Abstract
The RNase H activity of reverse transcriptase is essential for retroviral replication. RNA 5′ end-directed cleavages represent a form of RNase H activity that is carried out on RNA/DNA hybrids that contain a recessed RNA 5′. Previously, the distance from the RNA 5′ end has been considered the primary determinant for the location of these cleavages. Employing model hybrid substrates and the HIV-1 and M-MuLV reverse transcriptases, we demonstrate that cleavage sites correlate with specific sequences and that the distance from the RNA 5′ end determines the extent of cleavage. An alignment of sequences flanking multiple RNA 5′ end-directed cleavage sites reveals that both enzymes strongly prefer A or U at the +1 position and C or G at the -2 position, and additionally for HIV-1, A is disfavored at the -4 position. For both enzymes, 5′ end-directed cleavages occurred when sites were positioned between the 13th and 20th nucleotides from the RNA 5′ end, a distance termed the cleavage window. In examining the importance of accessibility to the RNA 5′ end, it was found that the extent of 5′ end-directed cleavages observed in substrates containing a free recessed RNA 5′ end was most comparable to substrates with a gap of 2 or 3 bases between the upstream and downstream RNAs. Together these finding demonstrate that the selection of 5′ end-directed cleavage sites by retroviral RNases H results from a combination of nucleotide sequence, permissible distance, and accessibility to the RNA 5′ end.
In reverse transcription, the singlestranded RNA genome of a retrovirus is transformed into double-stranded DNA by the viral-encoded reverse transcriptase (reviewed in (1,2)). This multifunctional enzyme carries out DNA synthesis, strand displacement synthesis, strand transfer, and degrades the RNA portion of RNA/DNA hybrids. The polymerase domain of reverse transcriptase represents the amino terminal two-thirds of the protein while the RNase H domain comprises the remaining C-terminal portion. Although the polymerase and RNase H activities are functionally separable, both activities are essential for retroviral replication. The reverse transcriptase of human immunodeficiency virus type 1 (HIV-1) functions as an asymmetric heterodimer composed of p66 and p51 subunits (3), while that of Moloney murine leukemia virus (M-MuLV) is a 76 kDa protein that may function as a homodimer or as a monomer (4-6).
RNase H contributes to reverse transcription in three distinct ways (reviewed in (1,7,8)). First, RNase H carries out degradation of the RNA genome both during and after minus-strand DNA synthesis to facilitate plus-strand DNA synthesis and strand transfers. Second, RNase H creates the polypurine tract (PPT) primer from the viral genome. And third, RNase H removes the PPT and tRNA primers used to prime plus-strand and minus-stand DNA synthesis, respectively. Given the vital roles of RNase H in retroviral replication, it is important to understand how RNase H recognizes the RNA/DNA hybrids created during reverse transcription as substrates for cleavage. Extensive studies have shown that both generation of the PPT primer and removal of the PPT and tRNA primers require specific sequences (9-19). By contrast, fewer studies have examined general degradation by RNase H (20-23), and less is known about the determinants that might influence RNase H specificity for this type of degradation during retroviral replication.
RNase H has both polymerization-dependent and polymerization-independent modes of cleavage. The polymerization-dependent mode accompanies RNAdependent DNA synthesis and the nascent DNA 3′ primer terminus positions RNase H cleavages. Such cleavages occur 17 to 20 nucleotides away on the RNA template strand but are not strictly coupled to DNA synthesis (20,24-26). This mode of cleavage likely functions at pause sites during minus-strand synthesis, when the 3′ terminus of the nascent DNA is recessed on the genomic template. However, the polymerization rate of reverse transcriptase is faster than the rate of RNase H cleavage (27) and the polymerization-dependent mode of RNase H cleavage does not completely degrade the template RNA (20,28). Thus significant amounts of RNA can remain annealed to the minus-strand DNA (22), and removal of these fragments by the polymerization-independent RNase H activity is likely necessary for efficient plus-strand DNA synthesis.
Polymerization-independent RNase H cleavages have been attributed to at least three distinct mechanisms which differ in how reverse transcriptase associates with the hybrid substrate (reviewed in (1,7,8)). First, reverse transcriptase can bind the RNA strand of a hybrid without positioning by either a DNA 3′ end or an RNA 5′ end, resulting in internal cleavages by the RNase H (23). Second, the polymerase domain can bind a recessed DNA 3′ primer terminus to facilitate DNA 3′ end-directed cleavages that occur approximately 17 to 20 nucleotides back on the RNA strand of a hybrid, similar to what happens at pause sites during polymerization. Third, the polymerase domain can associate with a recessed RNA 5′ end and RNase H can carry out 5′ end-directed cleavages that occur approximately 15 to 20 nucleotides downstream on the RNA strand. While the distance from the recessed RNA 5′ end has been characterized as the primary determinant for this mechanism (29), these cleavages have been observed as close as 12 to 15 nucleotides (30,31), and as far as 21 nucleotides (29,32) from the RNA 5′ end. Notably, this range of distances is much broader than the more fixed distance of 17 to 19 nucleotides that separates the polymerase and RNase H active sites as determined by crystallography studies or footprinting of reverse transcriptase with substrates (33-36).
A limited number of studies have suggested the possibility that sequence might influence 5′ end-directed cleavages (24,29,30), but this hypothesis has not been directly tested before. We recently reported that sequence specificity is an important determinant of internal RNase H cleavages, and that an RNA 5′ end at a nick does not promote 5′ end-directed RNase H cleavages (23). These results prompted us to ask three fundamental questions concerning RNA 5′ end-directed cleavages by retroviral RNases H. First, does sequence influence 5′ end-directed cleavages? Second, what distances from the RNA 5′ end are acceptable for 5′ end-directed cleavage sites? Third, how large of a gap is required between the 3′ terminus of an upstream RNA and the 5′ end of a downstream RNA to allow efficient 5′ end-directed cleavages in the downstream RNA? Our findings offer new insights into the mechanisms of RNA 5′ end-directed cleavages for M-MuLV and HIV-1 reverse transcriptases, and the role of RNase H in retroviral replication.
EXPERIMENTAL PROCEDURES
Enzymes and reagents—Recombinant HIV-1 reverse transcriptase was obtained from Worthington Biochemicals. Recombinant wild-type M-MuLV reverse transcriptase, T7 DNA polymerase, and calf intestinal alkaline phosphatase were purchased from Amersham Pharmacia Biotech. T4 polynucleotide kinase, T4 DNA ligase, T4 DNA polymerase, and all restriction enzymes were obtained from New England Biolabs. DNA oligonucleotides were obtained from Qiagen and Invitrogen. Oligonucleotides were gel-purified in denaturing polyacrylamide gels as described (23,37).
Preparation of RNAs—Preparation of RNAs Md1 - Md10, PPT20, PPT62, and NPPT has been described (23). To prepare R46 RNA, a 46-mer DNA oligonucleotide (5’-CAATGAAAGACCCCACCTGTAGGTTTG GCAAGCTAGCTTCGAGACG-3’) was annealed to a 54-mer DNA oligonucleotide (5’-AATTCGTCTCGAAGCTAGCTTGCCAAA CCTACAGGTGGGGTCTTTCATTGAGCT-3’), and this duplex was introduced into EcoRI- and Sac I-linearized pGEM9Zf(-) (Promega). The resulting plasmid was linearized with BsmBI and transcribed in vitro as described (23). All other RNAs were prepared by in vitro transcription of linearized plasmids as follows. To make 35-mer R7Z1, pGEM7Zf(+) (Promega) was linearized with XbaI. To make 28-mer R9Z1, pGEM9Zf(-) was linearized with XbaI. To make 32-mer RpKS1, pBluescript KSII(+) (Stratagene) was linearized with NotI. To make 30-mer R3Z1, pGEM3Zf(-) (Promega) was linearized with BamHI. Because RNA R3Z1 is identical in sequence to the 5′ end of a 41 nucleotide RNA that has been used extensively to map 5′ end-directed cleavages (29,38-43), we have used the cleavage sites for RNA R3Z1 as mapped in our own experiments.
To generate additional RNAs with different sequences, some pGEM plasmids were digested with single-cutting restriction enzymes downstream of the T7 promoter (as indicated below), the overhangs were filled in or removed by T4 DNA polymerase treatment, the blunt ends were ligated using T4 DNA ligase, and the resulting plasmids were transformed into XL1-Blue cells (Stratagene). The sequences of these plasmids were confirmed by DNA sequencing. For T4 DNA polymerase treatment, a digested plasmid was incubated in a 20 ml reaction containing 1 mM dNTPs and 3 units of T4 DNA polymerase for 5 min at 37 °C and then for 15 min at 12 °C. To make 25-mer R3Z2, pGEM3Zf(-) was digested by EcoRI and BamHI, and the resulting plasmid was linearized with HindIII prior to in vitro transcription. To make 27-mer R7Z2, pGEM7Zf(+) was digested by ApaI, treated with T4 DNA polymerase, ligated, digested with AatII, treated with T4 DNA polymerase, and ligated, and then the resulting plasmid was linearized with XbaI. To make 32-mer R7Z3, pGEM7Zf(+) was digested by ApaI and XbaI, and the resulting plasmid was linearized with Acc65I. To make 30-mer R7Z4, pGEM7Zf(+) was digested by ApaI and EcoRI, and the resulting plasmid was linearized with BstB I. To make 30-mer R7Z5, pGEM7Zf(+) was digested by ApaI and SmaI, and the resulting plasmid was linearized with HindIII.
5′ end-labeling, 3′ end-labeling, and 5′ endphosphorylation—5′ end-labeling or 3′ end-labeling of RNAs were carried out as described previously (23,37). Because cleavage by RNase H produces a 3′ hydroxyl group and a 5′ phosphate group, an unlabeled phosphate group was added to the 5′ ends of 3′ end-labeled RNAs as previously described (23).
