using Gene Cluster from M. B. Eisen [16]. The data were log-transformed, arrays and genes were median-centered and the transformed dataset was hierarchically clustered using uncentered correlational clustering. The results were displayed with TreeView, also from M. B. Eisen.
Identification of significantly over-represented gene ontologies
The annotations of the gene lists and their individual ontologies were first updated using the publicly available on-line tool DAVID EASE [17]. The null hypothesis that the clusters identified arose by chance was tested by determining whether any gene ontologies were over-represented above chance using the Gene Ontology Tree Machine (GOTM) [18,19].
Pathway analysis
Biologically relevant networks were assembled from identified clusters and groups of common genes by using Ingenuity Pathways Analysis (IPA). This web-delivered application [20] enables the visualization and analysis of direct and indirect interactions among genes. Selected data sets were uploaded as Excel files containing GenBank Accession numbers for genes of interest. Each gene identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base. Genes were not weighted by expression levels, and biological networks were built on this assumption.
Transcriptional regulatory element (TRE) analysis
One strategy for analyzing the significance of gene clusters identified by correlational clustering is based upon the assumption that co-expressed genes are likely to share common regulatory motifs [21]. This analysis was performed using the web-based program PAINT [22,23]. The results depend upon the reference chosen thereby reflecting Bayesian probability. If the entire genome is selected as the reference, the results will emphasize TREs common to bladder cells and would therefore identify tissue-specific TREs. Alternately, if the list of all genes expressed on a given matrix is selected as the reference, then the results should be enriched in those TREs responsible for the clustering rather than tissue-specific promoters. If the reference is the list of hypervariable genes, then significance of the partitioning of the hypervariable genes by clusters and TREs is identified. The PAINT analysis was performed against each of the indicated references, but only the reference against all hypervariable genes was reported.
Results
Identification of hypervariable genes
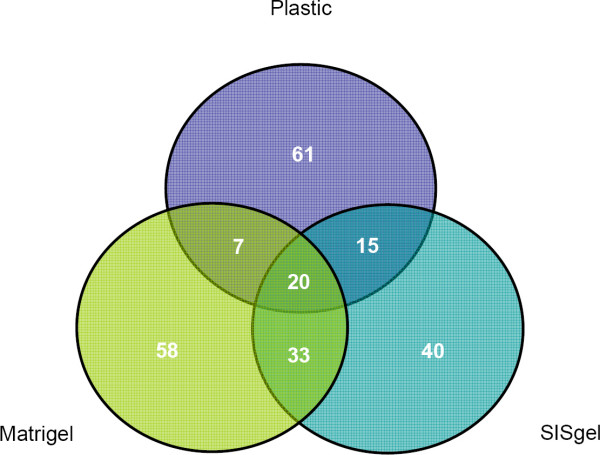
Cells grown on plastic expressed 422/1186 genes in at least 4/6 cell lines. On SISgel and Matrigel the corresponding numbers were 411 and 409 respectively. Overall, 371 genes were expressed in at least 12/18 conditions (6 cell lines grown under 3 conditions). The lists were further filtered by identifying the set of hypervariable genes. This yielded lists of approximately 100 genes from each set of experiments on an individual matrix. The results are expressed by a Venn diagram (Figure 1). The full gene lists and descriptions are available [see Additional file 1]. Figure 1 shows roughly half of each set of hypervariable genes was expressed uniquely on one ECM. Only 20 genes were expressed in common on all three ECMs (Table 1), and 33 genes were showed hypervariable expression on both SISgel and Matrigel (Table 2), whereas 15 and 7 were hypervariable on plastic and either SISgel and Matrigel, respectively. Full ontologic descriptions of these 20 and 33 genes are presented [see Additional file 2, Additional file 3].
Figure 1.
Venn diagram of hypervariable genes expressed on different matrixes. Each circle contains number of hypervariable genes unique and/or common for different matrixes. The size of circles does not represent relative number of genes in each group.
Table 1.
Hypervariable genes commonly expressed between Matrigel, Plastic and SISgel
| GENBANK | SYMBOL | GENENAME |
| U13696 | PMS2 | PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| M68520 | CDK2 | cyclin-dependent kinase 2 |
| X80692 | MAPK6 | mitogen-activated protein kinase 6 |
| X76104 | DAPK1 | death-associated protein kinase 1 |
| S85655 | PHB | prohibitin |
| AF000546 | P2RY5 | purinergic receptor P2Y, G-protein coupled, 5 |
| M97934 | STAT2 | signal transducer and activator of transcription 2, 113 kDa |
| X02851 | IL1A | interleukin 1, alpha |
| X86779 | FASTK | FAST kinase |
| U56390 | CASP9 | caspase 9, apoptosis-related cysteine protease |
| U60520 | CASP8 | caspase 8, apoptosis-related cysteine protease |
| X57766 | MMP11 | matrix metalloproteinase 11 (stromelysin 3) |
| M55172 | AGC1 | aggrecan 1 (chondroitin sulfate proteoglycan 1, large aggregating proteoglycan, antigen identified by monoclonal antibody A0122) |
| X03168 | VTN | vitronectin (serum spreading factor, somatomedin B, complement S-protein) |
| U65410 | MAD2L1 | MAD2 mitotic arrest deficient-like 1 (yeast) |
| X06374 | PDGFA | platelet-derived growth factor alpha polypeptide |
| L27943 | CDA | cytidine deaminase |
| X03124 | TIMP1 | tissue inhibitor of metalloproteinase 1 (erythroid potentiating activity, collagenase inhibitor) |
| A14844 | IL2 | interleukin 2 |
| M26326 | KRT18 | keratin 18 |
Table 2.
