Abstract
A potentially powerful approach for in vivo gene delivery is to target retrovirus to specific cells through interactions between cell surface receptors and appropriately modified viral envelope proteins. Previously, relatively large (>100 residues) protein ligands to cell surface receptors have been inserted at or near the N terminus of retroviral envelope proteins. Although viral tropism could be altered, the chimeric envelope proteins lacked full activity, and coexpression of wild-type envelope was required for production of transducing virus. Here we analyze more than 40 derivatives of ecotropic Moloney murine leukemia virus (MLV) envelope, containing insertions of short RGD-containing peptides, which are ligands for integrin receptors. In many cases pseudotyped viruses containing only the chimeric envelope protein could transduce human cells. The precise location, size, and flanking sequences of the ligand affected transduction specificity and efficiency. We conclude that retroviral tropism can be rationally reengineered by insertion of short peptide ligands and without the need to coexpress wild-type envelope.
Retroviral vectors are attractive vehicles for gene delivery. The retrovirus envelope protein provides the basis for specificity and cell entry. The Moloney murine leukemia virus (MLV) ecotropic envelope binds to an amino acid transporter (1) expressed only in mouse cells and closely related species and enters the cell by a pH-dependent endocytotic mechanism (15). The host range is determined by regions of variable sequences (VRA and VRB) within the extracellular domain (SU) of envelope.
Previous studies have examined targeting of pseudotyped MLV to human cells by using several strategies including antibody-streptavidin complexes that bridge the virus to target cell receptors (7, 21), display of variable antibody fragments as antibody-envelope fusion proteins (4, 22, 24, 31), and insertion of target ligands into wild-type envelope (5, 11, 13, 16, 25, 26-28). Typically, these modifications were at the N terminus of envelope, and viruses bearing such modified envelope derivatives were unable to transduce cells. These relatively large N-terminal modifications are believed to prevent envelope from undergoing the conformational change required for fusion and cell entry (31). The lack of full activity of N-terminally modified envelope derivatives necessitates the coexpression of wild-type envelope for production of transducing virus.
Integrin receptors are heterodimers composed of α and β subunits that play essential roles in cell-cell and cell-extracellular matrix interactions. Integrin receptor ligands contain the tripeptide RGD. RGD peptides as short as six amino acids (GRGDSP) can bind to integrin receptors and inhibit binding of full-length adhesion proteins (3, 6, 9, 29). Here we show that short RGD peptide ligands inserted at multiple locations within MLV envelope can direct an MLV retrovirus vector to transduce human cells. In addition, we demonstrate that the length and particular position of the inserted ligand can affect viral tropism.
We constructed >40 chimeric envelope derivatives containing in-frame insertions of either a 13- or a 21-amino-acid RGD peptide (RGD13 or RGD21, respectively; Table 1). The core of the RGD13 ligand is a six-amino-acid peptide, GRGDSP, which represents an RGD consensus sequence. The core of the RGD21 ligand is a 14-amino-acid sequence, QGATFALRGDNPQG, derived from the mouse laminin protein (3). Both the RGD13 and RGD21 peptides were flanked by cysteine residues to constrain the sequence within a loop (3, 12, 29), as cyclization of RGD peptides has been shown to increase affinity for integrin receptors (20). In some cases, chimeric envelope derivatives with multiple ligands in tandem were also generated. Several of the chimeric envelope derivatives had deletions of envelope sequences, in addition to ligand insertions, as a result of multiple restriction enzyme cleavages. In all, 26 chimeric envelope derivatives containing the RGD13 ligand, 16 chimeric envelope derivatives containing the RGD21 ligand, and 5 chimeric envelope derivatives containing an RGE21 ligand, a control nonbinding peptide (3, 10, 12, 23), were constructed.
TABLE 1.
