Abstract
Spermidine/spermine N1-acetyltransferase (SSAT) is a key enzyme in the control of polyamine levels in human cells, as acetylation of spermidine and spermine triggers export or degradation. Increased intracellular polyamine levels accompany several types of cancers as well as other human diseases, and compounds that affect the expression, activity, or stability of SSAT are being explored as potential therapeutic drugs. We have expressed human SSAT from the cloned cDNA in Escherichia coli and have determined high-resolution structures of wild-type and mutant SSAT, as the free dimer and in binary and ternary complexes with CoA, acetyl-CoA (AcCoA), spermine, and the inhibitor N1,N11-bis-(ethyl)-norspermine (BE-3-3-3). These structures show details of binding sites for cofactor, substrates, and inhibitor and provide a framework to understand enzymatic activity, mutations, and the action of potential drugs. Two dimer conformations were observed: a symmetric form with two open surface channels capable of binding substrate or cofactor, and an asymmetric form in which only one of the surface channels appears capable of binding and acetylating polyamines. SSAT was found to self-acetylate lysine-26 in the presence of AcCoA and absence of substrate, a reaction apparently catalzyed by AcCoA bound in the second channel of the asymmetric dimer. These unexpected and intriguing complexities seem likely to have some as yet undefined role in regulating SSAT activity or stability as a part of polyamine homeostasis. Sequence signatures group SSAT with proteins that appear to have thialysine Nε-acetyltransferase activity.
Keywords: GCN5-related N-acetyltransferase family, polyamines, symmetric and asymmetric dimer, self-acetylation, thialysine Nε-acetyltransferase
The polyamines, spermine, spermidine, and their diamine precursor, putrescine, are aliphatic cations essential to the normal physiology and growth of virtually all cells (1). Cellular polyamine levels are tightly regulated by import, export, synthesis, and degradation of the polyamines themselves and by synthesis and degradation of the enzymes of polyamine metabolism. The enzymes and pathways of human polyamine metabolism have been well characterized and are similar to those of other mammals (2). The first enzyme in the synthesis of polyamines is ornithine decarboxylase (ODC), a control point in the pathway that converts ornithine to putrescine. The resulting putrescine is converted to spermidine and spermine by the aminopropyl transferases spermidine synthase and spermine synthase, where the aminopropyl group is derived from the decarboxylation of S-adenosylmethionine (SAM) by SAM-decarboxylase (SAMDC). The key enzyme for reducing polyamine levels is spermidine/spermine N1-acetyltransferase (SSAT), which acetylates spermine and spermidine, thereby targeting them for export from the cell or degradation by N1-acetyl-polyamine oxidase (PAO).
Altered levels of intracellular polyamines have been observed in Alzheimer’s disease (3), cystic fibrosis (4, 5), and in a variety of tumors and tumor-derived cell lines, including non-small-cell lung cancer, pancreatic cancer, melanoma, and ovarian cancer (6–10). The association with cancer has led to the development of compounds that target polyamine metabolism as a strategy for disrupting tumor cell growth (11). Compounds that targeted the biosynthetic pathway, such as difluoromethyornithine (DFMO), proved disappointing because the cell compensated by increasing the uptake of polyamines as needed. However, polyamine analogs, designed with the expectation that they would down-regulate synthetic enzymes, were serendipitously found to affect the catabolic machinery (for review, see ref. 12). Symmetrically substituted polyamine analogs, including N1,N11-bis-(ethyl)-norspermine (BE-3-3-3), N1,N12-bis-(ethyl)-spermine (BE-3-4-3), and N1,N14-bis-(ethyl)-homospermine (BE-4-4-4), and nonsymmetric compounds such as N1-propargyl-N11-ethyl-norspermine (PENSpm) and N1-cyclopropyl-methyl-N11-ethylnorspermine (CPENSpm) (13) have been invaluable tools for understanding the cellular functions of the natural polyamines. For example, BE-3-3-3 is an inhibitor of SSAT activity and has been shown to suppress transport of natural polyamines, depress levels of polyamine biosynthetic enzymes, and superinduce SSAT levels (9, 14, 15).
SSAT is rapidly inducible by a polyamine-dependent pathway and typically has a short biological half-life, being efficiently ubiquitylated and subsequently degraded by the proteasome (16–19). Under normal conditions, SSAT protein levels are very low, and at least part of the increase in SSAT levels caused by BE-3-3-3 is due to an increase in half-life, presumably because the bound inhibitor reduces ubiquitylation and thus degradation. To help understand the enzymatic activity of SSAT and the effects of potential therapeutic drugs, we have determined the structures of wild-type and mutant SSAT and several complexes with cofactor, substrate, and the inhibitor BE-3-3-3.
Results and Discussion
Determination of Structures.
Wild-type and K26R-mutant (lysine-26 replaced by arginine) human SSAT were labeled with selenomethionine during expression in Escherichia coli, purified, and crystallized alone or in complexes with the cofactor acetyl-CoA (AcCoA), the product CoA, or combinations of CoA plus the substrate spermine or CoA plus the inhibitor BE-3-3-3. Structures were determined by single-wavelength anomalous dispersion (SAD). Most of the crystals diffracted to 2.0 Å or better, and the electron density maps were usually of sufficient quality that >70% of the backbone structure could be obtained by using autotracing tools (20). The data collection and refinement statistics for six independently determined structures are given in Table 1.