Preparation of hybrid substrates—To prepare the substrate without upstream RNA (No Upstream substrate), RNAs were annealed to the appropriate template DNAs at an RNA:DNA molar ratio of 1:2, and to prepare the Nick or Gap substrates, upstream PPT20 RNA was additionally included at a 3-fold molar excess over the first RNA. Annealings were carried out in 10 mM Tris-HCl, pH 8.0, and 200 mM KCl at 90 °C for 3 min followed by cooling to room temperature.
For the No Upstream substrates, the RNA 5′ end was recessed by 22 nucleotides from the DNA 3′ end and the RNA 3′ end extended two bases beyond the DNA 5′ end. Hybrids containing RNAs Md1, Md2, Md4, Md6, Md7, Md9, and Md10 were annealed to DNA strands D49, D491, D49△3, D49△5, D49△6, D49△8, and D49△9, respectively (23). Hybrids containing RNAs R3Z1, R3Z2, R7Z1, R7Z2, R7Z3, R7Z4, R7Z5, R9Z1, R46, and RpKS2 were similarly positioned on DNA strands containing the appropriate complementary sequences. RNA NPPT was annealed to D52N (23), and PPT62 was annealed to D+27temp4 (23).
For the Nick substrate containing upstream PPT20 and downstream Md1, the template strand was D49 (5′-CCAAACCTACAGGTGGGGTCTTTCATT T[ ]CCCC CCTTTTTCTGGAGACTAA-3′). The brackets in the sequence of D49 indicate the position that nucleotides were added to create the template strands for the corresponding Gap substrates, which are D49(1b) (5′...[T]...3′), D49(2b) (5′...[GT]...3′), D49(3b) (5′...[AGT]...3′), D49(4b) (5′...[CAGT]...3′), and D49(5b) (5′...[ACAGT]...3′).
For the Nick substrate containing upstream PPT20 and downstream Md10, the template strand was D49△9 (5′-GCTAGCTTGCCAAACCTACAGGTGGGG [ ]CCCCCC TTTTTCTGGAGACTAA-3′). The brackets in the sequence of D49△9 indicate the position that nucleotides were added to create the template strands for the corresponding Gap substrates, which are D49△9(1b) (5′...[T]...3′), D49△9(2b) (5′...[GT]...3′), D49△9(3b) (5′...[AGT]...3′), D49△9(4b) (5′...[CAGT]...3′), and D49△9(5b) (5′...[ACAGT]...3′).
Cleavage analysis of hybrid substrates—3 units (0.6 to 0.8 pmol) of M-MuLV reverse transcriptase or 0.5 units (0.2 pmol) of HIV-1 reverse transcriptase were incubated with 0.2 pmol (10 nM final concentration) of a hybrid substrate in 20 μ1 reactions containing 50 mM Tris-HCl, pH 8.0, 50 mM KCl, 6 mM MgCl2, and 5 mM DTT at 37 °C for the times indicated. Aliquots were added to formamide stop mix (95% formamide, 20 mM EDTA) and electrophoresed in denaturing 20% polyacrylamide gels (National Diagnostics). Products were visualized by PhosphorImager analysis (Amersham Biosciences).
Statistical analysis—For each position of the aligned sequences, the chi-square onedimensional test was used to determine the deviation from the random distribution of bases (44). The expected frequency for each base was determined by summing the nucleotide frequencies of the first 25 nucleotides from the 5′ end of each RNA used for HIV-1 (Fig. 5) and for M-MuLV (Fig. 6). For HIV-1, the expected frequencies were A = 0.233, C = 0.243, G = 0.319, and U = 0.205. For M-MuLV, the expected frequencies were A = 0.220, C = 0.251, G = 0.326, and U = 0.203.
Fig. 5.

Alignment of sequences flanking RNA 5′ end-directed cleavage sites recognized by HIV-1 RNase H. The left column lists the names of the RNAs used in this analysis, and asterisks denote those presented in prior studies. Cleavage sites mapped using RNAs in this study are described in the Materials and Methods. The 28 nt RNA is from (40). The 40-mer RNA from (25) was used as a blunt-end hybrid, but a later study has shown that while the extent of cleavage is diminished, the locations of 5′ end-directed cleavage sites are not affected by a blunt end (40). RNA D is from (29,32,42). The 20-mer RNA is from (45). In the center column, the sequence surrounding each cleavage site is given, with the location of the cleavage site represented as a gap. The right column gives the position of each cleavage site counting from the 5′ end of the RNA.
Fig. 6.

Alignment of sequences flanking RNA 5′ end-directed cleavage sites recognized by M-MuLV RNase H. The sequences are presented as described in the legend to Fig. 5. Cleavage sites mapped using RNAs in this study are described in the Materials and Methods.
Quantitative analysis of RNase H cleavage products—Band intensities of cleavage products were determined using ImageQuant Software (Amersham Biosciences). To compare the amount of product resulting from cleavage at the same sequence in different RNAs (for example, counting from the RNA 5′ end, site F is cleaved in Md1 between the 16th and 17th the 15th nucleotides, and in Md2 between and 16th nucleotides), the area integration function was used to quantitate the relative abundance of individual bands present in each sample. This analysis normalized the amount of cleavage product for a specific site as a percentage of the total cleavage products for each hybrid substrate. Quantitations using data from the 15 sec and 1 min time points were found to be essentially identical, and the results from 1 min time points are presented graphically in Fig. 4.
Fig. 4.
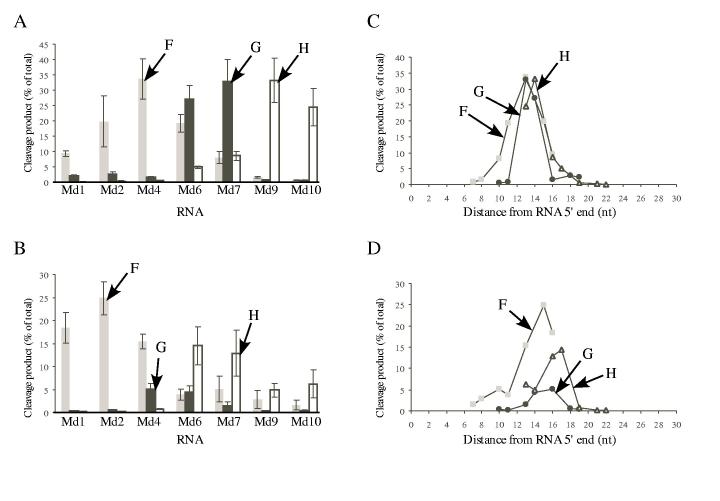
Extent of cleavage and optimal distances for cleavage at sites F, G, and H in RNAs Md1 through Md10 by HIV-1 and M-MuLV reverse transcriptases. The amount of product generated by cleavage (% of total) at sites F, G, and H in the indicated substrates was determined for HIV-1 (A) or M-MuLV (B) reverse transcriptase. Data from the 1 min time points in three (A) or four (B) independent experiments with 5′ end-labeled RNAs were used to determine the amount of product that resulted from the cleavages at site F (gray bars), site G (black bars), or site H (white bars) (± S.D.). These same data were also used to analyze the optimal distance for cleavage of each site relative to the 5′ RNA ends for HIV-1 (C) or M-MuLV (D) reverse transcriptase. The amount of product generated by cleavage (% of total) for sites F (gray squares), G (black circles), or H (open triangles) is plotted as a function of the cleavage site distance in nucleotides from the 5′ end of each substrate.