Hypervariable genes commonly expressed between Matrigel and SISgel
| GENBANK | SYMBOL | GENENAME |
| U13695 | PMS1 | PMS1 postmeiotic segregation increased 1 (S. cerevisiae) |
| M63167 | AKT1 | v-akt murine thymoma viral oncogene homolog 1 |
| M11810 | OAS1 | 2',5'-oligoadenylate synthetase 1, 40/46 kDa |
| X05360 | CDC2 | cell division cycle 2, G1 to S and G2 to M |
| M32865 | G22P1 | thyroid autoantigen 70 kDa (Ku antigen) |
| M57627 | IL10 | interleukin 10 |
| S74678 | HNRPK | heterogeneous nuclear ribonucleoprotein K |
| U69127 | FUBP3 | far upstream element (FUSE) binding protein 3 |
| D17516 | ADCYAP1R1 | adenylate cyclase activating polypeptide 1 (pituitary) receptor type I |
| U22398 | CDKN1C | cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
| L25081 | ARHC | ras homolog gene family, member C |
| M35416 | RALB | v-ral simian leukemia viral oncogene homolog B (ras related; GTP binding protein) |
| M11886 | HLA-C | major histocompatibility complex, class I, C |
| X74295 | MGC17301 | hypothetical protein MGC17301 |
| X02812 | TGFB1 | transforming growth factor, beta 1 (Camurati-Engelmann disease) |
| M18082 | SERPINB2 | serine (or cysteine) proteinase inhibitor, clade B (ovalbumin), member 2 |
| K02770 | IL1B | interleukin 1, beta |
| U60519 | CASP10 | caspase 10, apoptosis-related cysteine protease |
| D13866 | CTNNA1 | catenin (cadherin-associated protein), alpha 1, 102 kDa |
| U09579 | CDKN1A | cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
| U78095 | SPINT2 | serine protease inhibitor, Kunitz type, 2 |
| U53446 | DAB2 | disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) |
| X02811 | PDGFB | platelet-derived growth factor beta polypeptide (simian sarcoma viral (v-sis) oncogene homolog) |
| X91940 | WNT8B | wingless-type MMTV integration site family, member 8B |
| X74295 | ITGA7 | integrin, alpha 7 |
| M30938 | XRCC5 | X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining; Ku autoantigen, 80 kDa) |
| L07515 | CBX5 | chromobox homolog 5 (HP1 alpha homolog, Drosophila) |
| M34225 | KRT8 | keratin 8 |
| J05593 | TIMP2 | tissue inhibitor of metalloproteinase 2 |
| D83597 | LY64 | lymphocyte antigen 64 homolog, radioprotective 105 kDa (mouse) |
| M82882 | ELF1 | E74-like factor 1 (ets domain transcription factor) |
| J00209 | IFNA10 | interferon, alpha 10 |
| U09825 | TRIM26 | tripartite motif-containing 26 |
Clustering of hypervariable genes
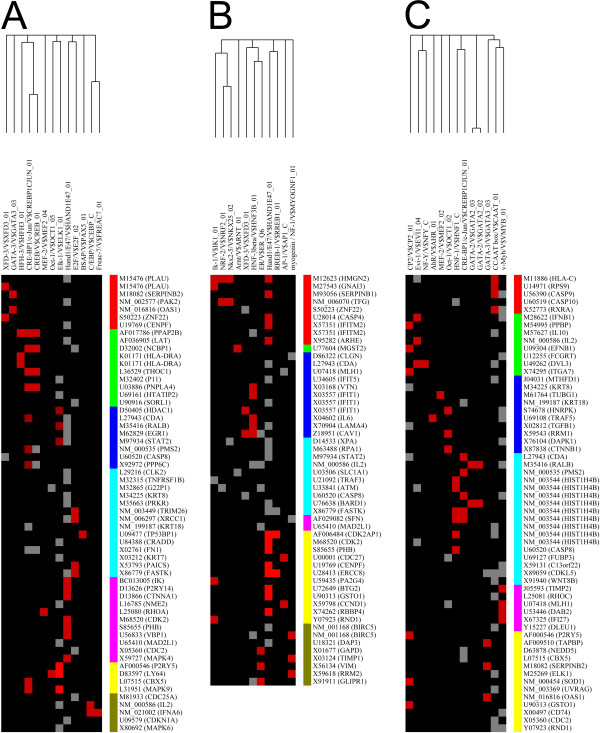
Hierarchical clustering by correlation of gene expression is presented in Figure 2. Gene expression from cells grown on Matrigel, where the bladder cancer cells are capable of fully expressing their malignant phenotype, clustered into seven main correlational clusters (M1–M7) (Figure 2A). The correlations in expression in some of the clusters, such as M2 and M4, are very high (>0.8). The clustering tended to be driven by expression in individual cell types rather than groups, such as cells derived from low-grade tumors. The driving signature (the cell line that drives organization of a cluster), and the over-represented ontologies are summarized in Table 3. The full statistical information as well as gene names associated with individual ontologies are listed [see Additional file 4].
