Abstract
Rotavirus NSP5 is a nonstructural protein that localizes in cytoplasmic viroplasms of infected cells. NSP5 interacts with NSP2 and undergoes a complex posttranslational hyperphosphorylation, generating species with reduced polyacrylamide gel electrophoresis mobility. This process has been suggested to be due in part to autophosphorylation. We developed an in vitro phosphorylation assay using as a substrate an in vitro-translated NSP5 deletion mutant that was phosphorylated by extracts from MA104 cells transfected with NSP5 mutants but not by extracts from mock-transfected cells. The phosphorylated products obtained showed shifts in mobility similar to what occurs in vivo. From these and other experiments we concluded that NSP5 activates a cellular kinase(s) for its own phosphorylation. Three NSP5 regions were found to be essential for kinase(s) activation. Glutathione S-transferase-NSP5 mutants were produced in Escherichia coli and used to determine phosphoacceptor sites. These were mapped to four serines (Ser153, Ser155, Ser163, and Ser165) within an acidic region with homology to casein kinase II (CKII) phosphorylation sites. CKII was able to phosphorylate NSP5 in vitro. NSP5 and its mutants fused to enhanced green fluorescent protein were used in transfection experiments followed by virus infection and allowed the determination of the domains essential for viroplasm localization in the context of virus infection.
Rotaviruses are double-stranded RNA viruses of the family Reoviridae causing diarrhea worldwide in several species, including humans (3, 4). Infective particles replicate in the cytoplasm of infected cells. Following infection, loss of the outer shell activates the virus transcriptase activity to produce viral mRNAs (11, 18). These RNAs direct the synthesis of virus-encoded proteins and serve as templates for the synthesis of genomic double-stranded RNA (replication). This step takes place in particular structures called viroplasms. Viroplasms also support the assembly of new particles in a process coupled to replication (21). NSP2 and NSP5 are two nonstructural proteins that localize in viroplasms, together with the structural proteins VP1, VP2, VP3, and VP6 (24, 32). NSP2 and NSP5 were shown to interact in virus-infected cells as well as in cells transfected with the two proteins. In the latter case, this interaction leads to the formation of viroplasm-like structures (VLS) (9) and enhancement of NSP5 phosphorylation (1). In addition, in virus-infected cells, the virus polymerase VP1 was found associated with NSP2 and NSP5 (1). NSP2, self-assembled into homomultimers (27, 30), has recently been associated with a nucleoside triphosphatase activity (28) and shown to have helix-destabilizing activity, suggesting a possible role in viral RNA unwinding (14, 29).
NSP5, encoded in genome segment 11, is a phosphorylated and O-glycosylated serine- and threonine-rich protein (2, 12) of 198 amino acids with a molecular mass of 26 to 28 kDa. This protein shows an extraordinary level of phosphorylation that confers different electrophoretic mobilities in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), producing isoforms ranging from 28 to 32-34 kDa (2, 5, 25). We have previously reported that in vivo hyperphosphorylation of NSP5 is up-regulated by NSP2 (1). Based on indirect evidence, we and others have suggested that NSP5 has autophosphorylation activity (1, 6, 9, 25, 31).
In this report, we address the phosphorylation, kinase activity, and viroplasm localization of NSP5. Five important aspects were characterized: (i) activation of an NSP5-specific cellular kinase(s) responsible for NSP5 hyperphosphorylation, (ii) NSP5 domains involved in kinase activation, (iii) NSP5 phosphorylation sites, (iv) NSP5 phosphorylation by casein kinase II (CKII), and (v) carboxy-terminal domains responsible for NSP5 localization to viroplasms.
MATERIALS AND METHODS
Cells, viruses, and transient transfections.
MA104 cells were routinely cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (Gibco). The simian rotavirus strain SA11 was propagated and grown in MA104 cells as described previously (8). To prepare cellular extracts, cells were transfected essentially as described by Afrikanova et al. (1). Briefly, 5 × 105 cells growing on a 35-mm-diameter petri dish were infected with T7 recombinant vaccinia virus (strain vTF7.3) (10) and 1 h later transfected with 5 μl of Transfectam reagent (Promega) containing 2 μg of plasmid DNA. At 16 h posttransfection, the cells were washed twice with phosphate-buffered saline (PBS), lysed in 60 μl of TNN lysis buffer (100 mM Tris-HCl [pH 8.0], 250 mM NaCl, 0.5% NP-40, and 1 mM phenylmethylsulfonyl fluoride) for 10 min at 4°C, and centrifuged at 10,000 × g for 5 min. The supernatants were used as cellular extracts in the kinase assays. For immunofluorescence with enhanced green fluorescent protein (EGFP), cells were grown on a 35-mm-diameter petri dish, transfected with 7.5 μl of Transfectam reagent containing 5 μg of plasmid DNA, and infected 48 h later with rotavirus for 5 h.
Immunofluorescence microscopy and antibodies.
For indirect immunofluorescence microscopy, cells were fixed in 3.7% paraformaldehyde in PBS for 10 min at room temperature. Coverslips were dehydrated in PBS and blocked with 1% bovine serum albumin in PBS for 30 min and incubated with guinea pig anti-NSP2 serum (1:100) in PBS-1% bovine serum albumin for 1 h in a moist chamber at room temperature. After three washings in PBS, the slides were stained for 45 min with rhodamine isothiocyanate-conjugated goat anti-guinea pig antibody (Sigma), washed, and mounted with ProLong mounting medium (Molecular Probes). Samples were analyzed by confocal microscopy (Axiovert; Carl Zeiss). Anti-NSP5 and anti-NSP2 sera were obtained by immunization of guinea pigs with glutathione S-transferase (GST)-NSP5 or GST-NSP2 fusion proteins essentially as described previously (12).
Plasmid constructs.