RESULTS
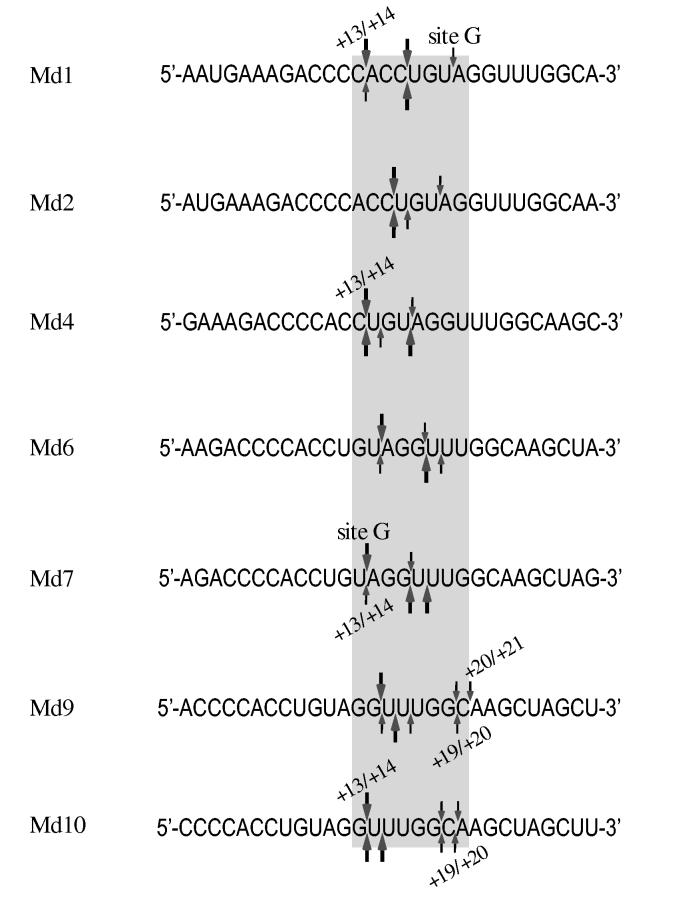
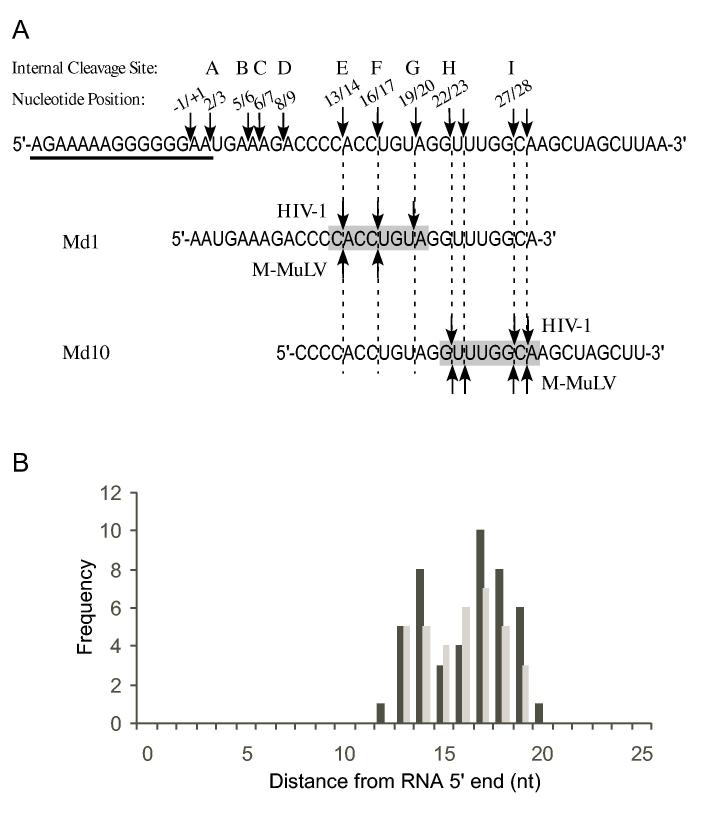
Sequence and distance are determinants of RNA 5′ end-directed cleavages—5′ end-directed cleavages by RNase H have been shown to occur 15 to 20 nucleotides from the RNA 5′ end. To address how RNase H cleaves at specific sites within this range, we have evaluated the roles of RNA sequence, distance, and accessibility to the RNA 5′ end as possible determinants of cleavage. As a model system, hybrid substrates with RNAs containing sequence from the M-MuLV PPT region were used. In this region of the M-MuLV genome, the cleavage that generates the 3′ end of the PPT primer is defined to occur between nucleotides - 1 and + 1, and is called the -1/+1 cleavage site. This numbering is also used to designate the positions of nucleotides in the model hybrid substrates, which contained seven different 29-mer RNAs, termed Md1 through Md10 (Fig. 1). These RNAs have 5′ ends beginning downstream of the PPT at positions +1, +2, +4, +6, +7, +9, or +10, and contain a total of 29 consecutive nucleotides of sequence, such that blocks of the same sequence are located at different distances from the RNA 5′ ends (shaded box in Fig. 1). These RNAs were used to ask if 5′ end-directed cleavages occur at identical distances from the RNA 5′ ends, and if sequence influences the positioning of 5′ end-directed cleavages.
Fig. 1.
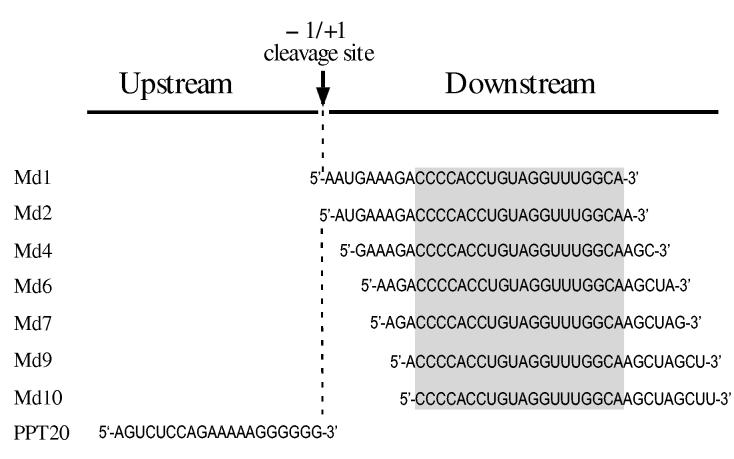
RNA oligonucleotides used in RNase H cleavage assays. All RNAs contain M-MuLV sequences from the PPT region, which contains the -1/+1 site where cleavage occurs to generate the plus-strand primer and which corresponds to nucleotides 7815 and 7816 in the M-MuLV sequence (55),. PPT20 is upstream of this site and spans nucleotides -20 to -1. The seven downstream RNAs are named as Moloney downstream (Md) followed by the nucleotide position of the 5′ terminus relative to the -1/+1 site. The sequence shared by all of these RNAs is indicated by a shaded box.
For RNase H cleavage assays, each RNA was 5′ or 3′ end-labeled, and annealed to a template DNA to generate a hybrid with a recessed RNA 5′ end. Hybrid substrates were incubated with HIV-1 or M-MuLV reverse transcriptase in time course assays; the products representing the initial 5′ end-directed cleavage events were found in the 15 sec and 1 min time points when most of the initial substrate remained uncleaved. These initial cleavage sites were mapped by comparing the mobilities of cleavage products to size ladders generated with nuclease P1 (data not shown). Depending upon whether an RNA was 5′ or 3′ end-labeled, the longest products represented the cleavages that occur the furthest from or the closest to the RNA 5′ end, respectively. Importantly, the location and extent of cleavage for each site was determined by using the data obtained for both 5′ and 3′ end-labeled RNAs. To facilitate comparison of the same sites in different substrates, the cleavage sites generating the observed products have been named A through I, as indicated in the relevant figures and as described previously (23).
When substrates containing 5′ end-labeled RNAs Md1 through Md10 were treated with HIV-1 reverse transcriptase, it was immediately apparent that the cleavage products were not of uniform size (Fig. 2A). Instead, the products had varied lengths indicating that cleavages occurred at different distances from the RNA 5′ ends. The expected products resulting from cleavage at the same sites in the different substrates (labeled E — I) are connected by lines in Fig. 2. Substrate Md1 was predominantly cleaved at sites E, F, and G (Fig. 2A, lanes 1-5). Counting from the RNA 5′ end, site E is between nucleotides 13 and 14, site F is between nucleotides 16 and 17, and site G, which represented the most distal 5′ end-directed cleavage site in Md1, falls between nucleotides 19 and 20. Cleavage of substrate Md2 was very similar to that of substrate Md1 except that all of the cleavage products were one nucleotide smaller (lanes 6-10). Importantly, site G remained the most distal 5′ end-directed cleavage, even though this site was now located between the 18th and 19th nucleotides from the Md2 RNA 5′ end and, unlike Md1, no cleavage occurred between the 19th and 20th nucleotides. While site H was not cleaved in Md1 or Md2, this site was detectably cleaved between the 19th and 20th nucleotides in substrate Md4 (lanes 11-15 and significantly cleaved between the 17th), 18th and nucleotides in substrate Md6 (lanes 16-20). No cleavage was observed at site I in substrates Md1 through Md7 (lanes 1-25), but this site was cleaved when located between the 19th and 20th nucleotides in substrate Md9, and between the 18th and 19th nucleotides in substrate Md10 (lanes 25-35).
Fig. 2.
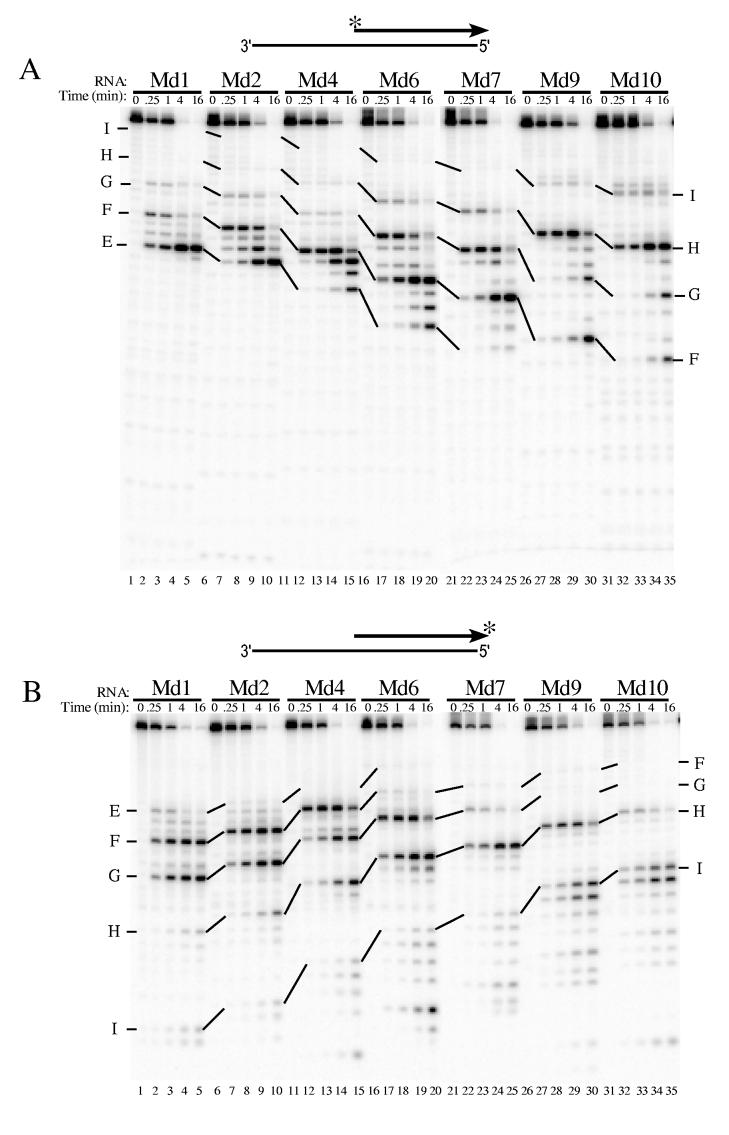
Analysis of 5′ end-directed RNase H cleavages in RNAs Md1 through Md10 using HIV-1 reverse transcriptase. The hybrid substrate (thick line RNA) shown schematically at the top of each panel contained the indicated 5′ end-labeled (A) or 3′ end-labeled (B) RNA. Substrates were treated with HIV-1 reverse transcriptase, aliquots were removed at the indicated times, and samples were analyzed in denaturing 20% polyacrylamide gels. Results were visualized using a Phosphorimager. Products resulting from cleavage at specific sites in Md1 are labeled as sites E — I, and solid lines track cleavage products corresponding to products from cleavage of the same sites in the other substrates. In Md1 (see Fig. 1), the cleavage sites are found between the following nucleotides from the RNA 5′ end: site E is +13/+14, site F is +16/+17, site G is +19/+20, site H is +22/+23, and site I is +27/+28. The cleavage sites identified in Md1 that give rise to the cleavage products shown for Md10 are indicated at the right.