Figure 2.
Hierarchical clustering of hypervariable genes across different cell lines on different matrixes. Hierarchical clustering of genes up- (red color) or downregulated (green color) on the given 6 cell lines. A, B and C show clustering of genes expressed on Matrigel, Plastic and SISgel, respectively. Colored bars outline individual clusters used for subsequent analysis.
Table 3.
Driving signatures and ontologies of gene clusters on different matrixes
| Cluster | Driving signature | Ontology |
| Matrigel_1 | J82 underexpressed | negative regulation of biological process |
| Matrigel_2 | JP underexpressed | mRNA processing |
| Matrigel_3 | HUC & JB overexpressed | mismatch repair; transcription factor activity |
| Matrigel_4 | HUC underexpressed | anti-apoptosis; induction of apoptosis by extracellular signals; protein binding; protein serine/threonine kinase activity |
| Matrigel_5 | JB underexpressed | regulation of cyclin dependent protein kinase activity; mitotic control; negative regulation of cell proliferation |
| Matrigel_6 | JB & J82 underexpressed | |
| Matrigel_7 | J82 & JP overexpressed | regulation of cyclin dependent protein kinase activity; anti-apoptosis; negative regulation of cell proliferation; positive regulation of cell proliferation; extracellular space |
| Plastic_1 | JP underexpressed | induction of apoptosis |
| Plastic_2 | JP underexpressed RT4 overexpressed |
immune response |
| Plastic_3 | TCCSUP & J82 overexpressed | negative regulation of cell cycle |
| Plastic_4 | TCCSUP underexpressed | protein kinase cascade; meiotic recombination; cell cycle arrest; anti-apoptosis; induction of apoptosis by extracellular signals; protein ubiquitination; nucleotide-excision repair; protein binding; nucleus |
| Plastic_5 | mitotic cell cycle; chromosome | |
| Plastic_6 | J82 underexpressed | cell cycle arrest; nucleus |
| Plastic_7 | HUC underexpressed | negative regulation of cellular process; antioxidant activity; extracellular matrix |
| SISgel_1 | J82 & JP overexpressed | cell cycle; induction of apoptosis; extracellular matrix |
| SISgel_2 | HUC underexpressed J82 overexpressed |
regulation of cell cycle; negative regulation of cell proliferation; inflammatory response |
| SISgel_3 | JB underexpressed | induction of apoptosis by extracellular signals; DNA replication; purine nucleotide biosynthesis; ligase activity, forming carbon-nitrogen bonds; cytoskeleton |
| SISgel_4 | JB overexpressed | |
| SISgel_5 | RT4 underexpressed | negative regulation of cell cycle |
| SISgel_6 | J82 underexpressed | cell division; protein complex assembly |
Analysis of biological pathways
Ingenuity Pathways Analysis identified specific biological pathways behind each given cluster on different ECMs. Cluster M4, containing 29 genes underexpressed in non-malignant HUC cells, showed two non-overlapping networks of 16 and 10 genes, respectively. The first pathway involved TGFβ1 signaling. The second pathway involved p53, IFNG and Fos-responsive genes. The details of the genes identified and their functions are listed in Table 4. The pathways showing the interconnections among the genes are shown [see Additional file 6]. Cluster P7, which consists of 16 genes underexpressed in HUC cells, showed a network of 12 genes involving TGFβ1 and p53 as central players (Table 4).
Table 4.