The constructs pEGFP-NSP5, pEGFP-Δ1, pEGFP-Δ2, pEGFP-Δ3, pEGFP-Δ4, pEGFP-Δ1/Δ3, and pEGFP-ΔT were obtained by PCR from previously described constructs (1, 9) by using specific primers to incorporate EcoRI and PstI restriction sites at the N- and C-terminal ends, respectively. Similar oligonucleotides were used to construct pEGFP-Δ1/Δ4T and pEGFP-Δ4T. All these fragments were cloned as EcoRI/PstI fragments in pEGFP-N1 (Clontech). Internal-deletion mutants were obtained by PCR using specific internal primers for the construction of pT7v-Δ1/Δ4T, pT7v-Δ1/Δ3, and pT7v-4T and were cloned as KpnI/BamHI fragments into pcDNA3 (Invitrogen). pT7v-His6-Δ1/Δ3 and pT7v-His6-Δ1 were obtained by inserting the His6 tag at the N terminus with the oligonucleotides 5′-AGCTTGTACCATGGGTCATCACCATCACCATCATGGTAC-3′ and 5′-CATGATGGTGATGGTGATGACCCATGGTACA-3′ ligated into HindIII/KpnI sites. pT7v-His6-Δ1/Δ3 (S→A) and the other serine mutants were obtained by double-step PCR using internal oligonucleotides and were cloned as KpnI/BamHI fragments into pT7v-(His)6. pEGFP-Δ1/Δ3(S→A) was obtained by insertion of a linker sequence into pEGFP-N1 (5′-AATTCCTGGTACCACACTGCAGGTAAGG-3′ and 5′-GATCCCTTA CCTGACGTGTGGTACCAGG-3′) followed by cloning of a KpnI/PstI fragment from pT7v-His6-Δ1/Δ3 (S→A). GST-NSP5 was obtained by cloning a BamHI fragment of NSP5 in pGex2T (Pharmacia). Similarly, GST-Δ2, -Δ3, -Δ4, -ΔT, -Δ4T, -Δ1/Δ3, and -4T were obtained by cloning the respective fragments (KpnI/BamHI) into a modified pGex2T vector to include the BamHI and KpnI sites.
Oligonucleotides.
The internal primers used for the construction of the pT7v mutants were 5′-AATGGTACCATGATTGGTAGGAG-3′ and 5′-GATCAGCGAGCTCTAGC-3′ for pT7v-Δ1/Δ3, 5′-CGGGGTACCATGGATAATAAAAAGGAG-3′ and 5′-GCGGGATCCTTACAAATCTTCGATC-3′ for pT7v-4T, and 5′-AATGGTACCATGATTGGTAGGAG-3′ and 5′-TTACTGCAGAGTTGAGATTGATAC-3′ for pT7v-Δ1/Δ4T. The oligonucleotides used for amplification of fragments inserted in pEGFP-NSP5, pEGFP-Δ2, pEGFP-Δ3, and pEGFP-Δ4 were 5′-CCGGAATTCATGTCTCTCAGCATTG-3′ and 5′-GATCCTTACTCGAGCAAATCTTCGATCAATTGCA-3′. The oligonucleotides for pEGFP-Δ1 and pEGFP-Δ1/Δ3 were 5′-CGGGAATTCATGATTGGTAGGAG-3′ and 5′-GATCCTTACTCGAGCAAATCTTCGATCAATTGCA-3′. The oligonucleotides for pEGFP-ΔT were 5′-CCGGAATTCATGTCTCTCAGCATTG-3′ and 5′-TTACTGCAGGTACTTTTTC-3′. The oligonucleotides for pEGFP-4T were 5′-CGGGAATTCATGGATAATAAAAAGGAGAAATCC-3′ and 5′-GATCCTTACTCGAGCAAATCTTCGATCAATTGCA-3′. The oligonucleotides for the construction of pT7v-His6-Δ1/Δ3 (S→A) and the other serine mutants were 5′-TCAGCATCTGCATCATCTAAAACATAATCTTC-3′ and 5′-GATGCTGACGCTGAAGATTATGTTTTAGATGA-3′.
Expression of GST fusion proteins.
GST fusion proteins were produced in Escherichia coli DH5α. Cultures were induced with 3 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 to 4 h at 37°C. The bacteria were centrifuged, the pellet was washed with ice-cold PBS and resuspended in 1.5% laurylsarcosine-PBS, and 0.1 μg of lysozime/μl, 0.1 μg of chymostatin, leupeptin, aprotinin, and pepstatin/μl, and 5 mM dithiothreitol (DTT) were added for sonication (six times for 10 s each time). The supernatant was supplemented with 1% Triton X-100 in PBS, and equilibrated slurry beads (Glutathione Sepharose 4 Fast Flow; Amersham Pharmacia Biotech) were added. After rolling for 1 h at 4°C, the sample was centrifuged at 1,000 × g for 5 min at 4°C and washed three times with 20 volumes of PBS-1%Triton X-100. Elution was performed with 2 volumes of elution buffer (50 mM Tris-HCl [pH 8], 150 mM NaCl, 5 mM DTT, 0.1% Triton X-100, 50 mM reduced glutathione).
Purification of His6-tagged proteins.
Transfected cellular extracts were incubated for 1 h at 4°C with nickel beads (nitrilotriacetic acid-agarose; Amersham Pharmacia Biotech) equilibrated in 5 volumes of loading buffer (20 mM imidazole-5 mM DTT in PBS). The beads were washed with 10 volumes of washing buffer (35 mM imidazole-5 mM DTT in PBS) and once with 35 mM imidazole, 400 mM NaCl, and 5 mM DTT in PBS. The protein was eluted with 2 volumes of elution buffer (250 mM imidazole-0.02% sodium azide-5 mM DTT in PBS) and dialyzed in PBS containing 5 mM DTT. The recovered protein was analyzed by Western blotting using guinea pig anti-NSP5 sera.
In vitro translation.
The in vitro-translated proteins were synthesized essentially as described by Afrikanova et al. (2) using the TNT-T7 Coupled Reticulocyte Lysate System (Promega). Briefly, 1 μg of plasmid construct was transcribed using T7 RNA polymerase, and the transcript was translated in rabbit reticulocyte lysates in the presence of 4 μl (1,000 Ci/mmol) of [35S]methionine and incubated for 1.5 h at 30°C.
In vitro phosphorylation assays.