Table 1.
Structural information
| Parameter | SSAT | SSAT + CoA + spermine | K26R SSAT | K26R SSAT +AcCoA | K26R SSAT +CoA +spermine | K26R SSAT +CoA +BE-3-3-3 |
|---|---|---|---|---|---|---|
| Unit cell, Å | a = b = 74.1, c = 63.1 | a = b = 74.0, c = 64.0 | a = b = 74.3, c = 63.3 | a = b = 63.0, c = 80.9 | a = b = 63.3, c = 81.8 | a = b = 73.1, c = 64.1 |
| Space group | P43 | P43 | P43 | P62 | P62 | P43 |
| Resolution, Å | 30.0–1.7 | 30.0–2.0 | 30.0–1.85 | 30.0–1.95 | 30.0–1.95 | 30.0–2.0 |
| No. of reflections | 36,988 | 22,575 | 28,155 | 12,884 | 13,228 | 21,641 |
| Rmerge (%) | 3.9 (11.5) | 4.7 (16.7) | 5.1 (27.9) | 4.2 (15.3) | 4.9 (13.0) | 4.6 (17.2) |
| Completeness (%) | 95.4 (77.4) | 97.6 (96.3) | 99.1 (99.6) | 96.6 (96.1) | 99.9 (100.0) | 95.9 (79.5) |
| Figure of merit | 0.46/0.63 | 0.46/0.58 | 0.37/0.58 | 0.47/0.61 | 0.52/0.66 | 0.43/0.54 |
| No. of reflections in free set | 1,497 | 1,127 | 1,404 | 651 | 658 | 1,085 |
| R-factor, % | 21.1 | 22.1 | 21.0 | 21.7 | 20.4 | 22.5 |
| Rfree, % | 24.1 | 25.5 | 24.6 | 25.5 | 25.2 | 26.5 |
| No. of protein | 2,667 | 2,802 | 2,697 | 1,380 | 1,380 | 2,794 |
| No. of N-acetyl on K26b | 3 | 3 | na | na | na | na |
| No. of CoA or AcCoA | na | 94 | na | 51 | 48 | 96 |
| No. of sulfate | 35 | 0 | 10 | 0 | 0 | 0 |
| No. of water | 323 | 75 | 157 | 59 | 82 | 83 |
| No. of spermine or BE-3-3-3 | na | weak density | na | na | not visible | 13 |
| PDB ID | 2B5G | 2B4D | 2B3U | 2B3V | 2B58 | 2B4B |
na, Not applicable.
Structure of ApoSSAT.
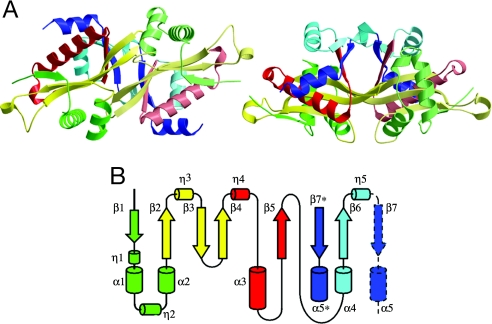
The structural model for wild-type SSAT contains a dimer in the asymmetric unit (Fig. 1A). The topology of secondary structure elements is given in Fig. 1B, and locations relative to the amino acid sequence are indicated in Fig. 2 above an alignment of SSAT with sequences of related proteins. The topology and central core structure of each SSAT monomer is typical of GCN5-related N-acetyltransferase (GNAT) superfamily members (21, 22) and is comprised of a β sheet flanked above and below by α helices. Dimer formation involves C-terminal arms, residues 143–171 that extend across the surface, incorporate strand β7 into the β sheet of the opposing monomer, and end in the amphipathic helix α5, which sits atop helix α3 of the opposing monomer at an angle of ≈45° and forms a part of its core structure (Fig. 1A). Dimer formation buries ≈1,300 Å2 of surface area, and the C-terminal arms contribute more than half of this surface.
Fig. 1.
Structure of apoSSAT. (A) Ribbon diagram showing two orthogonal views of the asymmetric dimer colored according to the topology diagram in B. (B) Topology of the secondary structure elements in chain A. From the N terminus, secondary structure elements are colored green (β1, η1, α1, η2, and α2), yellow (β2, η3, β3, and β4), red (η4, α3, and β5), cyan (α4, β6, and η5), and deep blue (β7 and α5). The C-terminal elements β7 and α5 (enclosed in a broken black line) form part of the core structure of the opposing monomer. The structure represented is that of PDB ID 2B5G5, in which density was not observed for residues 28–31 and 170–171 of chain A nor for residues 2–3, 47–50, 61–63, or 171 of chain B.
Fig. 2.