Assays with 3′ end-labeled RNAs and HIV-1 reverse transcriptase were used to confirm that the 5′ end-directed cleavages sites closest to the RNA 5′ ends in the above analysis represented the initial cleavage products and were not the result of secondary cleavages (Fig. 2B). In general, the locations were the same as for the 5′ end-labeled substrates, but the relative abundance of products sometimes varied because strong cleavage sites close to the labeled end diminished the detection of more distal sites. Site E was cleaved well in substrate Md1 at the earliest time points, but was only weakly cleaved in subsequent substrates (lanes 1-20). Site F was a strong cleavage site when located as close as between the 13th and 14th nucleotides from the RNA 5′ end (Md1-Md4, lanes 1-15), but was not cleaved efficiently when located closer than between the 11th 12th and nucleotides (Md6-Md10, lanes 15-35). Site G became the strongest cleavage site close to the RNA 5′ between the 14th end when and 15th located nucleotides in substrate Md6 (lanes 16-20) and between the 13th and 14th nucleotides in substrate Md7 (lanes 21-25). However this site was not cleaved when moved 2 bases closer to the RNA 5′ end in substrate Md9 (lanes 26-30). In substrates Md9 and Md10, the cleavage site recognized closest to the RNA 5′ end was site H (lanes 26-30). Site I was not cleaved until located between the 19th and 20th nucleotides in Md9 or the 18th and 19th nucleotides in Md10 (lanes 26-35).
These same substrates were also used to analyze 5′ end-directed cleavage by the RNase H of M-MuLV reverse transcriptase (Fig. 3). Most of the cleavage sites recognized in Md1 through Md10 by M-MuLV reverse transcriptase were identical to those recognized by the HIV-1 enzyme, but the extent of cleavage often varied. M-MuLV RNase H cleaved 5′ end-labeled Md1 at site D, between the 8th and 9th nucleotides, and at sites E and F, but notably, no 5′ end-directed cleavages occurred beyond site F (Fig. 3A, lanes 1-5). Faint cleavage of site G was apparent between the 18th and 19th nucleotides in substrate Md2, but was more distinct in substrate Md4, when this site was located between the 16th and 17th nucleotides (lanes 6-15). Adjacent cleavages beginning at site H were the most distal 5′ end-directed cleavages in substrates Md6 (between the 17th and 18th nucleotides) and Md7 (between the 16th and 17th nucleotides) (lanes 16-25). Similarly in substrates Md9 and Md10, a pair of cleavages between the 19th and 20th nucleotides or the 18th and 19th nucleotides, respectively, were the furthest 5′ end-directed cleavage sites and included site I (lanes 26-30).
Fig. 3.
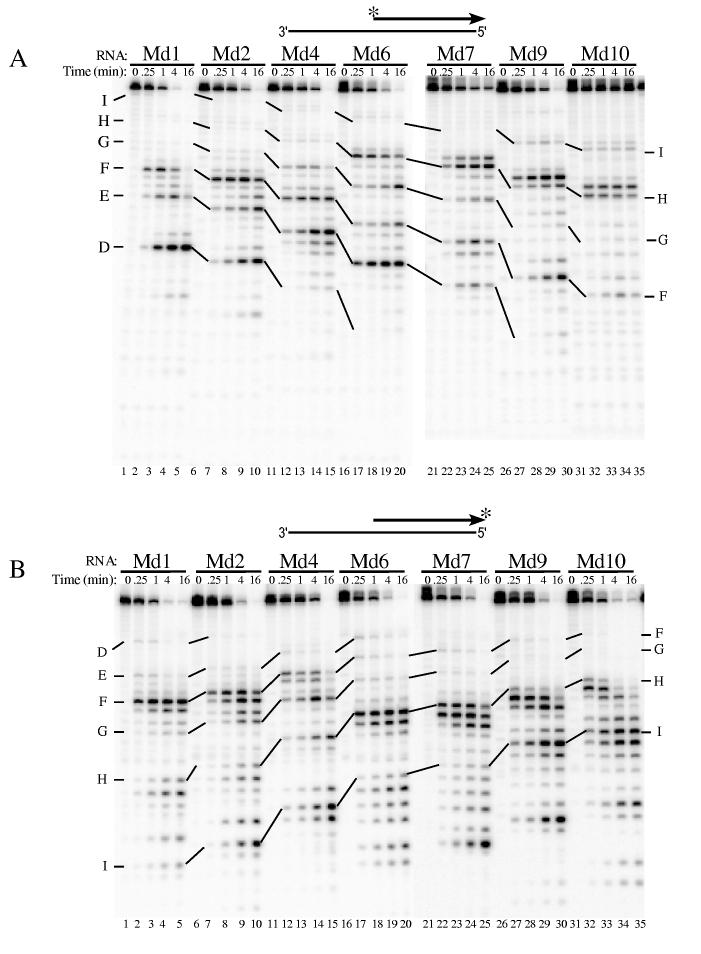
Analysis of 5′ end-directed RNase H cleavages in RNAs Md1 through Md10 using M-MuLV reverse transcriptase. The hybrid substrate shown schematically at the top of each panel contained the indicated 5′ end-labeled (A) or 3′ end-labeled (B) RNA. Substrates were treated with M-MuLV reverse transcriptase and analyzed as described in Fig. 2. In Md1, the cleavage sites are found between the following nucleotides from the RNA 5′ end: site D is +8/+9, site E is +13/+14, site F is +16/+17, site G is +19/+20, site H is +22/+23, and site I is +27/+28.
As described above, we used 3′ end-labeled RNAs in the hybrid substrates to map the 5′ end-directed cleavages close to the RNA 5′ ends for M-MuLV. In general, the strongest cleavages were observed at positions corresponding to site F in substrates Md1, Md2, and Md4 (Fig. 3B, lanes 1-15). In substrates Md6 through Md10, adjacent cleavages including site H were the strongest cleavages near the RNA 5′ end (lanes 16-35). The locations of these cleavages ranged from between the 17th and 18th nucleotides in Md6 to between the 13th and 14th nucleotides in Md10.
The short fragment resulting cleavage between the 8th and 9th from nucleotides at site D observed with the 5′ end-labeled substrate (Fig. 3A, lane 2) could have been a secondary cleavage product. However, at least some of this product was generated independent of other cleavages because the corresponding cleavage product was also observed using the 3′ end-labeled substrate Md1 (Fig. 3B, lanes 1-5). Limited but detectable independent cleavages occurred between the 8th and 9th also nucleotides of two other substrates, at site E in substrate Md6 and at site F in substrate Md9 (lanes 16-20 and 26-30, respectively). The relevance of these 5′ proximal cleavages to 5′ end-directed cleavage is considered in the Discussion.
Quantitative analysis of distance effects on extent of cleavage—To better understand how the extent of cleavage at a specific site is affected by the distance of the site relative to the RNA 5′ end, we carried out quantitative analyses of the cleavage products generated with the 5′ end-labeled substrates Md1 through Md10 shown in Figs. 2 and 3. Sites F, G, and H were chosen as representative cleavages because these sites were recognized by both HIV-1 and M-MuLV reverse transcriptases, and because they are present over the relevant range of distances in the various substrates.
First, the extent of cleavage at each site was evaluated in all substrates. For HIV-1 RNase H, cleavage at site F appeared to have a bell-shaped pattern, with the highest cleavage occurring with substrate Md4 (Fig. 4A). The pattern was similar for site G but cleavage was most abundant in substrates Md6 and Md7. The pattern for cleavage of site H was also similar, with the highest cleavage in substrates Md9 and Md10. For M-MuLV RNase H, the amount of cleavage at each site also appeared as non-overlapping, bell-shaped patterns distributed in substrates Md1 through Md10 (Fig. 4B). Cleavage of site F was maximal in substrate Md2, cleavage of site G was highest in substrates Md4 and Md6, and cleavage of site H was greatest in substrates Md6 and Md7. Interestingly, the greatest amount of cleavage for a particular site occurred in different substrates for M-MuLV and HIV-1 RNases H. For example, site F was cleaved to the greatest extent in substrate Md2 for M-MuLV RNase H and in substrate Md4 for HIV-1 RNase H (Fig. 4AB).
Next, we determined how the extent of cleavage at sites F, G, and H was affected by the distance from the RNA 5′ end. For HIV-1 RNase H (Fig. 4C), the plots of distance versus cleavage at each site overlapped, indicating that the overall effects of distance on the extent of cleavage were very similar. A plot of the equivalent data for M-MuLV RNase H also showed that distance from the RNA 5′ end similarly influenced the extent of cleavage at sites F, G, and H (Fig. 4D).