The in vitro phosphorylation assay was performed in a total volume of 50 μl containing 10 μl of in vitro-translated proteins, 15 μl of cellular extract, 5 μl of kinase buffer (500 mM Tris-HCl [pH 8], 15 mM spermidine, 80 mM MgCl2, 10 mM DTT, 5 mM ATP, and 50% glycerol). The reaction mixture was incubated for 25 min at 37°C, stopped with 5 μl of 50 mM EDTA, and immunoprecipitated with a guinea pig anti-NSP5 serum. For the assay with the GST fusion proteins, reactions were carried out in 50 μl of the same reaction buffer containing 0.2 μg of GST fusion protein, 1 μl of cellular extract, and 10 μCi of [γ-32P]ATP (3,000 Ci/mmol) (Amersham). For the CKII assay, the substrates were incubated in a total volume of 50 μl of a reaction buffer containing 50 mM HEPES (pH 7.8), 10 mM MgCl2, 150 mM NaCl, 0.5 mM DTT, 4 μM ATP supplemented with 1 μCi of [γ-32P]ATP (3,000 Ci/mmol) and 5 U of recombinant GST-CKIIα/GST-CKIIβ (26). Incubation was for 25 min at 30°C. β-Casein was used as a control at 100 ng per reaction. We established the correspondence between the activities of CKII and the Δ1/Δ3 cellular extract, using the GST-Δ1 fusion as a substrate: 1 μl of cellular extract corresponded to approximately 0.7 U of recombinant CKII, measured under the conditions of either the in vitro phosphorylation assay or the CKII assay. Where indicated, λ-phosphatase (λ-Ppase) treatment was performed on immunoprecipitates in 50-μl reaction volumes containing 50 mM Tris-HCl [pH 7.8], 5 mM DTT, 6 mM MnCl2, and 2 μl of λ-Ppase (400,000 U/ml; New England Biolabs) with incubation at 30°C for 2 h.
In vivo 32P labeling.
The 32P labeling of 5 × 105 transfected cells was performed 15 h posttransfection. The cells were washed three times in phosphate-free minimal essential medium and then starved for 30 min in 1 ml of the same medium. Then, 30 μl of carrier-free 32Pi (10 mCi/ml; Amersham Pharmacia Biotech) was added, incubation was continued for 1 h at 37°C, and the cells were lysed as described above.
Immunoprecipitations and PAGE analysis.
Kinase reaction mixtures (50 μl) and cellular extracts (50 μl) were immunoprecipitated by the addition of 1.5 μl of guinea pig anti-NSP5 serum, 1 μl of 100 mM phenylmethylsulfonyl fluoride, 50 μl of 50% protein A-Sepharose CL-4B beads (Pharmacia) in TNN buffer, and 50 μl of TNN buffer for 2 h at 4°C. The beads were washed three times with TNN buffer, and samples were analyzed by SDS-PAGE (17). Visualization of 35S-labeled proteins was enhanced by fluorography using Amplify (Amersham). Autoradiography was performed at −70°C using X-ray film (Kodak X-OMAT AR).
RESULTS
NSP5 activates cellular kinase(s).
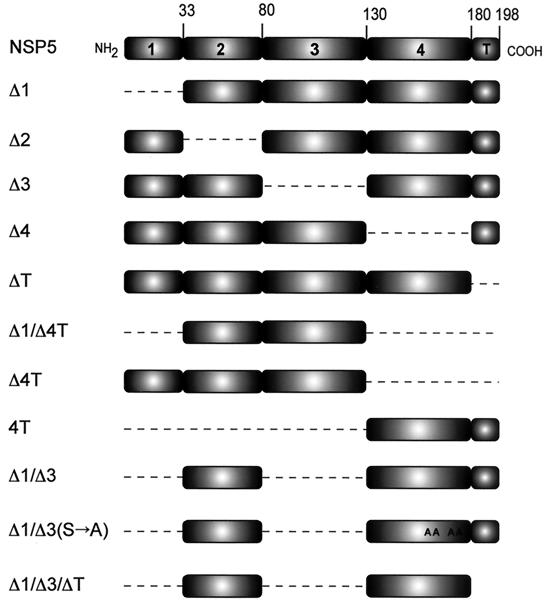
The complex pattern of NSP5 phosphorylation in virus-infected cells has been proposed to be due, in part, to autophosphorylation. However, no clear evidence of NSP5 enzymatic activity has been reported. To investigate the putative NSP5 kinase activity, we developed an in vitro phosphorylation assay which allowed us to obtain the characteristic PAGE mobility shift of hyperphosphorylated NSP5 (2). We selected as a substrate in this assay the in vitro-translated NSP5 mutant Δ1 (Fig. 1 provides a description of all mutants) for the following reasons: (i) it appears as a nonhyperphosphorylated band of ∼20 kDa in SDS-PAGE (Fig. 2A, lane 1), (ii) it becomes hyperphosphorylated when expressed in transfected MA104 cells (1, 9) in a way that is similar to wild-type NSP5 in infected cells, and (iii) it is the most efficiently phosphorylated of all mutants tested.
FIG. 1.
Schematic representation of NSP5 and mutants. The dashed lines correspond to deleted regions. A, Ser→Ala mutations.
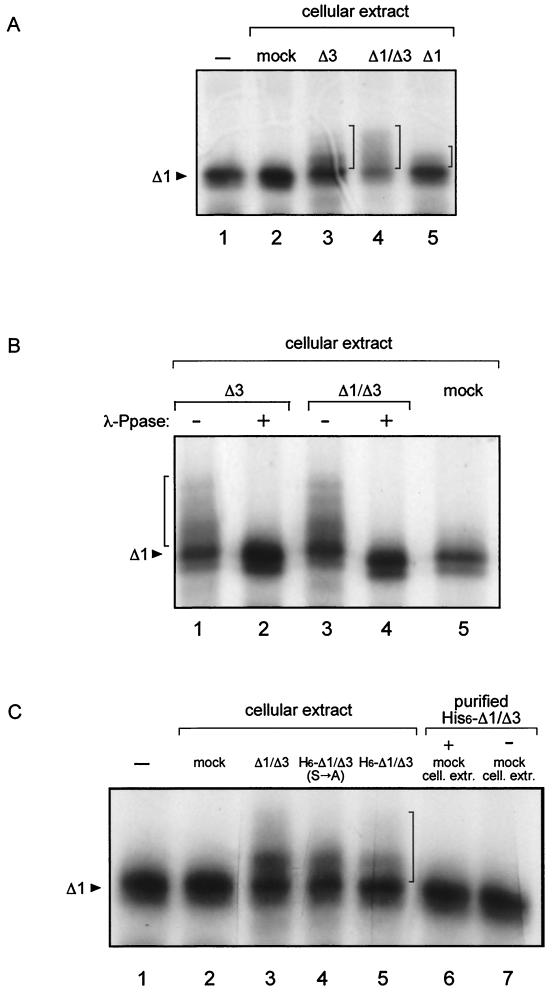
FIG. 2.