Alignment of SSAT with known TLAs and the PaiA polyamine N1-acetyltransferase of Bacillus subtilis, showing positions of secondary structure elements in the two protein structures. Dots above the SSAT sequence mark every tenth amino acid; helices of PaiA are numbered to correspond to those of SSAT, except that αC of PaiA has no corresponding helix in SSAT. The alignment shown summarizes the much more extensive alignment in Fig. 5, which is published as supporting information on the PNAS web site. Symbols above the SSAT sequence are as follows: A, residues involved in AcCoA binding; B, residues forming the alternate pocket for the acetyl group of AcCoA; K, residues involved in directing the acetyl group of AcCoA bound in channel 2 toward K26; P, residues implicated in binding polyamine or BE-3-3-3; W, residues that are simply part of the wall of the surface channel (as discussed in the text or shown in Figs. 3 and 4). Symbols that indicate similarities and differences in amino acid character between the SSAT-like sequences and PaiA-related sequences are as follows: *, the same amino acid in all proteins of the extensive alignment; C, conservation; S, similarity; D, difference. Potentially significant differences between conserved residues in vertebrate SSATs and in known or putative TLAs (apparent in the extensive alignment) occur at SSAT positions K26, Y29, M76, D82, P83, and A130.
The two monomers, although identical in amino acid sequence, have similar but not identical structures in the crystal form described here. The most pronounced difference between the structures of the two monomers is in the C-terminal arms, where a rigid-body divergence beginning at residue 158, near the end of β7, produces a difference of 3–4 Å in the relative positions of the C-terminal helices α5.
SSAT Can Self-Acetylate Lysine-26.
Mass spectroscopic analysis of purified wild-type SSAT produced in E. coli found that half of the monomers have an N-acetyl group on lysine-26, and the structure has acetyl density at K26 in chain B but not chain A. Replacement of lysine-26 by arginine, which cannot be acetylated, produced protein in which acetylation was not detected by mass spectroscopy. The structure of the unacetylated apoK26R SSAT is essentially identical to that of the acetylated wild-type protein (root-mean-square deviation 0.4 Å for all Cα atoms), and the two proteins were indistinguishable in their ability to acetylate polyamine substrates. Thus, within the limitations of these assays, acetylation of K26b appears neither to be required for nor to hinder the enzymatic activity of SSAT.
We found that the acetylation of wild-type SSAT produced in E. coli could be suppressed by the addition of 50 mM putrescine to the growth medium, yielding purified SSAT in which <10% of K26 residues were acetylated. Incubation of this purified, minimally acetylated enzyme with AcCoA produced acetylated protein in a reaction that could be followed by Western blotting by using antibody against acetyl-lysine. Preincubation with 1 mM of spermine before the addition of AcCoA prevented this self-acetylation. As discussed below, self-acetylation is most likely to occur through the action of AcCoA bound in channel 2 of the asymmetric dimer.
Channels in the Surface of the Dimer Provide Functionally Different Binding Sites.
Two channels run across the surface on opposite sides of the wild-type SSAT dimer. Each channel is lined with an equivalent set of residues from both monomer chains, a mixture of hydrophobic, aromatic, and charged residues, but the two channels are not identical (Fig. 3). In designated channel 1, residues L24, E92, D93, F94, and W132 from chain A and D82, W84, I85, E151, E152, and W154 from chain B create the inside surface of the channel, terminating in a cluster of arginine residues (R101, R142, and R143 from chain A) that have previously been implicated in cofactor binding (23). Channel 2, on the opposite surface of the dimer, is formed from the equivalent residues but from the opposite monomer chains. Because residues 28–31 of chain A are disordered, channel 1 appears to be exposed to solvent throughout its length. However, residues 28–31 of chain B are ordered to form a flap that covers residues E92, D93, and F94 of chain B in channel 2. Structures of binary and ternary complexes involving AcCoA or CoA, without or with spermine or BE-3-3-3, indicate that the two channels are functionally different.
Fig. 3.
Space-filling view of the two channels on opposite surfaces of the asymmetric dimer. The channels are at the dimer interface, with chain A mostly to the upper left in the view of channel 1 and to the lower right in the view of channel 2. The two views are related by a 180° rotation about the noncrystallographic 2-fold axis. Negatively charged residues that line the channels are colored red, positively charged residues are colored blue, and hydrophobic residues are colored yellow. The suffix a or b on the amino acid designations indicates whether the residue is from chain A or B. Arrows point to the parts of channel 1 that house AcCoA and polyamine. Channel 2 is occluded by residues 27–29 of chain B, which prevents polyamine but not CoA from binding.
Polyamine Binding.