Genes and Functions in Ingenuity Pathways
| Cluster/Network | Genes | Top Functions |
| Matrigel_4/1 | AKT1, CLK2, CSE1L, DAPK1, FN1, G22P1, KRT7, KRT8, KRT18, PARP1, RAC1, TGFB1, TIMP2, TNFRSF1B, XRCC1, XRCC5 | DNA replication, Recombination and Repair; Cell Death; Neurological Disease |
| Matrigel_4/2 | BAG1, CASP10, COPS5, CRADD, EIF2AK2, FASTIK, HNRPK, TP53BP1, TRAF6, XPO1 | Cell Cycle; Connective Tissue Disorders; Inflammatory Disease |
| Plastic_7/1 | BIRC5, CTPS, DAP3, GADP, GLIPR1, HLA-DMA, MMP7, MMP11, MT3, RRM2, TIMP1, TXNRD1 | Cancer; Cell Death; Respiratory Disease |
| Mat-Pla-SIS Common Genes/1 | AGC1, CASP8, CASP9, CDK2, DAPK1, FASTK, IL2, IL1A, KRT18, MAD2L1, MAPK6, MMP11, PDGFA, PHB, PMS2, STAT2, TIMP1, VNT | Cell Death; Cancer; Cell Cycle |
| Mat-SIS Common Genes/1 | ADCYAP1R1, AKT1, CDC2, CDKN1A, CDKN1C, HLA-C, IL10, IL1B, ITGA7, KRT8, PDGFB, PMS1, SERPINB2, SPINT2, TGFB1 | Cancer; Cell Morphology; Cell Cycle |
| Mat-SIS Common Genes/2 | CASP10, CBX5, CTNN1, DAB2, ELF1, G22P1, HNRPK, OAS1, RALB, TIMP2, XRCC5 | Cancer; Cell Death; Reproductive System Disease |
Pathway analysis of the 20 genes hypervariable under all growth conditions contained a network of 18 genes involved in TGFβ1 signaling. Contained in the 33 genes that were hypervariable on both ECM preparations but not on plastic were two networks of 15 and 11 genes respectively. The 15 gene network consisted of genes involved in TGFβ1 signaling, the AKT1 survival network, CDKN1A cell cycle regulation and IL1β signaling. The 11 gene network consisted of a c-Myc response network and EGF signaling. These genes are also listed in Table 4 and the pathways are presented [see Additional file 6].
Transcriptional regulatory element (TRE) analysis of correlational clusters
TRE analysis was used to determine whether the correlation clustering could be interpreted in terms of the actions of networks of transcription factors. Figure 3 shows the results of analysis of TREs with the PAINT program according to the clusters defined by the hierarchical clustering. Each cluster expressed a pattern that was distinct from other clusters confirming the results of hierarchical clustering. This can be seen on full TREs maps [see Additional file 5] or, more clearly, on filtered maps, see Figure 3.
Figure 3.
Filtered maps of TREs on gene clusters of hypervariable genes identified in cells growing on different matrixes. A, B and C show significantly significantly overrepresented (p < 0.05) TREs from cells grown on Matrigel, Plastic and SISgel, respectively. Colored bars represent individual gene clusters. The TREs and the transcription factors that bind to them (linked to Enterz Gene) are CCAAT/NFYC; CP2/TFCP2; CREB/CREBP; CRE-BP1/ATF2; E2F/E2F2; Elk-1/ELK1; ER/ESR1; GATA-3/GATA-3; Hand1/HAND1; HFH-3/FOXI1; HNF-1/TCF1; HNF-3/FOXA1; myogenin/MYOG; Oct-1/POU2F1; v-Myb/MYB; XFD-3/FOXA1.
Discussion
In this study we sought to identify genes that modulate the malignant phenotype of bladder cancer. The experimental design made genes modulating the phenotype hypervariable as six different cell lines with different inherent malignancies were grown on plastic and two different extracellular matrices. Because varying the substrate on which the cells are grown changes the malignant phenotype, genes that modulate the phenotype were therefore brought to the fore. Our approach for analysis of this noisy dataset first consisted of identifying the subset of hypervariable genes because this subset will contain the set of genes encapsulating the relevant biological effect [14]. This step is designed to minimize false negatives for further in silico analysis. The subsequent steps sought to generate testable hypotheses with minimal false positives. By finding gene ontologies, pathways and TRE motifs that were significantly enriched within individual clusters, genes that potentially functioned together in pathways were identified. Because microarray studies are weighted toward high expression genes, these approaches, particularly pathways and TRE analysis can identify effects due to low expression genes such as transcription factors whose expression may be too low to be detected with the microarray. Because these determinations are made with reference to statistical probability, the only step in the analysis process that does not contain a quantitative estimate of statistical significance is the clustering.
Except for clusters M4 and P7 the driving force for clustering appeared to represent the behavior of individual cell lines rather than overall malignancy. However, these two clusters identify genes generally expressed at higher levels in the malignant cells than in the HUC cells. Of particular interest is the apparent activation of pro-survival AKT1 signaling in the malignant cells on Matrigel [see Additional file 6]. Likewise, the receptor for TGFβ1 was expressed at high levels on all the malignant cells on Matrigel (Figure 2), which supports the finding of TGFβ1 signaling genes in several clusters by the pathway analysis tool. Other ontologies in these two clusters clearly showed connections to cancer and apoptosis. The gene ontology and pathway analysis agreed in identifying the p53 signaling and TGFβ1 pathways within these clusters. While the mechanisms identified as operating only within a single cell line are probably less interesting than those operating in several, identification of a range of behaviors in bladder cancers could become useful if pathway-specific drugs are developed.