Preferred nucleotides flank RNA 5′ end-directed cleavage sites—We next asked if preferred nucleotides could be identified near 5′ end-directed cleavage sites, since we recently showed that preferred nucleotides are found near internal cleavage sites recognized by the M-MuLV and HIV-1 RNases H (23). This analysis required the mapping of 5′ end-directed cleavage sites on a variety of different hybrid substrates containing RNAs with recessed 5′ ends. Three RNAs derived from viral sequences were used: PPT62, containing sequence from the M-MuLV genome (PPT62; (23)), and RNAs Md1 and Md10. To generate additional sequence diversity, several RNAs with unique sequences were made using in vitro transcription plasmids (see Materials and Methods). Finally, for HIV-1 reverse transcriptase, we have included 5′ end-directed cleavage sites reported previously by other investigators in studies where the exact positions of these sites were mapped (21,25,29,38-43,45).
To generate hybrid substrates with recessed 5′ RNA ends, RNAs were 5′ or 3′ end-labeled and annealed to DNA templates. These substrates were used in RNase H cleavage assays as shown above with HIV-1 or M-MuLV reverse transcriptase, and cleavage sites were mapped to the nucleotide level (data not shown). The relative extent of cleavage at the various sites in each substrate was classified as strong, medium or weak. As an example using substrate containing RNA Md1 and HIV-1 reverse transcriptase (Fig. 2AB, lanes 1-5), sites E and F were classified as strong, and site G was classified as medium. Using substrate Md1 with M-MuLV reverse transcriptase (Fig. 3AB, lanes 1-5), site E was classified as medium and site F was classified as strong. For both M-MuLV and HIV-1, cleavages between sites E and F represent weak sites. In our analysis, only strong and medium sites were considered. For the mapped cleavage sites observed in prior studies with HIV-1 (21,25,29,38-43,45), all of the identified RNase H cleavage sites were used.
To compare the sequences surrounding RNA 5′ end-directed cleavage sites, the cleavage was defined to occur between nucleotides -1 and +1 and the flanking sequences were aligned. These cleavage sites are presented for HIV-1 in Fig. 5 and for M-MuLV in Fig. 6. To statistically determine whether any base preferences correlated with the cleavage sites, the nucleotides from positions -10 to +4 were tabulated. The frequency of bases in the first 25 nucleotides of sequence beginning from each RNA 5′ end was calculated to determine the expected distribution of nucleotides. For a given cleavage site, the significance of any deviations from random nucleotide frequencies was determined by comparing the base distribution at each position with the expected distribution using the chi-square method (Materials and Methods). The resulting chi-square values were plotted against the nucleotide positions (Fig. 7).
Fig. 7.
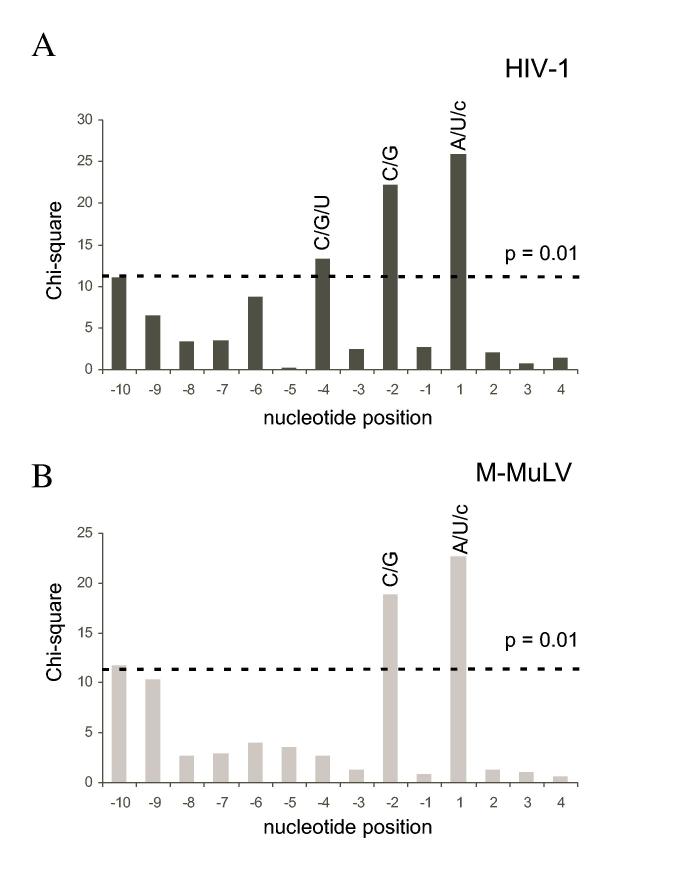
Base preferences at nucleotide positions flanking strong and medium RNA 5′ end-directed cleavages sites. The chi-square values of the nucleotide distribution from positions -10 to +4 were calculated by comparison to overall base frequencies and plotted as a function of nucleotide position, with the cleavage site located between positions -1 and +1. (A) Analysis of the 46 sites for HIV-1 shown in Fig. 5. (B) Analysis of the 35 sites for M-MuLV shown in Fig. 6. The dashed line indicates the chi-square value of 11.34, which has a p value of 0.01. Nucleotide positions with scores exceeding 11.34 are considered significant. The preferred nucleotides for each of the significant positions are indicated above the corresponding bar in upper case (strongly preferred) or lower case (acceptable) letters.
For both HIV-1 and M-MuLV, two nucleotide positions had p values less than 0.01 (X2 values > 11.34) and were considered strong deviations from random. Position +1 showed a strong preference for A or U, tolerated C, and strongly disfavored G. Position - 2 had a strong preference for G or C and especially disfavored U. In addition, the - 4 position for HIV-1 also had a p value less than 0.01 and disfavored A. Unlike the nucleotide preferences seen for internal cleavage sites (23), strong preferences for sequences further upstream were not observed (see Discussion).
Accessibility to a 5′ end affects RNA 5′ end-directed cleavages—We recently showed that the 5′ end of an RNA at a nick does not allow 5′ end-directed cleavages (23). To determine the gap size required for 5′ end recognition, the RNase H cleavage pattern was compared using substrates in which the same RNA was placed at different distances downstream from PPT20, an RNase H-resistant RNA containing nucleotides -20 to -1 of the M-MuLV PPT sequence (Fig. 1, (37)). The distances between PPT20 and the downstream RNAs were either a nick or a gap of 1 to 5 bases. For M-MuLV reverse transcriptase, 5′ end-labeled Md1 was used as the downstream RNA. In the No Upstream substrate, cleavages occurred predominantly at sites D, E and F in Md1 (Fig. 8, lanes 1-5 and 36-40). Addition of upstream PPT20 to create a nick reduced 5′ end-directed cleavages at these sites and promoted internal cleavages at sites closer to the nick, such as sites A and B (lanes 6-10). This cleavage pattern was identical to that observed using a continuous RNA containing the same sequence but lacking the nick (23); together these data first suggested that a nick was insufficient to direct 5′ end-directed cleavages. Introduction of a 1 or 2 base gap between the upstream and downstream RNAs decreased cleavage at the A and B sites, and increased cleavage at the E and F sites (Fig. 8, compare lanes 11-20 with lanes 6-10). The cleavage pattern with a 3 base gap most closely matched the No Upstream substrate (compare lanes 21-25 with 1-5 and 36-40). Interestingly, the highest extent of 5′ end-directed cleavages was observed in substrates with a gap size of 4 or 5 bases (lanes 26-35). Experiments performed using 3′ end-labeled Md1 confirmed these results (data not shown).
Fig. 8.
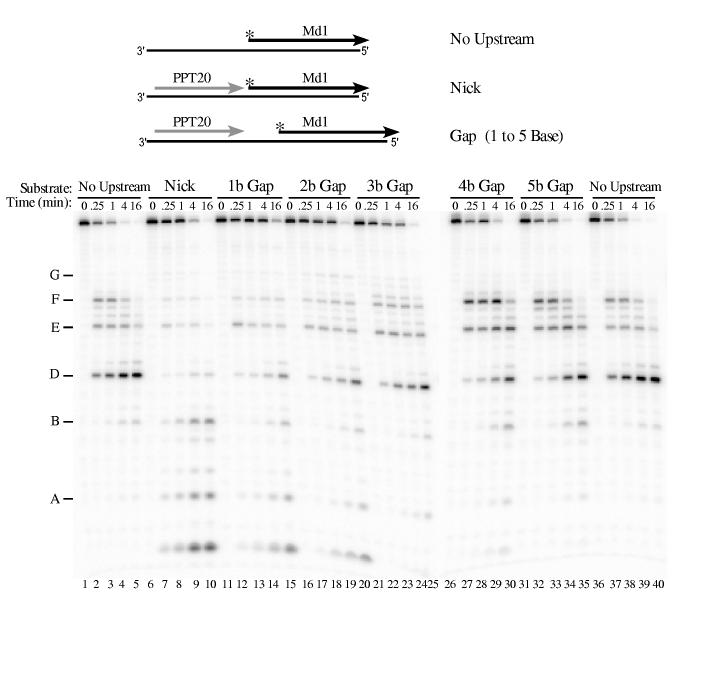
Influence of gap size on 5′ end-directed cleavage of Md1 by M-MuLV reverse transcriptase. 5′ end-labeled Md1 was used as a substrate without upstream RNA (No Upstream), as a substrate with PPT20 annealed immediately upstream (Nick), or as substrates with gaps of 1 to 5 bases between the 3′ terminus of upstream PPT20 and the 5′ end of Md1 (1 to 5 b Gap). Substrates were incubated with M-MuLV reverse transcriptase, and analyzed as described in Fig. 2. In Md1, cleavage site A is +2/+3, site B is +5/+6, and the other sites are as described in Fig. 3.