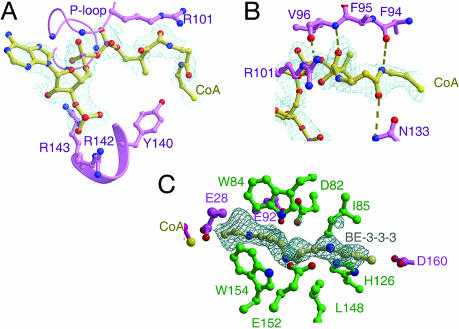
The structure of SSAT crystallized in the presence of CoA and spermine shows CoA bound in both channels of the SSAT dimer but only very weak density that could correspond to a spermine bound in channel 1. Channel 2 remains essentially unchanged, but residues 28–31 in chain A, which are disordered in the apoenzyme, are ordered in the ternary complex to form part of the surface of channel 1. The ternary complex of K26R SSAT, CoA, and BE-3-3-3 showed stronger, more ordered BE-3-3-3 density that more clearly delineates the polyamine binding site in channel 1 (Fig. 4C). The polyamine binding region has a repeating motif of acidic residues interspersed with large hydrophobic residues, a pattern that accommodates the interspersed basic and aliphatic regions of spermine, spermidine, and BE-3-3-3: Charges corresponding to spermine N1, N4, N9, and N12 would be neutralized by E28a, D82b, E92a, E152b, and D160a, whereas aliphatic carbons C5–C8 would be housed in the hydrophobic surface created by W84b and W154b, and carbons C10–C12 would be housed in the hydrophobic surface provided by M30a, I85b, and L148b (Figs. 3 and 4C). (Throughout, where assignments might be ambiguous, lowercase a or b as a suffix indicates whether a residue is in chain A or B.) Consistent with this interpretation, the substitutions E28Q and E152K both reduce enzyme activity (24). The ordered flap over channel 2 occludes D82 and E92 and apparently prevents spermine or BE-3-3-3 from binding in channel 2.
Fig. 4.
Simulated-annealing omit maps showing the electron density and interactions of bound CoA and BE-3-3-3. The electron density corresponding to CoA (A and B) and BE-3-3-3 (C) is drawn as light blue chicken wire. Oxygen atoms are red spheres, and nitrogen atoms are blue spheres. (A) Interactions of CoA that occur in both channel 1 and 2 of the asymmetric dimer. Backbone nitrogens from P-loop residues G102, F103, and G104 interact with the pyrophosphate moiety of CoA. R101, Y140, R142, and R143 side chains from the same monomer are drawn in ball-and-stick representation. (B) Hydrogen bond network with chain A residues that houses the pantetheine moiety of CoA in channel 1 of the asymmetric dimer. Side-chain residues of 94 and 95 beyond Cβ have been omitted for clarity. Hydrogen bonds are drawn as dashed lines. (C) Interactions of BE-3-3-3 in channel 1 of the asymmetric dimer. Ball-and-stick representations of residues from chain A and B of the asymmetric dimer are drawn in purple and green, respectively.
Binding and Functioning of AcCoA.
CoA (without the acetyl group) is bound in both channels 1 and 2 of the ternary complexes, but again the two channels differ. The adenosine and phosphates of CoA are well ordered in both channels and occupy equivalent positions. The pyrophosphate moiety forms interactions with the backbone amide nitrogens of the P-loop, G102, G104, G106, and S107, consistent with mutational analysis (25), whereas the 3′ phosphate is held in place by the guanidino groups of R142 and R143 (Fig. 4A). The 3′ and 5′ phosphates of CoA occupy the same positions as a pair of sulfates in each channel of the apoenzyme structure. The adenine of CoA sits on top of hydrophobic residues G102 and F103 and adopts an anticonformation with respect to the ribose ring. However, the pantetheine moiety adopts very different conformations in the two channels.
In channel 1, the pantetheine moiety of CoA extends toward the bound spermine or BE-3-3-3 and is pinned in place by a series of van der Waals and hydrogen bonding interactions involving residues Y27, F94, F95, V96, R101, and N133 of chain A (Figs. 3 and 4B). Consistent with the structure, replacement of F94 or V96 with a glutamic acid residue, expected to interfere with CoA binding in the channel 1 conformation (Fig. 4B), reduced SSAT activity to <1% of wild type (25). In channel 2, the flap occludes F94 and appears to block the path followed by the pantetheine moiety in channel 1. Instead, the pantetheine atoms of the CoA in channel 2, which are less well ordered, follow a path that folds back upon the ADP ribose moiety and makes van der Waals contact with P135, S136, and F139 of chain B. This conformation would put AcCoA in position to carry out the observed acetylation of K26b. Thus, channel 1 appears to be the active site for acetylation of polyamines, and AcCoA bound in channel 2 appears to be responsible for acetylation of K26b.
A binary complex between AcCoA and K26R SSAT, which is unable to self-acetylate, provided a surprise. Crystals formed in space group P62 instead of P43 (Table 1), and the protein structure is a symmetrical dimer with both surface channels approximating the channel 1 conformation but with residues 28–31 clearly ordered. Each channel of the symmetric dimer contains a well ordered AcCoA in the position observed for CoA in channel 1 of wild-type SSAT but with the acetyl group in either of two positions, with an estimated occupancy of 75% and 25%. The majority conformation has the β-methyl of the acetyl located in a hydrophobic pocket formed by the side chains of L91, F127, and L128, and the minor conformation has the β-methyl group in a hydrophobic pocket formed by the side chains of W132, Y27, and A130 from one monomer and W154 from the other. The majority conformation would be compatible with acetyl transfer to a bound polyamine substrate, but the minor conformation would not.
The majority conformation places the sulfur atom of AcCoA within hydrogen bonding distance of the conserved residue Y140 (Fig. 4A), a residue that is invariant in the SSAT-like proteins we have examined (Fig. 5). Structural and kinetic data for a range of GNAT family members support a mechanism of direct nucleophilic attack by the nearby N1 atom of the substrate on AcCoA (22, 26). It seems likely that Y140 of SSAT serves as the general acid that protonates the thiolate anion of CoA, and that AcCoA bound in the majority conformation in channel 1 transfers its acetyl group to the polyamine substrate. Residue E28, which is also invariant in putative SSAT homologs, is disordered in channel 1 of apoSSAT but becomes well positioned to serve as the general base upon binding of substrate or BE-3-3-3 (Fig. 4C). Consistent with such a mechanism, replacement of Y140 with phenylalanine reduced activity to <5% (our result), and replacement of E28 with glutamine reduced activity by 55% and increased Km three-fold (24).