Some 20 genes were identified as being hypervariable, regardless the substrate. These represent a group of genes that are not modulated by the ECM but are differentially expressed among the different cell lines. Examples include Prohibitin (PHB), a survival gene, which is entirely matrix-independent. The connection with AKT1 survival pathway is clear. In bladder cancer, activation of AKT1-mediated survival has only been shown in TCCSUP cells in culture [24]. AKT1 can cause resistance to therapy in other cancers [25]. Interestingly, genes coding for two polysaccharide-binding proteins were also present. That these two would be differentially expressed by different cell types growing on polysaccharide-containing gels and the minimal endogenous matrix secreted by cells growing on plastic is consistent with the role of these molecules in differentiation and growth [26]. Additionally, 33 genes were identified as being hypervariable on both Matrigel and SISgel, in spite of the suppression of many features of the malignant phenotype on the latter. These genes probably represent a "core" set of cancer genes. Ontologies for this cluster of genes were cancer, cell morphology and cell cycle networks. Pathway analysis showed fifteen of these genes functioned in TGFβ1 signaling, which is a potent regulator of extracellular matrix production and proliferation [27]. An important role for TGFβ1 in bladder cancer has been proposed [28]. Eleven of these 33 genes are interconnected within the MYC network. Deregulation of MYC genes is associated with several malignancies [29]. Although the ECM can have major effects on the biology of individual cancers, a core of oncogenes involved in the p53 and MYC networks is unaffected. Interestingly, a set of signature genes for the suppression of the malignant phenotype by SISgel was not observed. Either the correct probes are not on the array, or each cell type respond uniquely to a changing matrix. Previous results showed this response was not due to integrin signaling [9] as has been reported by Weaver, et. al [30] to govern phenotypic reversion of glandular breast cancers.
The TRE analysis confirmed the uniqueness of the clustering and suggests that a limited set of TREs may be driving gene expression in this study. Notable is Hand1, which appears in most of the genes. This gene has been identified as playing a role in heart development [31], but the abundance in these genes that are expressed in bladder suggests it may also act as a bladder-specific factor. Several genes overexpressed in malignant cell lines in cluster M4 carried the E2F TRE. The corresponding transcription factor has been shown to promote cell growth in common human carcinomas [32] and to be dependent on the p53 pathway [32,33] supporting its role in upregulating the cluster in cancer cells. A different set of TREs was identified in Cluster P7. An estrogen receptor (ER) and myogenin (MYOG) were separately associated within P7. This combination suggests loss of epithelial differentiation. Bladder cancer cells were earlier demonstrated to contain both functional androgen and estrogen receptors without regard to the sex of the donor [34,35] while myogenin was primary expressed in rhabdomyosarcomas [36,37]. Identification of myogenin expression in bladder cancer cell lines is novel, and further research will be needed to determine if this transcription factor is actually active.
Comparisons to other microarray studies are, in general, less informative because of wide differences among array technologies [38] and criteria to identify "significant" genes. Nonetheless the composition of Cluster M4, which is comprised of genes that are under-expressed in HUC cells in comparison to all the cancer cells, shows 3 of the 28 genes in the cluster corresponded to 3 of the 29 genes identified as being diagnostic for TCC in patient samples [39]. This association suggests this cluster might provide additional diagnostic genes which is strongly supported by the finding of the p53-responsive gene network. In addition, although the comparison was made difficult by a lack of accession numbers or universal gene symbols, considerable correspondence between the 32 genes identified as hypervariable in 3-dimensional growth was noted with the set of genes reported to differentially expressed between superficial and invasive TCC [40].
Conclusion
The extracellular matrix on which cancer cells are grown has a major effect on gene expression. A core of 20 malignancy-related genes were not affected by matrix, and 33 were differentially expressed on 3-dimensional culture as opposed to plastic. Activation of the AKT1 survival pathway on Matrigel suggests this pathway could be relevant to clinical bladder cancer. The TGFβ1 signaling pathway plays an important role in the expression of the malignant phenotype. The p53 response network is confirmed to play a central role, as does c-MYC signaling. Evidence suggests the E2F TF drives some malignancy-related genes, with several other TFs being implicated as well. The identified pathways may provide new targets for detection and treatment therapies.