In vitro phosphorylation assay. Analysis of immunoprecipitates of in vitro-translated, [35S]methionine-labeled mutant Δ1. (A) Substrate was incubated with cellular extracts from cells transfected with the indicated mutants; mock, extracts from cells transfected with the same plasmid without the insert. −, no addition. (B) λ-Ppase treatment as indicated (+, treated; −, untreated). (C) Purified His6-Δ1/Δ3 was obtained by nickel column purification, and the same amount was used in lanes 5, 6, and 7. cell. extr., cellular extracts. The arrowheads indicate the unphosphorylated Δ1 substrate, and the vertical brackets indicate the positions of mobility-shifted phosphorylated forms. −, no addition.
The kinase activity, determined as the capability to induce PAGE mobility shift of 35S-labeled Δ1 protein, was investigated in cellular extracts derived from either untransfected cells or cells transfected with NSP5 or NSP5 deletion mutants. Extracts containing mutants Δ1, Δ3, and Δ1/Δ3, but not extracts from mock-transfected cells, had phosphorylation activity (Fig. 2A). In agreement with this result, these three mutants are hyperphosphorylated in vivo in transfected cells (9) (see Fig. 4C). Conversely, extracts from cells transfected with wild-type NSP5 showed a marginal effect, consistent with its low phosphorylation in transfected cells, in the absence of viral replication (1).
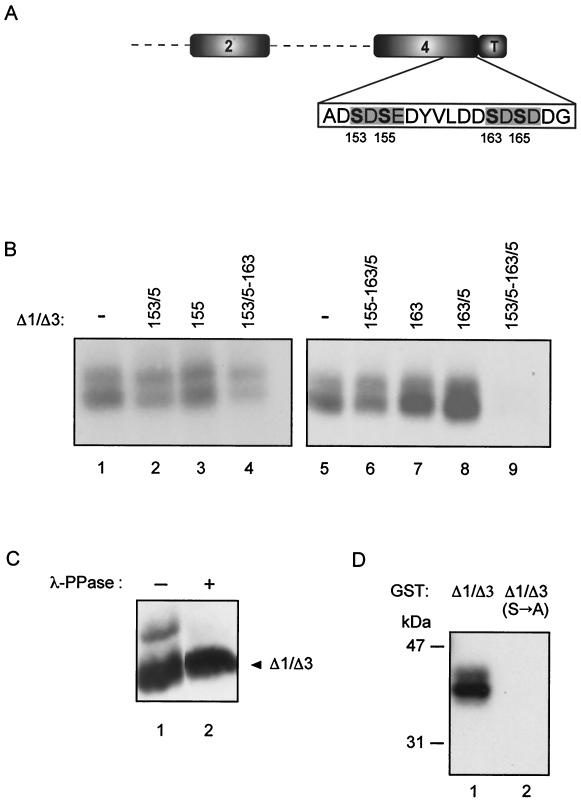
FIG. 4.
Characterization of Ser→Ala mutants. (A) Scheme of Δ1/Δ3 mutant showing positions of the four serines mutated to alanine within region 4. The residue numbers correspond to the wild-type protein. (B) Immunoprecipitates of in vivo 32Pi-labeled MA104 cells transfected with nonmutated (−) or with the indicated mutant constructs. The numbers refer to mutated Ser residues. (C) Western immunoblot of λ-Ppase-treated (+) transfected Δ1/Δ3 mutant. (D) Phosphorylation with [γ-32P]ATP of GST-Δ1/Δ3 and GST-Δ1/Δ3(S→A) as for Fig. 3.
Treatment with λ-Ppase confirmed that PAGE mobility changes corresponded to hyperphosphorylated forms of the substrate (Fig. 2B). Interestingly, extracts transfected with the two mutants that lack region 3 (Δ3 and Δ1/Δ3) showed the highest phosphorylation activities. Extracts from cells transfected with other mutants, such as Δ2, Δ4, ΔT, or Δ1/Δ3/ΔT, did not show significant activity. Table 1 shows the activities of all mutants tested. In all cases, similar amounts of transfected proteins were used, as judged by Western blot analysis. From these results, it appears that domains 2, 4, and T are absolutely required for extract activities. Also, other mutants were tested as substrates in this assay (Table 1).
TABLE 1.
Summary of NSP5 properties
| Protein | Presence of propertya
|
||
|---|---|---|---|
| Localization to viroplasms | Activity of cellular extractb | Activity as substratec | |
| NSP5 | + | − | ± |
| Δ1 | + | + | + |
| Δ2 | + | − | + |
| Δ3 | + | + | + |
| Δ4 | − | − | − |
| ΔT | − | − | − |
| Δ1/Δ4T | − | − | − |
| Δ4T | − | ND | − |
| 4T | + | − | − |
| Δ1/Δ3 | + | + | − |
| Δ1/Δ3(S→A) | + | + | − |
| Δ1/Δ3/ΔT | ND | − | − |
+, present; −, absent; ±, marginal effect; ND, not determined.
NSP5-Δ1 used as a substrate.
Δ1/Δ3 cellular extract used as a source of cellular kinases(s).
The phosphorylated shifted forms of Δ1 were only achieved with extracts from cells transfected with NSP5 mutants that showed hyperphosphorylation in vivo. This result suggested that either the activity resides in the transfected protein itself or the transfected protein induces or activates an otherwise-inactive cellular kinase(s).
To discriminate between these two hypotheses, a His6-tagged version of the Δ1/Δ3 protein was purified from transfected cellular extracts on a nickel column. The purified protein was analyzed by Western immunoblotting (not shown). As shown in Fig. 2C, the cellular extract containing His6-Δ1/Δ3 showed phosphorylation activity (lane 5) while the same amount of the purified protein did not (lane 7).