Asymmetric and Symmetric Dimers.
Wild-type and K26R SSAT both crystallized in space group P43, with the asymmetric unit containing the biological dimer, as did wild type in the presence of CoA and spermine and K26R SSAT in the presence of CoA and BE-3-3-3 (Table 1). However, K26R SSAT in the presence of AcCoA or in the presence of both CoA and spermine crystallized in space group P62, in which the asymmetric unit contains half of a perfectly symmetric biological dimer. It seems likely that the asymmetric and symmetric dimers are in equilibrium in solution, and that the form observed in crystals reflects some combination of the equilibrium distribution of the two forms and the relative stabilities of the two crystal packing configurations under conditions where the crystals formed. Crystal-packing interactions involve slightly different contacts between R119 of one molecule and residues 27–30 of a symmetry-related molecule in the two space groups and seem unlikely by themselves to be responsible for the differences between the asymmetric and symmetric dimers.
The structures of the asymmetric dimer and its complexes imply that channel 1 is the site of polyamine acetylation and that channel 2 cannot bind substrate but that it positions AcCoA for acetylation of K26b. Both channels of the symmetric dimer seem accessible to cofactor and substrate, and cofactor binds in both channels in an orientation suitable for acetylation of substrate. However, spermine has not been observed bound simultaneously with CoA in the structures determined so far. Binding of spermine to wild-type SSAT (acetylated on K26b) or binding of BE-3-3-3 to unacetylated K26R SSAT was measured in solution in the absence of cofactor by following the change in intrinsic fluorescence of SSAT (data not shown). Binding appears to saturate at 2 molecules per dimer, suggesting that substrate binding in the absence of cofactor stabilizes the symmetric dimer. This interpretation would be consistent with the observation that preincubation with an excess of spermine prevents acetylation of K26b, which should occur only on asymmetric dimers.
Potential Biological Implications.
The two structural forms of SSAT and the unexpected self-acetylation reaction may have a biological role in regulating the level or activity of intracellular SSAT, an important element in polyamine homeostasis. As polyamine levels in the cell decrease to the point where SSAT dimers remain unbound to polyamine for long enough, self-acetylation might affect SSAT activity or trigger SSAT degradation. Our assays did not detect an effect of self-acetylation on the activity of purified SSAT, although it is possible that activity could be modulated through interaction with one or more additional factors present in vivo. An attractive idea would be that self-acetylation somehow directs SSAT to the known pathway for degradation of SSAT by the proteasome (18). However, initial tests in vitro by using a rabbit reticulocyte system (19) did not detect a difference in stability between wild-type and K26R SSAT. More work will be required to define the catalytic capabilities of the two structural forms of SSAT and to determine whether or how self-acetylation or transition between asymmetric and symmetric forms might affect SSAT activity or stability.
Relationship of SSAT to Other GNAT Proteins.
The sequence of human SSAT from amino acids ≈71–145, encompassing β3 through α4 in the structure, corresponds to the sequence signature of the GNAT family acetyltransferases in the Pfam database (27), which is shared by >5,000 proteins. In addition, human SSAT shares with a subset of 33 GNAT family members a conserved sequence signature corresponding to residues 6–58 (Prodom domain PD012661 (28); Pfam B accession number PB001901), encompassing β1 through β2 in the structure. We extended this limited set of SSAT-like proteins with a few more proteins of similar size and sequence, identified by blast (29) sequence comparisons.
The structure of Bacillus subtilis PaiA protein has recently been determined, and the protein was found to belong to the GNAT family and to have polyamine N1-acetyltransferase activity (30). Amino acid sequences of PaiA and nine bacterial proteins of related sequence show little resemblance to SSAT-like proteins by blast comparisons, but a sequence and structure alignment of SSAT and PaiA is given in Fig. 2, along with three SSAT-like thialysine Nε-acetyltransferases (TLAs) discussed below. This alignment is derived from an alignment of 9 vertebrate SSATs, 21 known and putative TLAs from vertebrates, lower eukaryotes, archaea and bacteria, and the 10 bacterial PaiA-type proteins (Fig. 5). The two structures have some relatively well conserved residues and similar topology from the N terminus through helix α2, and especially in the GNAT core region of β3 through α4; however, they are highly divergent in sequence and topology in the region between α2 and β3 and in the C-terminal region beyond approximately SSAT residue G144, which is involved in dimerization and polyamine binding in SSAT. The PaiA protein is a monomer and seems to differ at most of the SSAT positions that appear to be involved in polyamine binding. Only the positions corresponding to SSAT G104, N133, and Y140, all involved in AcCoA binding, are completely invariant in all of the proteins of the more extensive alignment. The structural and catalytic details of the active site seem likely to differ considerably between SSAT and PaiA.