To test how a gap influences 5′ end-directed cleavages by HIV-1 reverse transcriptase, 5′ end-labeled Md10 was used as a downstream RNA (Fig. 9). In the No Upstream substrate, the intial cleavages in Md10 occurred at sites H and I (lanes 1-5 and 36-40). Addition of upstream PPT20 decreased cleavage at sites H and I, and increased internal cleavages at site E among others (lanes 6-10). For substrate with a 1 Base Gap, cleavages closer to the RNA 5′ end such as site E decreased while cleavages at sites H and I slightly increased (lanes 11-15). In the 2 Base and 3 Base Gap substrates, RNase H cleavages were most similar to those in substrate lacking upstream RNA (compare lanes 16-25 with lanes 1-5 and 36-40). Experiments performed with 3′ end-labeled Md10 and HIV-1 reverse transcriptase generated comparable results (data not shown).
Fig. 9.
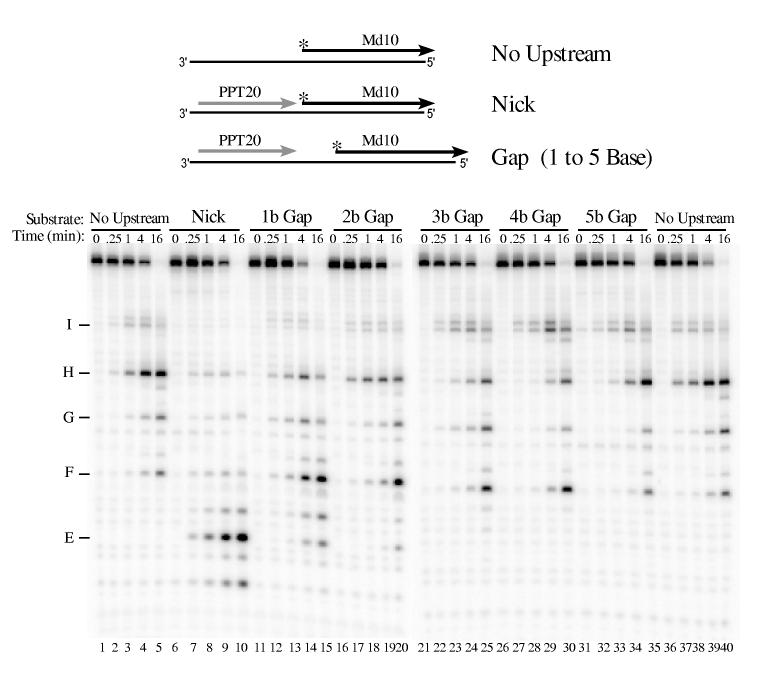
Influence of gap size on 5′ end-directed cleavage of Md10 by HIV-1 reverse transcriptase. 5′ end-labeled Md10 was used as a substrate without upstream RNA (No Upstream), in substrate with PPT20 annealed immediately upstream (Nick), or in substrates with gaps of 1 to 5 bases between the 3′ terminus of upstream PPT20 and the 5′ end of Md10 (1 to 5 b Gap). Substrates were incubated with HIV-1 reverse transcriptase, and analyzed as described in Fig. 2. Products resulting from cleavage at specific positions in Md10 are indicated at left, as described in Figs. 2 and 3. The smallest cleavage product (most distinct in lanes 7 - 10 below site E) corresponds to cleavage between the first and second nucleotides of Md10.
RNase H assays were also carried out with M-MuLV or HIV-1 reverse transcriptase and substrate containing downstream Md10 or Md1, respectively (date not shown). In both cases and similar to the data shown in Figs. 8 and 9, the pattern of RNase H cleavages in substrates with a 2 or 3 base gap most clearly matched the No Upstream substrate.
DISCUSSION
The ability to cleave the RNA strand of hybrids formed during reverse transcription is essential for retroviral replication. RNase H cleavages positioned by an RNA 5′ end represent a very robust form of RNase H activity that likely provides an important role in general degradation of the viral genome. The data in this study demonstrate that multiple determinants affect the specificity of RNA 5′ end-directed cleavage by retroviral RNases H.
Previous work has shown that reverse transcriptase does not efficiently utilize the RNA 5′ end at a nick to position 5′ end-directed cleavages (23). In this study, we addressed how much space between the 3′ and 5′ ends of RNA is required to render the RNA 5′ end accessible for directing cleavage in the downstream RNA. By increasing the distance between an upstream RNA 3′ end and a downstream RNA 5′ end in single nucleotide increments, we found that gaps of 2 or 3 bases permit 5′ end-directed cleavages at a level comparable to that observed for a recessed, free RNA 5′ end. It seems likely that the binding of reverse transcriptase to a recessed RNA 5′ end in a hybrid involves recognition of the discontinuity in the substrate structure at the junction between the single strand and the duplex. A similar discontinuity is offered by a typical primer-template, where the recessed end is a DNA 3′ terminus rather than an RNA 5′ end. Thus, perhaps the primer-template binding cleft and in particular the primer grip region (34) of the polymerase domain positions the recessed RNA 5′ end of a hybrid for 5′ end-directed cleavage by recognizing some of the same structural features presented by a primer terminus. In substrates containing a nick or a 1 base gap, the RNA 5′ end is obscured and RNase H cleavages are limited to the internal model of cleavage. A substrate with a gap of 2 or more bases offers a sufficient distance between the upstream and downstream RNAs to allow the access and recognition required for the 5′ end-directed mode of cleavage, and additionally may offer contact sites in the upstream RNA that facilitate enzyme binding. During general degradation of the genome, it seems likely that gaps are important to promote 5′ end-directed cleavages.
To initially test how sequence might influence RNA 5′ end-directed cleavages, RNase H assays were carried out with hybrid substrates that contained portions of the same sequence located at different positions relative to an RNA 5′ end. A comparison of the cleavages in these substrates (Fig. 10) reveals several interesting features for the 5′ end-directed mode of cleavage that are shared between the reverse transcriptases of HIV-1 (arrows above sequences) and M-MuLV (arrows below sequences). First, cleavages are not restricted to a discrete distance from the RNA 5′ end. Counting from the RNA 5′ end for HIV-1, strong or medium cleavage sites were as close as between positions +13 and +14 (Md1, Md4, Md7, Md10) and as far as between positions +20 and +21 (Md9). For M-MuLV, strong and medium cleavage sites ranged from between positions +13 and +14 (Md1, Md4, Md7, Md10) to between positions +19 and +20 (Md9, Md10). Second, the same substrate can be cleaved at multiple sites, and independent cleavages can be distributed over a span of up to 8 nucleotides (for example, Md9 and Md10). Third, cleavages occur at the same sites in the sequence even though these sites are located at different distances from the RNA 5′ ends. As an example for HIV-1 RNase H, site G is cleaved in substrate Md1 between the 19th and 20th nucleotides, but this site is also cleaved when located 6 nucleotides closer to the RNA 5′ end in substrate Md7 (indicated in Fig. 10). Fourth, some positions are consistently resistant to cleavage irrespective of distance from the RNA 5′ end, even when located between other strong 5′ end-directed cleavage sites. Finally, the strong and medium 5′ end-directed cleavage sites fall between the 13th and 20th nucleotides from the RNA 5′ end (Fig. 10, indicated with a grey box). These data indicate that 5′ end-directed cleavage sites are not randomly chosen, and that both sequence and distance determine the cleavage positions of retroviral RNases H.
Fig. 10.
Comparison of HIV-1 and M-MuLV RNase H 5′ end-directed cleavages in the sequences of RNAs Md1 - Md10. The sequences of the 29-mer RNAs Md1 through Md10 are aligned by the RNA 5′ ends to compare the positions of 5′ end-directed cleavage sites. In each sequence, the extent of cleavage at a site is indicated as strong (large arrows) or medium (small arrows) for HIV-1 reverse transcriptase (above) or M-MuLV reverse transcriptase (below). As described in the Discussion, the range of the closest and furthest independent 5′ end-directed cleavage sites is indicated by the positions of the bordering nucleotides from the RNA 5′ end, the position of site G in substrates Md1 and Md7 is indicated, and the grey box highlights nucleotide positions +13 and +20 that include the range of distances where the 5′ end-directed cleavages occur.
Since some early studies suggested that sequence might influence RNA 5′ end-directed cleavages (24,29,30), it is somewhat surprising that this possibility has not been investigated more thoroughly until now. Most likely this is because prior studies utilized a limited number of RNAs and thus had very little variation in sequence (21,29,32,38-43). Also for several studies, the precise mapping of RNA 5′ end-directed cleavage sites was often difficult or not required by the experimental design (21,29,32,46). Most recently, RNA 5′ end-directed cleavages have been examined with regard to the order of cleavage that occurs on an RNA/DNA hybrid with a recessed RNA 5′ end (38-40). From these studies, it was proposed that 5′ end-directed cleavages constitute an initial or primary cleavage ∼ 18 nucleotides from the RNA 5′ end, and are followed by independent, secondary cleavages that are 8 to 9 nucleotides from the RNA 5′ end and occur at a slower rate. While our findings demonstrate that sequence is an important determinant of 5′ end-directed cleavages, it remains to be investigated how sequence influences secondary cleavages.
By mapping multiple 5′ end-directed cleavage sites in hybrids containing RNAs with different sequences, we observed a statistically significant bias for specific nucleotides at positions flanking these sites. For both HIV-1 and M-MuLV RNases H, A or C, but not G, was preferred at position +1, and C or G, but not U, was preferred at position -2. In addition, HIV-1 RNase H disfavored A at position -4. Precisely how the preferred nucleotides facilitate RNase H cleavage is unknown, but it has been proposed that the structures associated with some hybrid sequences might affect cleavage specificity (35). Several features influenced by sequence are the base composition, the trajectory of the helical axis, and the width of the major and minor grooves (35,47,48). While it is possible that the preferred nucleotides flanking a cleavage site reflect interactions in the DNA strand instead of or in addition to the RNA strand, the co-crystal structure of HIV-1 reverse transcriptase and a PPT-containing RNA/DNA hybrid reveals potential hydrogen-bonding contacts between the +1 RNA base and Arg448, and between the -2 RNA base and Gln475 (35) that might contribute to the observed sequence preferences. Our observation that an A is disfavored at the -4 position does not correspond to any base contacts in the co-crystal structure, but there are phosphate contacts from -4 to -9 in the DNA strand and the RNase H primer grip region of the HIV-1 enzyme (35). Notably, the -4 position in the co-crystal structure falls within an unusual structural deformation consisting of a region of unpaired bases that may be PPT specific and consequently may not reflect the binding situation for hybrids containing other sequences. Importantly, our data suggest that hybrid substrates containing the preferred nucleotides at the most critical positions of -2 and +1 may facilitate the generation of co-crystals of reverse transcriptase with bound substrate in future crystallographic analyses.