The possibility that the transfected NSP5 mutant is complexed with a cellular kinase, thus activating phosphorylation of NSP5 itself, seems unlikely, since addition of purified His6-Δ1/Δ3 to a mock cellular extract did not reconstitute the activity (Fig. 2C, lane 6). This suggests that expression of the protein is necessary to promote activation of the cellular kinase(s). Hyperphosphorylation of NSP5 in virus-infected cells also occurs in the presence of actinomycin D, indicating that transcription of cellular genes is not required for the emergence of the kinase activity (data not shown). Phosphorylation and the capacity to activate the cellular kinase(s) appear to be distinct characteristics of NSP5. Indeed, mutation of four serine residues within domain 4 [mutant Δ1/Δ3(S→A)], which completely abolished phosphorylation of the protein expressed in vivo (see below), did not affect its ability to activate the cellular kinase(s) (Fig. 2C).
Mapping phosphorylation sites of NSP5.
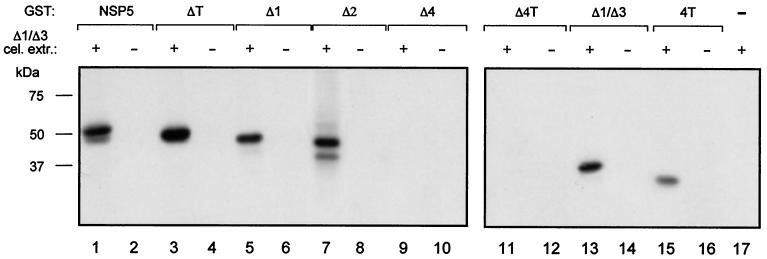
In order to map the phosphorylation sites on NSP5, we constructed a variety of NSP5 deletion mutants as GST fusion proteins. None of these proteins produced in bacteria showed any phosphorylation activity (Fig. 3) nor were they phosphorylated by His6-Δ1/Δ3 purified from transfected cells (data not shown). This evidence supports the idea that NSP5 does not have kinase activity. However, some of the mutants, as well as wild-type NSP5, served as substrates when incubated in vitro with a cellular extract from Δ1/Δ3-transfected cells as a source of enzyme. As shown in Fig. 3, only mutants containing region 4 were phosphorylated, indicating that most of the phosphorylation occurs within this domain and suggesting that in vivo phosphorylation sites could reside in this region. We have previously reported that NSP5 phosphorylation takes place in Ser and Thr residues (2). To further characterize the phosphorylation sites, we mutated to alanine (in mutant Δ1/Δ3) four serines (Ser153, Ser155, Ser163, and Ser165) within region 4 present (boldface) in a particular acidic amino acid context (ADSDSEDYVLDDSDSDDG) (Fig. 4A) and tested them for the ability to be phosphorylated in vivo. Figure 4B shows immunoprecipitates of several transfected Δ1/Δ3 Ser→Ala mutants following labeling with 32Pi. The four serines appeared to be sites of phosphorylation. The only mutant that showed no phosphorylation was the one with all four serines mutated (Fig. 4B, lane 9). All of the lanes in Fig. 4B contained comparable amounts of transfected protein as determined by Western immunoblotting (not shown). The double band corresponded to low-mobility hyperphosphorylated forms, as judged by λ-Ppase sensitivity (Fig. 4C). Further confirmation that the four serines within region 4 are the main phosphorylation sites was obtained with a GST-Δ1/Δ3 fusion protein with all four serines mutated [GST-Δ1/Δ3(S→A)] which was not phosphorylated by Δ1/Δ3 cellular extracts (Fig. 4D).
FIG. 3.
Mapping phosphorylation sites of NSP5. SDS-PAGE analysis of immunoprecipitated, in vitro-phosphorylated GST-NSP5 mutant proteins (0.2 μg) incubated with (+) and without (−) cellular extract (cel. extr.) from Δ1/Δ3-transfected cells. GST protein negative control (−) (lane 17) was loaded without immunoprecipitation.
NSP5 is a substrate of CKII.
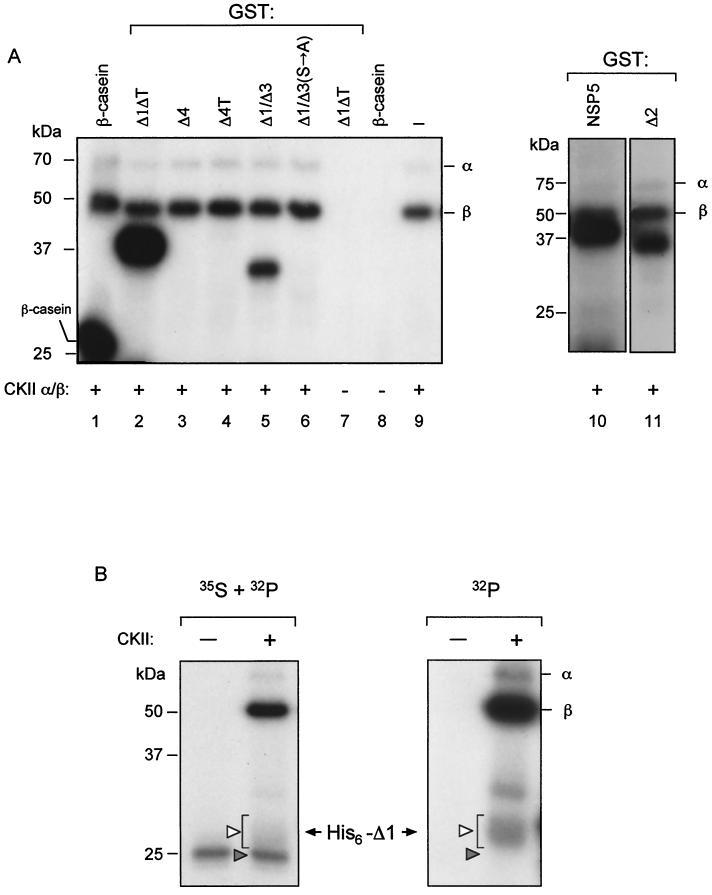
The sequences of NSP5 region 4 containing the four phosphorylated serines (boldface), SDSE and SDSD, are characteristic of substrates of CKII 19; http://www.ebi.ac.uk [Prosite-EMBL]). We therefore tested whether CKII was able to phosphorylate the different GST fusions. As shown in Fig. 5A, CKII was able to phosphorylate NSP5 and all mutants containing region 4, namely, Δ1/ΔT, Δ1/Δ3, and Δ2. This phosphorylation appeared to be restricted to serines 153, 155, 163, and 165, since the mutant GST-Δ1/Δ3(S→A) was not phosphorylated by CKII. This result is in complete agreement with the lack of phosphorylation of this mutant in vivo (Fig. 4B). CKII was also able to phosphorylate the in vitro-translated His6-Δ1 substrate, as demonstrated by the results shown in Fig. 5B. The left panel shows both the 35S label of the in vitro-translated substrate and the 32P label from [γ-32P]ATP. In the right panel, only the 32P radioactivity was detected. Under these conditions, CKII was able to convert part of the substrate into mobility-shifted forms, similarly to the transfected cellular extracts. Phosphorylation of the GST fusions with CKII was also performed under the conditions used with the cellular extracts with similar results. These results suggested that a CKII-like enzymatic activity is involved in NSP5 phosphorylation.