One catalytic activity in addition to SSAT has been identified in the limited subset of GNAT proteins most similar to SSAT: The human protein originally named SSAT2 and similar proteins of Schizosaccharomyces pombe, Caenorhabditis elegans, and Leishmania major have all been shown to be much more active as TLAs than as SSATs, and may have other biologically meaningful substrates (31–33). The 9 vertebrate SSATs in the extended alignment are well conserved throughout their sequences, as are the 5 vertebrate TLAs; the 16 nonvertebrate SSAT-like proteins are much more diverse, but many positions are well conserved in the entire set of SSAT-like proteins. Six positions have conserved differences between the vertebrate SSATs and all of the other proteins in the SSAT-like subset (Figs. 2 and 5), indicating that all of those other proteins are likely to be TLAs. Conserved TLA residues corresponding K or R at Y29, S at D82, T at P83, and L at A130 of SSAT have all been shown to be important for the TLA activity of L. major (33). Interestingly, K26, which can be self-acetylated by human SSAT and is invariant in vertebrate SSATs, is one of the residues that is completely different in TLAs and also is not conserved in PaiA-like polyamine acetyltransferases.
Implications for Drug Design.
The SSAT structures provide some insight into relative affinities of various polyamine analogs. For example, BE-3-3-3 binds with higher affinity than does N1,N12-bis-(ethyl)-spermine (BE-3-4-3) (13). Although the first ten nitrogen and carbon atoms in these two compounds are the same, E152b is well positioned to neutralize the nitrogen at position 11 of BE-3-3-3, whereas the nitrogen at position 12 of BE-3-4-3 would be less well accommodated in a more hydrophobic environment. The acetyl group of AcCoA and the terminal ethyl group of BE-3-3-3 occupy the same binding pocket on the protein (Fig. 4C), and both groups can also occupy a nearby alternative pocket, an observation that may explain the enhanced affinity of asymmetric polyamine analogs that have a bulkier group substituted at one end (12). The reduced SSAT activity resulting from the L156F mutation selected in Chinese hamster ovary cells as giving increased resistance to BE-3-3-3 (34) appears likely to be because of a change in the binding surface for the aliphatic carbons at positions 8–10.
The structures we have determined should be helpful in understanding and improving the current generation of inhibitors of SSAT and in the design of new types of therapeutic drugs that affect SSAT activity or stability. Compounds that affect self-acetylation or the transition between symmetric and asymmetric dimer conformations would be particularly interesting in helping to define the biological significance of these unexpected features of SSAT and might provide another path to useful therapeutics.
Materials and Methods
Cloning, Expression, and Purification of Human SSAT.
The coding sequence for human SSAT (SwissProt P21673) (1) was amplified by reverse transcriptase and PCR from total RNA prepared from the human cell line A549 (ATCC no. DDL-185) (the kind gift of P. Freimuth) by using SSAT-specific primers. The amplified coding sequence was cloned by placing the initiation codon in the NdeI site of pET-13a (35) under control of the T7lac promoter and upstream translation signals of the T7 capsid protein. Mutation of lysine-26 to arginine (K26R) and tyrosine-140 to phenylalanine (Y140F) were accomplished by using the Quikchange mutagenesis kit (Stratagene). The correctness of the clones was confirmed by DNA sequencing. SSAT was expressed in E. coli B834(DE3)RIL by autoinduction in ZYP-5052 medium for unlabeled protein and in PASM-5052 for selenomethionine-labeled protein (36). Purification to near homogeneity was achieved by DEAE, Q, and Superdex 75 size-exclusion chromatography, and the fractions containing SSAT protein were identified by gel electrophoresis and staining. The elution position on size-exclusion chromatography was consistent with the protein being a dimer, in agreement with previous studies (23). Typical yields of either unlabeled or selenomethionine-labeled SSAT were 25–50 mg of purified SSAT per liter of autoinduced culture.
Acetylation.
Analysis by MALDI-TOF and electrospray ionization mass spectrometry determined that purified wild-type SSAT had approximately equal amounts of two peaks. One peak had the mass expected for a monomer of SSAT lacking the initiating methionine residue, residues 2–171 in Fig. 1. The second was heavier by the mass of a single acetyl group. Mass spectrometry of tryptic fragments established that the acetyl group was located specifically on lysine-26. Minimally acetylated SSAT was prepared by isopropyl β-d-thiogalactoside induction of the expression clone in ZYP-5052 medium containing 50 mM of putrescine. Self-acetylation of purified, minimally acetylated SSAT was carried out at a concentration of 10 μM SSAT and 100 μM AcCoA in 100 mM NaCl/10 mM Tris·HCl, pH 7.5/1 mM DTT at 20°C and followed by Western blots by using anti-acetyl-lysine (rabbit antiserum) (Upstate Biotechnology). Acetylation of spermidine or spermine by SSAT was assayed in a coupled reaction originally developed to measure histone acetyltransferase activity (37). The CoA generated in the acetyltransferase reaction is used by pyruvate dehydrogenase to oxidize pyruvate, a reaction that is accompanied by the reduction of NAD+ to NADH, which is measured spectrophotometrically at 340 nm.
Crystallization.