Very recently, the first co-crystal structure of a prokaryotic RNase H with the hybrid substrate positioned in the enzyme active site has been reported (49). The authors note that four residues (Asn77, Asn106, Gln134, Asn105) donate hydrogen bonds to bases close to the scissile phosphate, and one residue (Arg195) has a sequence-specific contact with the DNA strand at the +5 G residue. This latter residue is homologous to Arg557 in HIV-1 reverse transcriptase, but thus far our sequence analyses of internal and 5′ end-directed cleavage sites have not suggested a preference for C in the RNA strand at the +5 position ((23); data not shown).
Both the 5′ end-directed and internal modes of RNase H cleavage prefer similar nucleotides at positions +1 and -2 for M-MuLV and positions +1, -2, and -4 for HIV-1 (this work; (23)). Internal cleavages also exhibit nucleotide preferences further upstream from the cleavage site (at positions -6 and -11 for M-MuLV and positions -7, -12, and -14 for HIV-1; (23)), but no equivalent positions were identified from the statistical analyses of 5′ end-directed cleavage sites. The absence of obvious nucleotide preferences further upstream of 5′ end-directed cleavage sites may derive from the nature of the upstream sequences found in many of our hybrid substrates, where the first 8 to 10 nucleotides of RNA sequence are dictated by the in vitro transcription vectors. Although these sequences may not offer the optimal nucleotides that would otherwise be preferred for 5′ end-directed cleavage, the specificity determinant contributed by binding the RNA 5′ end may substitute or compensate for additional upstream preferences. This consideration suggests that preferred nucleotide positions located most proximal to the cleavage site are the most important in determining the positioning for both modes of RNase H cleavage.
As would be predicted from the overlap between the preferred nucleotide specificity of 5′ end-directed and internal cleavage sites, it appears that the 5′ end-directed sites are a subset of the possible internal sites that are only cleaved when they meet certain distance constraints. In support of this prediction, all of the 5′ end-directed cleavage sites we have observed thus far are also observed as internal cleavage sites on longer RNA/DNA hybrids that do not require a nucleic acid end for positioning ((23) and data not shown). For example, the predominant 5′ end-directed sites in substrates Md1 through Md10 are also recognized as internal cleavage sites in longer RNAs containing the sequence +1 through +29 beyond the M-MuLV PPT origin (Figs. 1 and 2; (23)). As indicated in Fig. 11A, only some of the possible internal cleavage sites in the +1 to +29 sequence (indicated by arrows above the upper sequence) are recognized as 5′ end-directed sites in substrates Md1 and Md10 (indicated by vertical dashed lines). Importantly, the internal sites located close to the 5′ or 3′ ends of substrates Md1 or Md10 are not cleaved efficiently by the RNA 5′ end-directed mechanism because of the restrictions imposed by the distance from the RNA 5′ end.
Fig. 11.
“Cleavage Window” model for sequence and distance as determinants of 5′ end-directed cleavage. (A) The sequence of the M-MuLV region surrounding the -1/+1 site is shown, beginning at -20 and ending at +40, with the PPT underlined. In this sequence, the locations of the most prominent internal cleavage sites recognized by M-MuLV or HIV-1 reverse transcriptase (23) are indicated by arrows. Above each arrow is the 5′ and 3′ nucleotide positions bordering the cleavage site and the letter designation given to that site. Site C is an internal cleavage site recognized by HIV-1 RNase H that is not observed in 5′ end-directed cleavages, possibly because it is too close to the 5′ end. The sequences of Md1 and Md10 are aligned below with vertical dashed lines indicating the positions of 5′ end-directed cleavage sites. The “cleavage window” that defines eligible sites for 5′ end-directed cleavage is indicated by the shaded box. (B) A frequency distribution plot relative to specific positions that are used for 5′ end-directed cleavages for HIV-1 (black bars) or M-MuLV (grey bars), based upon the 46 sequences shown for HIV-1 in Fig. 5 and the 35 sequences shown for M-MuLV shown in Fig. 6.
The internal mode of RNase H cleavage primarily recognizes sites according to sequence (23). Our findings that a combination of an accessible RNA 5′ end, proper sequence context, and an appropriate distance from an RNA 5′ end are all determinants of RNA 5′ end-directed cleavages suggest a revision in the model for how retroviral RNases H carry out 5′ end-directed cleavages. In this model, 5′ end-directed cleavage sites are not chosen randomly at a measured distance from the RNA 5′ end. Instead, to be eligible for cleavage, a site must conform to the preferred nucleotide sequence and the site must fall between a minimum and maximum range from an accessible RNA 5′ end. We term this acceptable range the “cleavage window”. A bar graph depicting how frequently each position relative to the RNA 5′ end is utilized in the strong and medium cleavage sites in Figs. 5 and 6 reveals the breadth of the cleavage window (Fig. 11B). This graph indicates that 5′ end-directed sites can occur at all positions thwithin tha cleavage window between the 13th and 20th nucleotides from the RNA 5′ end. As illustrated in Fig. 11A, the observed pattern of cleavages in substrates Md1 and Md10 result from recognition of cleavage sites only positioned within this window.
Our quantitative data with substrates containing 5′ end-labeled RNAs (Fig. 4) also support the view that recognition of 5′ end-directed cleavage sites is optimal within a defined cleavage window. By defining that the extent of cleavage was significant if the cleavage product represented ≥5% of all bands, cleavages were optimal when the sites were located between the 13th and 18th nucleotides for HIV-1, and between the 13th and 17th nucleotides for M-MuLV in these substrates. Because these experiments used substrates with labeled RNA 5′ ends, these data were biased somewhat against the more distal cleavages in the cleavage window.
This cleavage window model accounts for the range of distances observed in RNA 5′ end-directed cleavages assays. While cleavage sites are often 15 to 19 nucleotides from the RNA 5′ end, (for example, see (25,27,29,50-53)), cleavages from 10 to 21 nucleotides from the RNA 5′ end have been reported in this study and others (29-32). These cleavage distances are broader than the 17 or 18 nucleotides that separate the polymerase and RNase H active sites in HIV-1 reverse transcriptase, based upon co-crystal structures of the enzyme with duplex DNA or RNA/DNA hybrids in which a DNA 3′ end fits in the polymerase active site (33-35). It is possible that contacts between the polymerase domain and a DNA 3′ primer terminus more tightly anchor the RNase H domain on the substrate while an RNA 5′ end allows more flexibility in positioning. This would predict that the cleavage window for DNA 3′ end-directed cleavages is smaller than that for RNA 5′ end-directed cleavages, and experiments are underway to test this possibility. It is also possible that the co-crystal structures reflect the active sites distance when the enzyme initially binds the substrate, and that, in the case of RNA 5′ end-directed cleavages, the enzyme can slide on the substrate after the initial contacts with the RNA 5′ end are released or adjusted.
Secondary RNase H cleavages have been proposed to occur independently of primary, 5′ end-directed cleavages and result from an initial binding and more extensive sliding of reverse transcriptase on its substrate (38-40). For M-MuLV, independent cleavages in this report were observed as close as between the 8th and 9th nucleotides from the RNA 5′ end (for example, site D in Md1). At this time, we cannot conclude whether these cleavages result from a secondary cleavage or a 5′ end-directed mechanism. It may be that the definition of 5′ end-directed versus secondary cleavages is difficult to distinguish at sites closer to the RNA 5′ end, and more experiments are required to determine the nature of these cleavages.
After minus-strand synthesis, extensive degradation of the RNA genome is required to facilitate plus-strand synthesis and strand transfers, and much of this general degradation appears to proceed by the polymerization-independent mode of RNase H activity (20,22,25,28). The remaining RNA template that requires further degradation will likely contain nicks generated during polymerization of the minus-strand. Since the RNA 5′ end at a nick does not efficiently promote 5′ end-directed RNase H cleavages, this form of RNase H activity would be restricted to initially act at gaps, but whether gaps of sufficient size are generated during minus-strand synthesis remains to be determined. Internal cleavages do not require positioning by an RNA or DNA end, so at least initially the polymerization-independent activity of RNase H can carry out internal cleavages. If recognition sites are located closely together and cleaved by the internal mode, this would create gaps that are sufficient for 5′ end-directed cleavages. Because 5′ end-directed cleavages are kinetically favored over internal cleavages (54), the multitude of possible internal cleavage sites combined with robust 5′ end-directed cleavages would together facilitate rapid and thorough degradation of the retroviral genome.
Acknowledgments
We thank all members of the Champoux laboratory for helpful discussions during the course of this work and expecially Heidrun Intherthal for critical review of the manuscript.
Footnotes
This work was supported by National Institutes of Health grant CA51605.
The abbreviations used are: HIV-1, human immunodeficiency virus type 1; M-MuLV, Moloney murine leukemia virus; PPT, polypurine tract; ribonuclease H, RNase H.