FIG. 5.
NSP5 is a substrate of CKII. Shown is SDS-PAGE analysis of an in vitro CKII phosphorylation assay with [γ-32P]ATP. (A) The indicated GST-NSP5 mutants were used as substrates. Positive and negative controls are shown in lanes 1, 7, 8, and 9. −, no substrate added. (B) Purified, in vitro-translated 35S-His6-Δ1 was used as a substrate in the presence or absence of CKII. The two panels show autoradiography of the same gel, detecting 35S plus 32P (left) and 32P (right). The solid and open arrowheads indicate His6-Δ1 precursor and hyperphosphorylated forms, respectively. Autophosphorylated GST-α and GST-β CKII subunits are indicated. +, present; −, absent.
NSP5 localization to viroplasms.
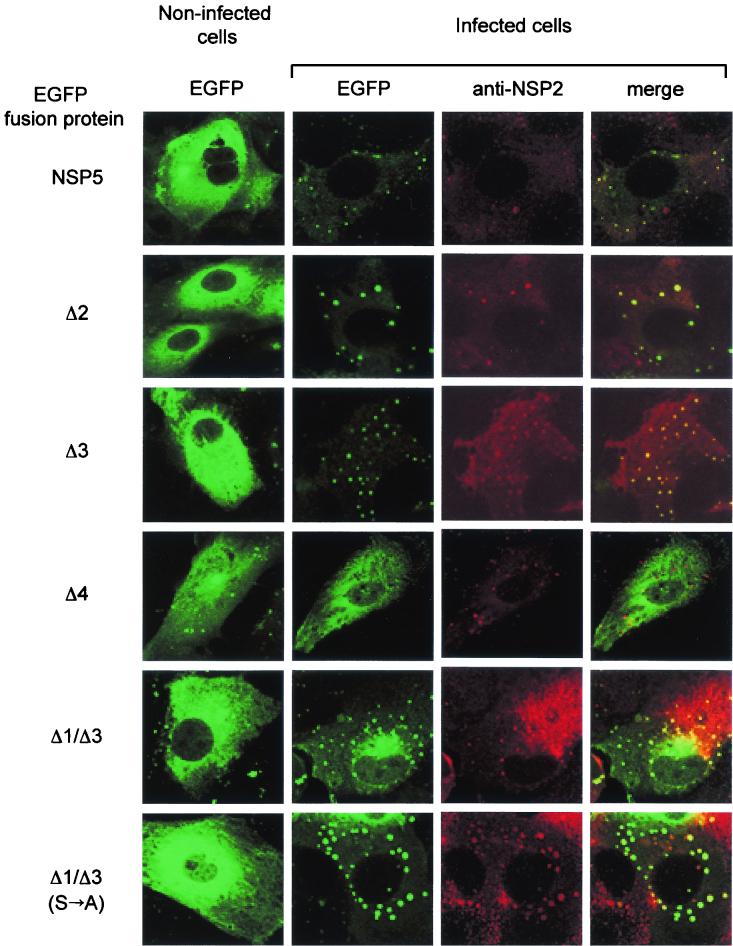
NSP5 and NSP2 are the two rotavirus nonstructural proteins that localize in viroplasms in the cytoplasm of infected cells. We had previously shown that the two transfected proteins directly interact in vivo, producing VLS (9). In order to investigate the requirements of NSP5 to localize in true viroplasms in infected cells, we analyzed the fates of several NSP5 mutants fused to EGFP. Cells were transfected with different constructs, infected with rotavirus (strain SA11) 48 h later, and fixed 6 h postinfection. Immunofluorescence was performed on a confocal microscope, and viroplasms were revealed with anti-NSP2 sera.
Figure 6 shows that, in noninfected cells, NSP5 wild type and mutants have a diffused distribution in the cytoplasm. Following infection, this distribution was highly affected, both for NSP5 and for some of the deletion mutants, such as Δ1, Δ2, Δ3, and Δ1/Δ3, which became rapidly relocalized in viroplasms. On the other hand, mutants lacking region 4 (such as Δ4) or the carboxy-terminal tail (ΔT) (not shown) did not localize to viroplasms. Table 1 summarizes the results obtained with all mutants tested. Interestingly, mutant Δ1/Δ3(S→A) with all four serines in region 4 mutated, which we previously demonstrated not to be phosphorylated in vivo (see above), showed a clear colocalization in viroplasms. This result suggested that, although the presence of the carboxy-terminal regions 4 and T is required, phosphorylation is not essential for NSP5 localization to viroplasms in the context of infected cells.
FIG. 6.
Confocal immunofluorescence microscopy. MA104 cells were transfected with NSP5 mutants fused to EGFP followed by infection with rotavirus. Viroplasms were detected with an anti-NSP2 (red) antibody. The rightmost column is the superimposition of the two independently acquired images.
An interesting observation regards mutant Δ2, which does not get phosphorylated in vivo and does not form VLS (9) whereas it localized to viroplasms (Fig. 6). This is most likely due to interaction with wild-type NSP5, which was shown to depend on the last 10 carboxy-terminal amino acids (31). We have now determined that localization to VLS was only obtained when Δ2-EGFP was cotransfected with NSP2 into a cell line stably expressing NSP5 (MA104-C7) (2) and not when it was cotransfected into a cell not expressing NSP5 (data not shown).