Purified SSAT dialyzed against 10 mM Tris·HCl, pH 7.5/100 mM NaCl/1 mM DTT/0.1 mM EDTA was used in crystallization trials. Crystals of selenomethionine-labeled wild-type or K26R SSAT grew over a range of conditions, between 20% and 30% polyethylene glycol 5,000 monomethyl ether (PEG MME 5,000) in 0.1 M ammonium sulfate/50 mM Mes between pH 5.5 and 6.3. A typical crystal grew over a period of days as a tetragonal bipyramid. Binary and ternary complexes were obtained by cocrystallization under similar conditions by using 5 mg/ml SSAT together with 2 mM CoA or AcCoA, 1 mM spermine or BE-3-3-3. In all cases, 40% PEG MME 5,000 was used as a cryoprotectant allowing crystals to be flash cooled by direct transfer into the cryostream.
Data Collection, Structure Determination, and Refinement.
X-ray crystallographic data were collected at 99 K by using beamlines X12C and X26C at the National Synchrotron Light Source. For each crystal, a single data set was collected at 0.98 Å, the peak of the selenium anomalous signal, by using the Brandeis B4 detector and processed by using the hkl2000 program suite (38). Structures were solved by using the program solve (20, 39) to locate selenium atoms and calculate experimental phases to the limit of the data. resolve (20) was used to perform density modification and automatic fitting of the majority of the residues, and phases were improved by using solvent flattening and histogram matching. Gaps, turns, and side chains were fitted manually by using the program turbo-frodo (40, 41). Methionine residues in the native protein were modeled as selenomethionine, and the initial model was refined by using cns (Crystallography & NMR System) (42) by using noncrystallographic symmetry restraints where appropriate. Further cycles of refinement and model building were performed until the model could not be improved, as judged by a decrease in Rfree. The data collection and refinement statistics for six different structures are given in Table 1. In all cases, the conformations of all nonglycine residues were found to lie within the allowed regions of the Ramachandran plot. The structures have been deposited in the Protein Data Bank (PDB) (accession codes in Table 1).
Note Added in Proof.
After completion of this work, we became aware that other groups recently deposited in the Protein Data Bank structures for His-tagged human SSAT (PDB ID code 2F5I) and human TLA (SSAT2) in a complex with AcCoA (PDB ID code 2BEI).
Supplementary Material
Acknowledgments
Work at Brookhaven National Laboratory was supported by the Office of Biological and Environmental Research of the U.S. Department of Energy. A.E.P. acknowledges support from National Institutes of Health Grant CA018138.
Glossary
Abbreviations:
- AcCoA
acetyl CoA
- BE-3-3-3
N1,N11-bis-(ethyl)-norspermine
- GNAT
GCN5-related N-acetyltransferase
- SSAT
spermidine/spermine N1-acetyltransferase
- K26R
mutant SSAT with lysine-26 replaced by arginine
- TLA
thialysine Nε-acetyltransferase.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2B5G, 2B4D, 2B3U, 2B3V, 2B58, and 2B4B).
References
- 1.Casero R. A., Jr., Celano P., Ervin S. J., Applegren N. B., Wiest L., Pegg A. E. J. Biol. Chem. 1991;266:810–814. [PubMed] [Google Scholar]
- 2.Pegg A. E. Biochem. J. 1986;234:249–262. doi: 10.1042/bj2340249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morrison L. D., Kish S. J. Neurosci Lett. 1995;197:5–8. doi: 10.1016/0304-3940(95)11881-v. [DOI] [PubMed] [Google Scholar]
- 4.Russell D. H., Rosenblum M. G., Beckerman R. C., Durie B. G., Taussig L. M., Barnett D. R. Pediatr. Res. 1979;13:1137–1140. doi: 10.1203/00006450-197910000-00011. [DOI] [PubMed] [Google Scholar]
- 5.Rosenblum M. G., Durie B. G., Beckerman R. C., Taussig L. M., Russell D. H. Science. 1978;200:1496–1497. doi: 10.1126/science.663632. [DOI] [PubMed] [Google Scholar]
- 6.Russell D. H. Nat. New Biol. 1971;233:144–145. doi: 10.1038/newbio233144a0. [DOI] [PubMed] [Google Scholar]
- 7.Russell D. H. Cancer Res. 1972;32:2459–2462. [PubMed] [Google Scholar]
- 8.Janne J., Poso H., Raina A. Biochim. Biophys. Acta. 1978;473:241–293. doi: 10.1016/0304-419x(78)90015-x. [DOI] [PubMed] [Google Scholar]
- 9.Casero R. A., Jr., Ervin S. J., Celano P., Baylin S. B., Bergeron R. J. Cancer Res. 1989;49:639–643. [PubMed] [Google Scholar]
- 10.Bernacki R. J., Bergeron R. J., Porter C. W. Cancer Res. 1992;52:2424–2430. [PubMed] [Google Scholar]