Description of RGD virusesa
| Envelope no. (sequence) | Location (aa)b | No. of insert(s) | Nucleotides deleted from envelope |
|---|---|---|---|
| RGD13 (CAAA-GRGDSP-TRC) | |||
| 1 | 1 | 1 | |
| 2 | 1 | 2 | |
| 3 | 1 | 4 | |
| 4 | 38 | 1 | |
| 5 | 38 | 3 | |
| 6 | 38 | 1 | 5990-6082 |
| 7 | 68 | 1 | |
| 8 | 68 | 2 | |
| 9 | 68 | 1 | 6082-6191 |
| 10 | 120 | 1 | |
| 11 | 120 | 2 | 6238-6281 |
| 12 | 120 | 3 | |
| 13 | 185 | 1 | |
| 14 | 230 | 1 | |
| 15 | 230 | 2 | |
| 16 | 235 | 1 | |
| 17 | 235 | 4 | |
| 18 | 310 | 1 | |
| 19 | 310 | 2 | |
| 20 | 321 | 1 | |
| 21 | 321 | 2 | |
| 22 | 382 | 1 | |
| 23 | 382 | 2 | |
| 24 | 382 | 3 | |
| 25 | 388 | 1 | |
| 26 | 388 | 2 | |
| RGD21 (CAAA- QGATFALRGDNPQG-TRC) | |||
| 1 | 1 | 1 | |
| 2 | 38 | 1 | |
| 3 | 38 | 1 | 5990-6082 |
| 4 | 68 | 1 | |
| 5 | 68 | 1 | 6082-6191 |
| 6 | 120 | 1 | |
| R | 120 | 1 | 6238-6281 |
| 8 | 185 | 1 | |
| 9 | 230 | 1 | |
| 10 | 235 | 1 | |
| 11 | 310 | 1 | |
| 12 | 321 | 1 | |
| 13 | 382 | 1 | |
| 14 | 388 | 1 | |
| 15 | 168 | 1, 1 | |
| 16 | 1230 | 1, 1 | |
| RGE21 (CAAA- QGATFALRGENPQG-TRC) | |||
| 1 | 1 | 1 | |
| 2 | 38 | 1 | 5990-6082 |
| 3 | 68 | 1 | |
| 4 | 68 | 1 | 6082-6191 |
| 5 | 230 | 1 |
The sequences of the RGD13, RGD21, and RGE21 ligands are shown. For the chimeric envelope derivatives RGD13 1 to 26, RGD21 1 to 16, and RGE21 1 to 5, the position of the ligand insertion, the number of inserts, and any additional modifications are indicated. For the construction of chimeric envelope derivatives, the extracellular domain (gp70) of ecotropic MLV envelope was linearized at random locations by partial digestion with blunt-end restriction endonucleases in the the presence of 50 to 400 ng of ethidium bromide per ml. The 13-amino-acid RGD sequence (CAAAGRGDSPTRC; RGD13) was derived by annealing two oligonucleotides, RGD13-A (TGCGCGGCCGCTGGCCGTGGCGATTCTCCCACGCGTTGT) and RGD13-B (ACAACGCGTGGGAGAATCGCCACGGCCAGCGGCCGCGCA). The annealed sequence was ligated into the linearized envelope plasmid, and subclones were screened for insert position and orientation. The resultant chimeric envelope derivatives were cloned into the envelope expression vector, pCEE (14). The RGD13 1 to 3 chimeric envelope derivatives were constructed by insertion of an NaeI linker at the C terminus of the signal sequence of wild-type envelope, and the annealed RGD oligonucleotides were cloned into the NaeI site. Chimeric envelope derivatives with the 21 amino acid RGD sequence (CAAAQGATFALRGDNPQGTRC; RGD21) were constructed by restriction endonuclease digestion of RGD13 envelopes with NotI and MluI and insertion of the RGD21 annealed oligonucleotides RGD21-A(GGCCGCTCAAGGCGCAACGTTCGCGCTCAGAGGCGATAATCCACAGGGGA) and RGD21-B (CGCGTCCCCTGTGGATTATCGCCTCTGAGCGCGAACGTTGCGCCTTGAGC). The RGD21 envelope derivatives were cloned into an expression plasmid that contained a zeocin selection marker (Invitrogen). RGE21 was constructed analogous to RGD21. Chimeric envelope derivatives expressing two RGD sequences, RGD21-15 and RGD21-16, were constructed by removal of the BstEII/ClaI fragment of RGD21-1 and insertion of the BstEII/ClaI region from RGD21-4 and RGD21-9, respectively.
aa, amino acid.