DISCUSSION
Phosphorylation of NSP5 is a complex process which generates different phosphorylated isoforms with apparent molecular masses ranging from 26 to 32-34 kDa. These events, which take place in the context of virus replication where NSP5 appears as a main band of 28 kDa, are much less evident in NSP5-transfected cells. In that case, the main product corresponds to the 26-kDa isoform with almost complete absence of the hyperphosphorylated forms (higher than 28 kDa) (2, 5, 25). O-glycosylation also modifies NSP5 posttranslationally (12), mainly involving isoforms of 26 and 28 kDa (2). We have previously demonstrated that in vivo NSP5 phosphorylation changes when NSP5 is cotransfected with NSP2, producing a pattern that resembles the one obtained in virus-infected cells (1). However, we now show that cell extracts containing NSP5 deletion mutants lacking either domain 1 or 3 or both (Δ1, Δ3, or Δ1/Δ3, respectively) efficiently induce PAGE mobility shift in an in vitro-translated NSP5-derived substrate. Consistent with these results, these same mutants were previously shown to undergo mobility shift phosphorylation in vivo (9). Similarly, cell extracts of mutants lacking region 2, 4, or T, which were inactive in the assay, were unable to be phosphorylated in vivo (9) in spite of the fact that two of them (Δ2 and ΔT) contain region 4. Interestingly, a cellular extract of cells transfected with the histidine-tagged version of mutant Δ1/Δ3 (His6-Δ1/Δ3) showed high activity while the purified protein did not. Moreover, GST fusion proteins showed a complete lack of kinase activity, whereas they were good substrates. Taken together, these results indicate that NSP5 is not itself a kinase but rather that it activates a cellular kinase(s), otherwise inactive in the untransfected cellular extracts, for its own phosphorylation.
The marginal phosphorylation activity of extracts containing wild-type NSP5 is not surprising, since when expressed alone, NSP5 is very little phosphorylated, producing mainly the 26-kDa polypeptide (1, 9), while upon coexpression with NSP2 it becomes hyperphosphorylated (1). Regions 1 and 3 appear to have an inhibitory effect (region 3 being stronger than region 1 [Fig. 2A]) on the capacity of NSP5 to activate the cell kinase(s), as revealed by the activities of mutants Δ1, Δ3, and Δ1/Δ3. One could speculate that in the replicative cycle, interaction with NSP2 may neutralize the inhibition of these two regions.
Region 4 contains the main phosphoacceptor sites, mapped to serines 153, 155, 163, and 165, located in an acidic region with homology to CKII phosphorylation sites (SDSE and SDSD) (19). In fact, we showed that CKII was able to phosphorylate NSP5 in vitro precisely in those positions, although this does not demonstrate that CKII is the kinase responsible for NSP5 phosphorylation in vivo or that other kinases may not be involved. These results suggest that the cellular kinase(s) activated by NSP5 is a CKII-like enzyme. Up to now, CKII has been described as phosphorylating several viral proteins of both DNA and RNA viruses (7, 13, 15, 16, 20, 22, 23), with the exception of viruses of the family Reoviridae.
It is noteworthy that phosphorylation of the four serines in region 4 appears not to be required for kinase activation, since mutant Δ1/Δ3(Ser→Ala) was still able to induce the cellular kinase(s).
Many questions concerning the phosphorylation of NSP5 and its ability to localize in viroplasms await the development of a reverse genetic system. We have previously demonstrated that NSP5 and NSP2 can form VLS independently of the viral context. The formation of these particular cytoplasmic structures reflects the abilities of the two nonstructural proteins to interact with each other. On the other hand, in the context of viral infection, the situation is more complex and localization can also involve the interaction of mutants with wild-type NSP5 (31). Contrary to what was observed with VLS formation, all NSP5 mutants containing regions 4 and T localized to viroplasms independently of the phosphorylation state. Interaction with wild-type NSP5 could explain this behavior, since multimerization of NSP5 was shown to depend on the last 10 carboxy-terminal amino acids (31). In fact, mutant Δ2 is not phosphorylated in vivo and does not form VLS (9), whereas in this report we show localization to viroplasms. This attribute was dependent on complementation with wild-type NSP5, since VLS were formed only when Δ2-EGFP was cotransfected with NSP2 into a cell line stably expressing NSP5.
Even though the biological function of NSP5 remains to be elucidated, the data presented here delineate meaningful aspects of NSP5 phosphorylation in the context of virus replication.
Acknowledgments
C. Eichwald and F. Vascotto contributed equally to this work.
C.E., F.V., and E.F. were supported by SISSA predoctoral fellowships.
We are grateful to Francisco Romero-Oliva for providing the recombinant CKII and to Mauro Sturnega and Giancarlo Lunazzi for excellent technical assistance.