- 11.Wallace H. M., Fraser A. V. Biochem. Soc. Trans. 2003;31:393–396. doi: 10.1042/bst0310393. [DOI] [PubMed] [Google Scholar]
- 12.Casero R. A., Jr., Woster P. M. J. Med. Chem. 2001;44:1–26. doi: 10.1021/jm000084m. [DOI] [PubMed] [Google Scholar]
- 13.Wu R., Saab N. H., Huang H., Wiest L., Pegg A. E., Casero R. A., Jr., Woster P. M. Bioorg. Med. Chem. 1996;4:825–836. doi: 10.1016/0968-0896(96)00072-7. [DOI] [PubMed] [Google Scholar]
- 14.Wallace H. M., Mackarel A. J. Biochem. Soc. Trans. 1998;26:571–575. doi: 10.1042/bst0260571. [DOI] [PubMed] [Google Scholar]
- 15.Porter C. W., Ganis B., Libby P. R., Bergeron R. J. Cancer Res. 1991;51:3715–3720. [PubMed] [Google Scholar]
- 16.Fogel-Petrovic M., Shappell N. W., Bergeron R. J., Porter C. W. J. Biol. Chem. 1993;268:19118–19125. [PubMed] [Google Scholar]
- 17.Wang Y., Xiao L., Thiagalingam A., Nelkin B. D., Casero R. A., Jr. J. Biol. Chem. 1998;273:34623–34630. doi: 10.1074/jbc.273.51.34623. [DOI] [PubMed] [Google Scholar]
- 18.Coleman C. S., Pegg A. E. J. Biol. Chem. 1997;272:12164–12169. doi: 10.1074/jbc.272.18.12164. [DOI] [PubMed] [Google Scholar]
- 19.Coleman C. S., Pegg A. E. Biochem. J. 2001;358:137–145. doi: 10.1042/0264-6021:3580137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terwilliger T. C. Methods Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- 21.Neuwald A. F., Landsman D. Trends Biochem. Sci. 1997;22:154–155. doi: 10.1016/s0968-0004(97)01034-7. [DOI] [PubMed] [Google Scholar]
- 22.Vetting M. W., de Carvalho L., Yu M., Hegde S. S., Magnet S., Roderick S. L., Blanchard J. S. Arch. Biochem. Biophys. 2005;433:212–226. doi: 10.1016/j.abb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Coleman C. S., Huang H., Pegg A. E. Biochem. J. 1996;316:697–701. doi: 10.1042/bj3160697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coleman C. S., Huang H., Pegg A. E. Biochemistry. 1995;34:13423–13430. doi: 10.1021/bi00041a020. [DOI] [PubMed] [Google Scholar]
- 25.Lu L., Berkey K. A., Casero R. A., Jr. J. Biol. Chem. 1996;271:18920–18924. doi: 10.1074/jbc.271.31.18920. [DOI] [PubMed] [Google Scholar]
- 26.Draker K. A., Northrop D. B., Wright G. D. Biochemistry. 2003;42:6565–6574. doi: 10.1021/bi034148h. [DOI] [PubMed] [Google Scholar]
- 27.Bateman A., Birney E., Cerruti L., Durbin R., Etwiller L., Eddy S. R., Griffiths-Jones S., Howe K. L., Marshall M., Sonnhammer E. L. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corpet F., Gouzy J., Kahn D. Nucleic Acids Res. 1998;26:323–326. doi: 10.1093/nar/26.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 30.Forouhar F., Lee I. S., Vujcic J., Vujcic S., Shen J., Vorobiev S. M., Xiao R., Acton T. B., Montelione G. T., Porter C. W., et al. J. Biol. Chem. 2005;280:40328–40336. doi: 10.1074/jbc.M505332200. [DOI] [PubMed] [Google Scholar]
- 31.Coleman C. S., Stanley B. A., Jones A. D., Pegg A. E. Biochem. J. 2004;384:139–148. doi: 10.1042/BJ20040790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abo-Dalo B., Ndjonka D., Pinnen F., Liebau E., Luersen K. Biochem. J. 2004;384:129–137. doi: 10.1042/BJ20040789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luersen K. FEBS Lett. 2005;579:5347–5352. doi: 10.1016/j.febslet.2005.08.063. [DOI] [PubMed] [Google Scholar]
- 34.McCloskey D. E., Pegg A. E. J. Biol. Chem. 2003;278:13881–13887. doi: 10.1074/jbc.M205689200. [DOI] [PubMed] [Google Scholar]
- 35.Gerchman S. E., Graziano V., Ramakrishnan V. Protein Expression Purif. 1994;5:242–251. doi: 10.1006/prep.1994.1037. [DOI] [PubMed] [Google Scholar]
- 36.Studier F. W. Protein Expression Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 37.Kim Y., Tanner K. G., Denu J. M. Anal. Biochem. 2000;280:308–314. doi: 10.1006/abio.2000.4546. [DOI] [PubMed] [Google Scholar]
- 38.Otwinowski Z., Minor W. Methods in Enzymology. In: Carter C. W., Sweet R. M., editors. Vol. 276. New York: Academic; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 39.Terwilliger T. C., Berendzen J. Acta Crystallogr. D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones T. A. Methods Enzymol. 1985;115:157–171. doi: 10.1016/0076-6879(85)15014-7. [DOI] [PubMed] [Google Scholar]
- 41.Roussel A., Fontecilla-Camps J. C., Cambillau C. J. Mol. Graphics. 1990;8:86–88, 91. doi: 10.1016/0263-7855(90)80087-v. [DOI] [PubMed] [Google Scholar]
- 42.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.