List of abbreviations
ECM – extracellular matrix
GO – gene ontology
SIS – small intestine submucosa
TCC – transitional cell carcinoma
TF – transcription factor
TRE – transcription regulatory element
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
MD performed overall analysis of hypervariable genes, prepared figures, tables and additional material and helped to draft and finalize the manuscript; KDK made the microarray studies, including isolation of mRNA and amplification and performed the preliminary data analysis; REH wrote the first draft of the manuscript and directed the overall study; RS assisted with the interpretation of the TRE analysis; NK performed preliminary clustering and identification of hypervariable genes; ID performed the data normalization and assisted with the interpretation of results; MBC directed the overall bioinformatics program and helped with the data interpretation. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Table 1. Description and ontology of each hypervariable gene grouped by clusters on different matrixes
Table 2. Hypervariable genes commonly expressed between different matrixes and their ontologies. GOTM results for this group of genes are shown at the bottom of the table. O:Observed gene number in the GO category; E:Expected gene number in the GO category; R:Ratio of enrichment for the GO category; P:Significance of enrichment for the GO category
Table 3. Hypervariable genes commonly expressed between SISgel and Matrigel and their ontologies. GOTM results for this group of genes are shown at the bottom of the table. O:Observed gene number in the GO category; E:Expected gene number in the GO category; R:Ratio of enrichment for the GO category; P:Significance of enrichment for the GO category
Table 4. Genes driving significant ontologies on clusters on different matrixes. O:Observed gene number in the GO category; E:Expected gene number in the GO category; R:Ratio of enrichment for the GO category; P:Significance of enrichment for the GO category
Figure 2. Gene networks identified by Ingenuity analysis. Pathways for Matrigel, cluster 4, Plastic, cluster 7, common genes between all three matrixes, common genes between Matrigel and SISgel are shown. Focus genes that map to the Global Molecular Network are displayed with bold text. User input genes are emphasized by gray color. For pathway legend see bottom of the figure.
Figure 1A, 1B, 1C. Complete TREs maps for different matrixes. A, B and C show all promoters for a given set of genes expressed on Matrigel, Plastic and SISgel, respectively. Colored bars represent individual gene clusters.
Acknowledgments
Acknowledgements
This work was supported in part by NIH grants CA75322, DK069808 and a grant from Cook Biotech (REH) and P20 RR1557, P20 RR17703, and P20 RR16478 (MBC).
Contributor Information
Mikhail G Dozmorov, Email: Mikhail-Dozmorov@ouhsc.edu.
Kimberly D Kyker, Email: Kimberly-Kyker@ouhsc.edu.
Ricardo Saban, Email: Ricardo-Saban@ouhsc.edu.
Nicholas Knowlton, Email: Nicholas-Knowlton@omrf.ouhsc.edu.
Igor Dozmorov, Email: Igor-Dozmorov@omrf.ouhsc.edu.
Michael B Centola, Email: Michael-Centola@omrf.ouhsc.edu.
Robert E Hurst, Email: Robert-Hurst@ouhsc.edu.
References
- Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, Feuer EJ, Thun MJ. Cancer statistics, 2005. CA Cancer J Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- Amling CL. Diagnosis and management of superficial bladder cancer. Curr Probl Cancer. 2001;25:219–278. doi: 10.1067/mcn.2001.117539. [DOI] [PubMed] [Google Scholar]
- Heney NM, Ahmed S, Flanagan MJ, Frable W, Corder MP, Hafermann MD, Hawkins IR. Superficial bladder cancer: progression and recurrence. J Urol. 1983;130:1083–1086. doi: 10.1016/s0022-5347(17)51695-x. [DOI] [PubMed] [Google Scholar]
- Smith BA, Kennedy WJ, Harnden P, Selby PJ, Trejdosiewicz LK, Southgate J. Identification of genes involved in human urothelial cell-matrix interactions: implications for the progression pathways of malignant urothelium. Cancer Res. 2001;61:1678–1685. [PubMed] [Google Scholar]
- Syrigos KN, Harrington KJ, Pignatelli M. Role of adhesion molecules in bladder cancer: an important part of the jigsaw. Urology. 1999;53:428–434. doi: 10.1016/S0090-4295(98)00527-5. [DOI] [PubMed] [Google Scholar]
- Rebel JM, Thijssen CD, Vermey M, Zwarthoff EC, van der Kwast TH. Modulation of intra-epithelial expansion of human T24 bladder-carcinoma cells in murine urothelium by growth factors and extracellular-matrix components. Int J Cancer. 1995;60:707–711. doi: 10.1002/ijc.2910600523. [DOI] [PubMed] [Google Scholar]