REFERENCES
- 1.Arts EJ, LeGrice SFJ. Prog. Nucleic Acid Res. Mol. Biol. 1998;58:339–393. doi: 10.1016/s0079-6603(08)60041-0. [DOI] [PubMed] [Google Scholar]
- 2.Coffin JM, Hughes SH, Varmus HE. Retroviruses. Cold Spring Harbor Laboratory Press; Plainview, N. Y.: 1997. [PubMed] [Google Scholar]
- 3.di Marzo Veronese F, Copeland TD, DeVico AL, Rahman R, Oroszlan S, Gallo RC, Sarngadharan MG. Science. 1986;231:1289–1291. doi: 10.1126/science.2418504. [DOI] [PubMed] [Google Scholar]
- 4.Telesnitsky A, Goff SP. Proc. Natl. Acad. Sci. U.S.A. 1993;90:1276–1280. doi: 10.1073/pnas.90.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Misra HS, Pandey PK, Pandey VN. J. Biol. Chem. 1998;273:9785–9789. doi: 10.1074/jbc.273.16.9785. [DOI] [PubMed] [Google Scholar]
- 6.Guo J, Wu W, Yuan ZY, Post K, Crouch RJ, Levin JG. Biochemistry. 1995;34:5018–5029. doi: 10.1021/bi00015a013. [DOI] [PubMed] [Google Scholar]
- 7.Champoux JJ. In: Reverse Transcriptase. Skalka AM, Goff SP, editors. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1993. pp. 103–118. [PubMed] [Google Scholar]
- 8.Rausch JW, Le Grice SF. Int J Biochem Cell Biol. 2004;36:1752–1766. doi: 10.1016/j.biocel.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 9.Fuentes GM, Rodriguez-Rodriguez L, Fay PJ, Bambara RA. J. Biol. Chem. 1995;270:28169–28176. doi: 10.1074/jbc.270.47.28169. [DOI] [PubMed] [Google Scholar]
- 10.Furfine ES, Reardon JE. Biochemistry. 1991;30:7041–7046. doi: 10.1021/bi00243a001. [DOI] [PubMed] [Google Scholar]
- 11.Huber HE, Richardson CC. J. Biol. Chem. 1990;265:10565–10573. [PubMed] [Google Scholar]
- 12.Pullen KA, Ishimoto LK, Champoux JJ. J Virol. 1992;66:367–373. doi: 10.1128/jvi.66.1.367-373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rattray AJ, Champoux JJ. J Virol. 1987;61:2843–2851. doi: 10.1128/jvi.61.9.2843-2851.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith JS, Roth MJ. J. Biol. Chem. 1992;267:15071–15079. [PubMed] [Google Scholar]
- 15.Schultz SJ, Whiting SH, Champoux JJ. J. Biol. Chem. 1995;270:24135–24145. doi: 10.1074/jbc.270.41.24135. [DOI] [PubMed] [Google Scholar]
- 16.Finston WI, Champoux JJ. J Virol. 1984;51:26–33. doi: 10.1128/jvi.51.1.26-33.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo GX, Sharmeen L, Taylor J. J Virol. 1990;64:592–597. doi: 10.1128/jvi.64.2.592-597.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pullen KA, Champoux JJ. J Virol. 1990;64:6274–6277. doi: 10.1128/jvi.64.12.6274-6277.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Powell MD, Levin JG. J Virol. 1996;70:5288–5296. doi: 10.1128/jvi.70.8.5288-5296.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeStefano JJ, Mallaber LM, Fay PJ, Bambara RA. Nucleic Acids Res. 1994;22:3793–3800. doi: 10.1093/nar/22.18.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuentes GM, Fay PJ, Bambara RA. Nucleic Acids Res. 1996;24:1719–1726. doi: 10.1093/nar/24.9.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelleher CD, Champoux JJ. J. Biol. Chem. 2000;275:13061–13070. doi: 10.1074/jbc.275.17.13061. [DOI] [PubMed] [Google Scholar]
- 23.Schultz SJ, Zhang M, Champoux JJ. J Mol Biol. 2004;344:635–652. doi: 10.1016/j.jmb.2004.09.081. [DOI] [PubMed] [Google Scholar]
- 24.Furfine ES, Reardon JE. J. Biol. Chem. 1991;266:406–412. [PubMed] [Google Scholar]
- 25.Gopalakrishnan V, Peliska JA, Benkovic SJ. Proc. Natl. Acad. Sci. U.S.A. 1992;89:10763–10767. doi: 10.1073/pnas.89.22.10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kati WM, Johnson KA, Jerva LF, Anderson KS. J. Biol. Chem. 1992;267:25988–25997. [PubMed] [Google Scholar]
- 27.Gotte M, Fackler S, Hermann T, Perola E, Cellai L, Gross HJ, Le Grice SF, Heumann H. EMBO J. 1995;14:833–841. doi: 10.1002/j.1460-2075.1995.tb07061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeStefano JJ, Buiser RG, Mallaber LM, Myers TW, Bambara RA, Fay PJ. J. Biol. Chem. 1991;266:7423–7431. [PubMed] [Google Scholar]
- 29.DeStefano JJ, Mallaber LM, Fay PJ, Bambara RA. Nucleic Acids Res. 1993;21:4330–4338. doi: 10.1093/nar/21.18.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu TB, Taylor J. J Virol. 1992;66:4271–4278. doi: 10.1128/jvi.66.7.4271-4278.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ben-Artzi H, Zeelon E, Amit B, Wortzel A, Gorecki M, Panet A. J. Biol. Chem. 1993;268:16465–16471. [PubMed] [Google Scholar]
- 32.DeStefano JJ. Nucleic Acids Res. 1995;23:3901–3908. doi: 10.1093/nar/23.19.3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang H, Chopra R, Verdine GL, Harrison SC. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 34.Jacobo-Molina A, Ding J, Nanni RG, Clark AD, Jr., Lu X, Tantillo C, Williams RL, Kamer G, Ferris AL, Clark P, Hizi A, Hughes SH, Arnold E. Proc. Natl. Acad. Sci. U.S.A. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarafianos SG, Das K, Tantillo C, Clark AD, Jr., Ding J, Whitcomb JM, Boyer PL, Hughes SH, Arnold E. EMBO J. 2001;20:1449–1461. doi: 10.1093/emboj/20.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gotte M, Maier G, Onori AM, Cellai L, Wainberg MA, Heumann H. J. Biol. Chem. 1999;274:11159–11169. doi: 10.1074/jbc.274.16.11159. [DOI] [PubMed] [Google Scholar]
- 37.Schultz SJ, Zhang M, Champoux JJ. J Virol. 2003;77:5275–5285. doi: 10.1128/JVI.77.9.5275-5285.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wisniewski M, Balakrishnan M, Palaniappan C, Fay PJ, Bambara RA. Proc. Natl. Acad. Sci. U.S.A. 2000;97:11978–11983. doi: 10.1073/pnas.210392297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wisniewski M, Balakrishnan M, Palaniappan C, Fay PJ, Bambara RA. J. Biol. Chem. 2000;275:37664–37671. doi: 10.1074/jbc.M007381200. [DOI] [PubMed] [Google Scholar]
- 40.Wisniewski M, Chen Y, Balakrishnan M, Palaniappan C, Roques BP, Fay PJ, Bambara RA. J. Biol. Chem. 2002;277:28400–28410. doi: 10.1074/jbc.M201645200. [DOI] [PubMed] [Google Scholar]
- 41.Palaniappan C, Wisniewski M, Jacques PS, Le Grice SF, Fay PJ, Bambara RA. J. Biol. Chem. 1997;272:11157–11164. doi: 10.1074/jbc.272.17.11157. [DOI] [PubMed] [Google Scholar]
- 42.Palaniappan C, Kim JK, Wisniewski M, Fay PJ, Bambara RA. J. Biol. Chem. 1998;273:3808–3816. doi: 10.1074/jbc.273.7.3808. [DOI] [PubMed] [Google Scholar]
- 43.Palaniappan C, Fuentes GM, Rodriguez-Rodriguez L, Fay PJ, Bambara RA. J. Biol. Chem. 1996;271:2063–2070. [PubMed] [Google Scholar]
- 44.Capranico G, Kohn KW, Pommier Y. Nucleic Acids Res. 1990;18:6611–6619. doi: 10.1093/nar/18.22.6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hang JQ, Rajendran S, Yang Y, Li Y, In PW, Overton H, Parkes KE, Cammack N, Martin JA, Klumpp K. Biochem Biophys Res Commun. 2004;317:321–329. doi: 10.1016/j.bbrc.2004.03.061. [DOI] [PubMed] [Google Scholar]
- 46.Gao HQ, Sarafianos SG, Arnold E, Hughes SH. J Virol. 2001;75:11874–11880. doi: 10.1128/JVI.75.23.11874-11880.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kopka ML, Lavelle L, Han GW, Ng HL, Dickerson RE. J Mol Biol. 2003;334:653–665. doi: 10.1016/j.jmb.2003.09.057. [DOI] [PubMed] [Google Scholar]
- 48.Han GW, Kopka ML, Langs D, Sawaya MR, Dickerson RE. Proc. Natl. Acad. Sci. U.S.A. 2003;100:9214–9219. doi: 10.1073/pnas.1533326100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 50.Schatz O, Mous J, Le Grice SF. EMBO J. 1990;9:1171–1176. doi: 10.1002/j.1460-2075.1990.tb08224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wohrl BM, Moelling K. Biochemistry. 1990;29:10141–10147. doi: 10.1021/bi00496a001. [DOI] [PubMed] [Google Scholar]
- 52.Gao G, Orlova M, Georgiadis MM, Hendrickson WA, Goff SP. Proc. Natl. Acad. Sci. U.S.A. 1997;94:407–411. doi: 10.1073/pnas.94.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao G, Goff SP. J Virol. 1998;72:5905–5911. doi: 10.1128/jvi.72.7.5905-5911.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schultz SJ, Zhang M, Kelleher CD, Champoux JJ. J. Biol. Chem. 1999;274:34547–34555. doi: 10.1074/jbc.274.49.34547. [DOI] [PubMed] [Google Scholar]
- 55.Shinnick TM, Lerner RA, Sutcliffe JG. Nature. 1981;293:543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]