REFERENCES
- 1.Afrikanova, I., E. Fabbretti, M. C. Miozzo, and O. R. Burrone. 1998. Rotavirus NSP5 phosphorylation is up-regulated by interaction with NSP2. J. Gen. Virol. 79:2679-2686. [DOI] [PubMed] [Google Scholar]
- 2.Afrikanova, I., M. C. Miozzo, S. Giambiagi, and O. R. Burrone. 1996. Phosphorylation generates different forms of rotavirus NSP5. J. Gen. Virol. 77:2059-2065. [DOI] [PubMed] [Google Scholar]
- 3.Ball, J. M., P. Tiang, C. Q.-Y. Zeng, A. P. Morris, and M. K. Estes. 1996. Age-depedent diarrhea induced by a rotaviral non-structural glycoprotein. Science 272:101-104. [DOI] [PubMed] [Google Scholar]
- 4.Bern, C., J. Martines, I. de Zoysa, and R. I. Glass. 1992. The magnitude of the global problem of diarrheal disease: a ten year update. Bull. W. H. O. 70:705-714. [PMC free article] [PubMed] [Google Scholar]
- 5.Blackhall, J., A. Fuentes, K. Hansen, and G. Magnusson. 1997. Serine protein kinase activity associated with rotavirus phosphoprotein NSP5. J. Virol. 71:138-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackhall, J., M. Muñoz, A. Fuentes, and G. Magnusson. 1998. Analysis of rotavirus nonstructural protein NSP5 phosphorylation. J. Virol. 72:6398-6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dupuy, L. C., S. Dobson, V. Bitko, and S. Barik. 1999. Casein kinase 2-mediated phosphorylation of respiratory syncytial virus phosphoprotein P is essential for the transcription elongation activity of the viral polymerase: phosphorylation by casein kinase 1 occurs mainly at Ser215 and is without effect. J. Virol. 73:8384-8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Estes, M. K., D. Y. Graham, C. P. Gerba, and E. M. Smith. 1979. Simian rotavirus SA11 replication in cell cultures. J. Virol. 31:810-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fabbretti, E., I. Afrikanova, F. Vascotto, and O. Burrone. 1999. Two non-structural rotavirus proteins, NSP2 and NSP5, form viroplasm-like structures in vivo. J. Gen. Virol. 80:333-339. [DOI] [PubMed] [Google Scholar]
- 10.Fuerst, R. T., G. E. Niles, W. F. Studier, and B. Moss. 1986. Eucaryotic transient-expression system based on recombinant vaccinia virus that synthesized bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillies, S., S. Bullivant, and A. R. Bellamy. 1971. Viral RNA polymerases: electron microscopy of reovirus reaction core. Science 174:694-696. [DOI] [PubMed] [Google Scholar]
- 12.Gonzáles, S. A., and O. Burrone. 1991. Rotavirus NS26 is modified by addition of single O-linked residues of N-acetylglucosamine. Virology 182:8-16. [DOI] [PubMed] [Google Scholar]
- 13.Haneda, E., T. Furuya, S. Asai, Y. Morikawa, and K. Ohtsuki. 2000. Biochemical characterization of casein kinase II as a protein kinase responsible for stimulation of HIV-1 protease in vitro. Biochem. Biophys. Res. Commun. 275:434-439. [DOI] [PubMed] [Google Scholar]
- 14.Kattoura, M. D., L. L. Clapp, and J. T. Patton. 1992. The non-structural protein NS35 is a non-specific RNA-binding protein. Virology 191:698-708. [DOI] [PubMed] [Google Scholar]
- 15.Kim, J., D. Lee, and J. Choe. 1999. Hepatitis C virus NS5A protein is phosphorylated by casein kinase II. Biochem. Biophys. Res. Commun. 257:777-781. [DOI] [PubMed] [Google Scholar]
- 16.Kohl, A., V. di Bartolo, and M. Bouloy. 1999. The rift valley fever virus nonstructural protein NSs is phosphorylated at serine residues located in casein kinase II consensus motifs in the carboxy-terminus. Virology 263:517-525. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 18.Lawton, J. A., M. K. Estes, and B. V. V. Prasad. 1997. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat. Struct. Biol. 4:118-121. [DOI] [PubMed] [Google Scholar]
- 19.Marin, O., F. Meggio, S. Sarno, L. Cesaro, M. A. Pagano, and L. A. Pinna. 1999. Tyrosine versus serine/threonine phosphorylation by protein kinase casein kinase-2. J. Biol. Chem. 274:29260-29265. [DOI] [PubMed] [Google Scholar]
- 20.Matsushita, Y., K. Hanazawa, K. Yoshioka, T. Oguchi, S. Kawakami, Y. Watanabe, M. Nishiguchi, and H. Nyunoya. 2000. In vitro phosphorylation of the movement protein of tomato mosaic tobamovirus by a cellular kinase. J. Gen. Virol. 81:2095-2102. [DOI] [PubMed] [Google Scholar]
- 21.Mattion, N. M., J. Cohen, and M. K. Estes. 1994. The rotavirus proteins, p. 169-249. In A. Kapikian (ed.), Viral infections of the gastrointestinal tract, 2nd ed. Marcel Dekker, New York, N.Y.
- 22.Mirigou, V., L. Stevanato, R. Manservigi, and P. Mavromara. 2000. The C-terminal cytoplasmic tail of herpes simplex virus type 1 gE protein is phosphorylated in vivo and in vitro by cellular enzymes in the absence of other viral proteins. J. Gen. Virol. 81:1027-1031. [DOI] [PubMed] [Google Scholar]
- 23.Morrison, E. E., Y.-F. Wang, and D. M. Meredith. 1998. Phosphorylation of structural components promotes dissociation of the herpes simplex virus type 1 tegument. J. Virol. 72:7108-7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petrie, B. L., H. B. Greenberg, D. Y. Graham, and M. K. Estes. 1984. Ultrastructural localization of rotavirus antigens using colloidal gold. Virus Res. 1:133-152. [DOI] [PubMed] [Google Scholar]
- 25.Poncet, D., P. Lindenbaum, R. L'Haridon, and J. Cohen. 1997. In vivo and in vitro phosphorylation of rotavirus NSP5 correlates with its localization in viroplasm. J. Virol. 71:34-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romero-Oliva, F., and J. E. Allende. 2001. Protein p21WAF1/CIP1 is phosphorylated by protein kinase CK2 in vitro and interacts with the amino terminal end of the CK2 beta subunit. J. Cell. Biochem. 81:445-452. [DOI] [PubMed] [Google Scholar]
- 27.Schuck, P., Z. Taraporewala, P. McPhie, and J. T. Patton. 2001. Rotavirus nonstructural protein NSP2 self-assembles into octamers that undergo ligand-induced conformational changes. J. Biol. Chem. 276:9679-9687. [DOI] [PubMed] [Google Scholar]
- 28.Taraporewala, Z., D. Chen, and J. Patton. 1999. Multimers formed by the rotavirus nonstructural protein NSP2 bind to RNA and have nucleoside triphosphatase activity. J. Virol. 73:9934-9943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taraporewala, Z. F., and J. T. Patton. 2001. Identification and characterization of the helix-destabilizing activity of rotavirus nonstructural protein NSP2. J. Virol. 75:4519-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taraporewala, Z. F., D. Chen, and J. T. Patton. 2001. Multimers of the bluetongue virus non-structural protein NS2 possess nucleotidyl phosphatase activity: similarities between NS2 and rotavirus NSP2. Virology 280:221-231. [DOI] [PubMed] [Google Scholar]
- 31.Torres-Vega, M. A., R. A. González, M. Duarte, D. Poncet, S. López, and C. F. Arias. 2000. The C-terminal domain of rotavirus NSP5 is essential for its multimerization, hyperphosphorylation and interaction with NSP6. J. Gen. Virol. 81:821-830. [DOI] [PubMed] [Google Scholar]
- 32.Welch, S. K., S. E. Crawford, and M. K. Estes. 1989. Rotavirus SA11 genome segment 11 protein is a nonstructural phosphoprotein. J. Virol. 63:3974-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]