- Stoehr R, Zietz S, Burger M, Filbeck T, Denzinger S, Obermann EC, Hammerschmied C, Wieland WF, Knuechel R, Hartmann A. Deletions of chromosomes 9 and 8p in histologically normal urothelium of patients with bladder cancer. Eur Urol. 2005;47:58–63. doi: 10.1016/j.eururo.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Bohle A, Jurczok A, Ardelt P, Wulf T, Ulmer AJ, Jocham D, Brandau S. Inhibition of bladder carcinoma cell adhesion by oligopeptide combinations in vitro and in vivo. J Urol. 2002;167:357–363. doi: 10.1097/00005392-200201000-00101. [DOI] [PubMed] [Google Scholar]
- Hurst RE, Kyker KD, Bonner RB, Bowditch RG, Hemstreet GP. Matrix-Dependent Plasticity of the Malignant Phenotype of Bladder Cancer Cells. Anticancer Res. 2003;23:3119–3128. [PMC free article] [PubMed] [Google Scholar]
- Kyker KD, Culkin DJ, Hurst RE. A model for 3-dimensional growth of bladder cancers to study mechanisms of phenotypic expression. Urologic Oncology. 2003;21:255–261. doi: 10.1016/s1078-1439(02)00279-x. [DOI] [PubMed] [Google Scholar]
- Dinney CP, Fishbeck R, Singh RK, Eve B, Pathak S, Brown N, Xie B, Fan D, Bucana CD, Fidler IJ. Isolation and characterization of metastatic variants from human transitional cell carcinoma passaged by orthotopic implantation in athymic nude mice. J Urol. 1995;154:1532–1538. doi: 10.1097/00005392-199510000-00087. [DOI] [PubMed] [Google Scholar]
- Hurst RE, Kamat CD, Kyker KD, Green DE, Ihnat MA. A novel multidrug resistance phenotype of bladder tumor cells grown on Matrigel or SIS gel. Cancer Lett. 2005;217:171–180. doi: 10.1016/j.canlet.2004.07.043. [DOI] [PubMed] [Google Scholar]
- Seth D, Gorrell MD, McGuinness PH, Leo MA, Lieber CS, McCaughan GW, Haber PS. SMART amplification maintains representation of relative gene expression: quantitative validation by real time PCR and application to studies of alcoholic liver disease in primates. J Biochem Biophys Methods. 2003;55:53–66. doi: 10.1016/S0165-022X(02)00177-X. [DOI] [PubMed] [Google Scholar]
- Dozmorov I, Knowlton N, Tang Y, Shields A, Pathipvanich P, Jarvis JN, Centola M. Hypervariable genes--experimental error or hidden dynamics. Nucleic Acids Res. 2004;32:e147. doi: 10.1093/nar/gnh146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozmorov I, Knowlton N, Tang Y, Centola M. Statistical monitoring of weak spots for improvement of normalization and ratio estimates in microarrays. BMC Bioinformatics. 2004;5:53. doi: 10.1186/1471-2105-5-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVID EASE. 2005.
- GOTM. 2005.
- Zhang B, Schmoyer D, Kirov S, Snoddy J. GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinformatics. 2004;5:16.:16. doi: 10.1186/1471-2105-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingenuity Systems. 2005.
- Chiang DY, Brown PO, Eisen MB. Visualizing associations between genome sequences and gene expression data using genome-mean expression profiles. Bioinformatics. 2001;17 Suppl 1:S49–S55. doi: 10.1093/bioinformatics/17.suppl_1.s49. [DOI] [PubMed] [Google Scholar]
- Vadigepalli R, Chakravarthula , Zak DE, Schwaber JS, Gonye GE. PAINT: A Promoter Analysis and INteraction Network Generation Tool for Gene Regulatory Network Identification. OMICS: A Journal of Integrative Biology. 2003;7:235–252. doi: 10.1089/153623103322452378. [DOI] [PubMed] [Google Scholar]
- PAINT. 2005.
- Kim J, Adam RM, Freeman MR. Trafficking of nuclear heparin-binding epidermal growth factor-like growth factor into an epidermal growth factor receptor-dependent autocrine loop in response to oxidative stress. Cancer Res. 2005;65:8242–8249. doi: 10.1158/0008-5472.CAN-05-0942. [DOI] [PubMed] [Google Scholar]
- Kirkegaard T, Witton CJ, McGlynn LM, Tovey SM, Dunne B, Lyon A, Bartlett JM. AKT activation predicts outcome in breast cancer patients treated with tamoxifen. J Pathol. 2005;207:139–146. doi: 10.1002/path.1829. [DOI] [PubMed] [Google Scholar]
- Iozzo RV. Matrix proteoglycans: from molecular design to cellular function. Annu Rev Biochem. 1998;67:609–652. doi: 10.1146/annurev.biochem.67.1.609. [DOI] [PubMed] [Google Scholar]
- Kim JH, Shariat SF, Kim IY, Menesses-Diaz A, Tokunaga H, Wheeler TM, Lerner SP. Predictive value of expression of transforming growth factor-beta(1) and its receptors in transitional cell carcinoma of the urinary bladder. Cancer. 2001;92:1475–1483. doi: 10.1002/1097-0142(20010915)92:6<1475::AID-CNCR1472>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Ignotz RA, Massague J. Transforming growth factor-beta stimulates the expression of fibronectin and collagen and their incorporation into the extracellular matrix. J Biol Chem. 1986;261:4337–4345. [PubMed] [Google Scholar]
- Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- Weaver V, Lelievre S, Lakins J, Chrenek M, Jones J, Giancotti F, Werb Z, Bissell M. beta4 integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer Cell. 2002;2:205. doi: 10.1016/S1535-6108(02)00125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entrez Gene. 2005. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene&cmd=Retrieve&dopt=full_report&list_uids=9421
- Zacharatos P, Kotsinas A, Evangelou K, Karakaidos P, Vassiliou LV, Rezaei N, Kyroudi A, Kittas C, Patsouris E, Papavassiliou AG, Gorgoulis VG. Distinct expression patterns of the transcription factor E2F-1 in relation to tumour growth parameters in common human carcinomas. J Pathol. 2004;203:744–753. doi: 10.1002/path.1582. [DOI] [PubMed] [Google Scholar]
- Steinhoff C, Prior A, Reichmann G, Seifert HH, Schulz WA. Activity of E2F-dependent promoters in bladder carcinoma cells and their use for tumour-specific targeting of p53-induced apoptosis. Int J Oncol. 2002;21:1033–1040. [PubMed] [Google Scholar]
- Croft PR, Lathrop SL, Feddersen RM, Joste NE. Estrogen receptor expression in papillary urothelial carcinoma of the bladder and ovarian transitional cell carcinoma. Arch Pathol Lab Med. 2005;129:194–199. doi: 10.5858/2005-129-194-EREIPU. [DOI] [PubMed] [Google Scholar]
- Waliszewski P, Waliszewska MK, Hemstreet GP, Hurst RE. Expression of Sex Steroid Receptor Genes and Co-Modulation with Retinoid Signaling in Normal Human Uroepithelial Cells and Bladder Cancer Cell Lines. Urol Oncol. 1997;3:141–147. doi: 10.1016/S1078-1439(98)00011-8. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Sakai H, Hirata A, Yonemaru K, Yanai T, Watanabe K, Yamazoe K, Kudo T, Masegi T. Expression of myogenic regulating factors, Myogenin and MyoD, in two canine botryoid rhabdomyosarcomas. Vet Pathol. 2004;41:275–277. doi: 10.1354/vp.41-3-275. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Baba K, Saito A, Hoshi N, Suzuki T. Pseudosarcomatous myofibroblastic tumor and myosarcoma of the urogenital tract. Arch Pathol Lab Med. 2001;125:1070–1073. doi: 10.5858/2001-125-1070-PMTAMO. [DOI] [PubMed] [Google Scholar]
- Tan PK, Downey TJ, Spitznagel ELJ, Xu P, Fu D, Dimitrov DS, Lempicki RA, Raaka BM, Cam MC. Evaluation of gene expression measurements from commercial microarray platforms. Nucleic Acids Res. 2003;31:5676–5684. doi: 10.1093/nar/gkg763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor O, Nativ O, Stein A, Novak L, Lehavi D, Shiboleth Y, Rozen A, Berent E, Brodsky L, Feinstein E, Rahav A, Morag K, Rothenstein D, Persi N, Mor Y, Skaliter R, Regev A. Molecular analysis of transitional cell carcinoma using cDNA microarray. Oncogene. 2003;22:7702–7710. doi: 10.1038/sj.onc.1207039. [DOI] [PubMed] [Google Scholar]
- Modlich O, Prisack HB, Pitschke G, Ramp U, Ackermann R, Bojar H, Vogeli TA, Grimm MO. Identifying superficial, muscle-invasive, and metastasizing transitional cell carcinoma of the bladder: use of cDNA array analysis of gene expression profiles. Clin Cancer Res. 2004;10:3410–3421. doi: 10.1158/1078-0432.CCR-03-0134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1. Description and ontology of each hypervariable gene grouped by clusters on different matrixes
Table 2. Hypervariable genes commonly expressed between different matrixes and their ontologies. GOTM results for this group of genes are shown at the bottom of the table. O:Observed gene number in the GO category; E:Expected gene number in the GO category; R:Ratio of enrichment for the GO category; P:Significance of enrichment for the GO category
Table 3. Hypervariable genes commonly expressed between SISgel and Matrigel and their ontologies. GOTM results for this group of genes are shown at the bottom of the table. O:Observed gene number in the GO category; E:Expected gene number in the GO category; R:Ratio of enrichment for the GO category; P:Significance of enrichment for the GO category
Table 4. Genes driving significant ontologies on clusters on different matrixes. O:Observed gene number in the GO category; E:Expected gene number in the GO category; R:Ratio of enrichment for the GO category; P:Significance of enrichment for the GO category
Figure 2. Gene networks identified by Ingenuity analysis. Pathways for Matrigel, cluster 4, Plastic, cluster 7, common genes between all three matrixes, common genes between Matrigel and SISgel are shown. Focus genes that map to the Global Molecular Network are displayed with bold text. User input genes are emphasized by gray color. For pathway legend see bottom of the figure.
Figure 1A, 1B, 1C. Complete TREs maps for different matrixes. A, B and C show all promoters for a given set of genes expressed on Matrigel, Plastic and SISgel, respectively. Colored bars represent individual gene clusters.