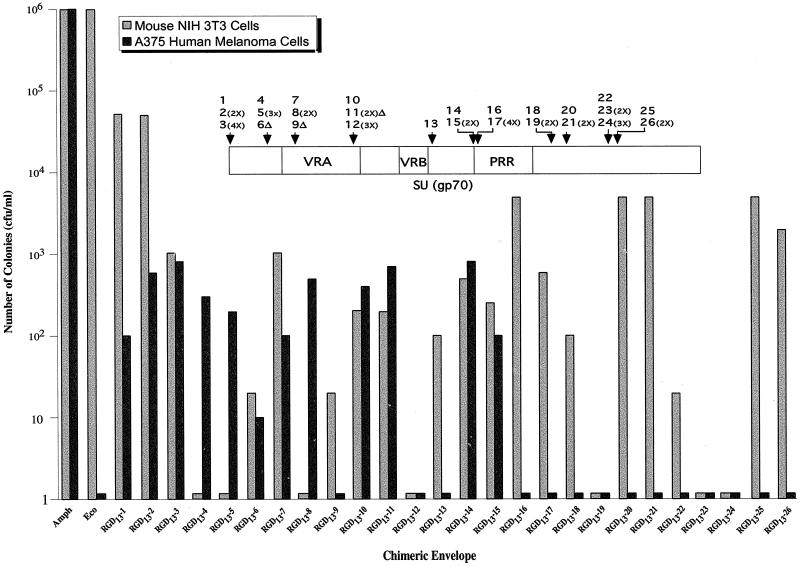
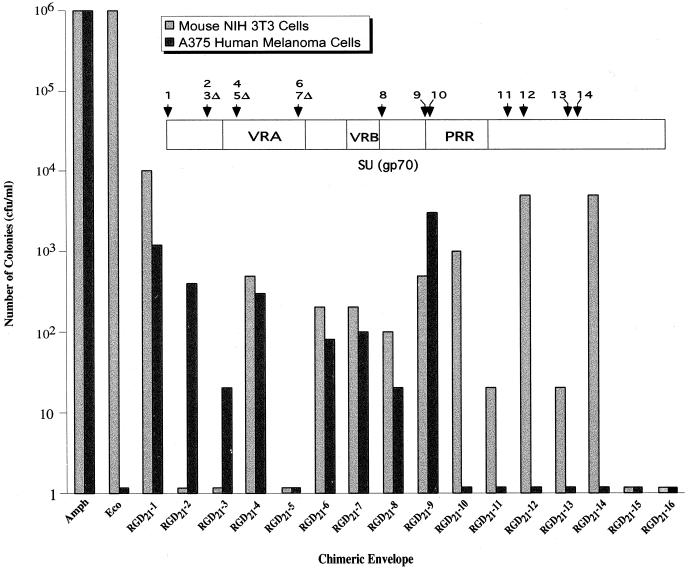
Pseudotyped virus containing chimeric envelope derivatives (RGD viruses) was generated by using a human 293T-cell-based packaging cell line, Anjou 65 (18). Immunoblotting of purified virions indicated that in all cases tested the chimeric envelope derivatives were incorporated into the virion and correctly processed (data not shown). The RGD viruses were initially tested for their ability to transduce mouse NIH 3T3 cells and, on the basis of these results (Fig. 1 and 2), we can draw several conclusions. First, many of the RGD viruses retained their ability to transduce mouse cells, but those bearing insertions within the N terminus (RGD13-4,5 and RGD21-2,3), the VRA (RGD13-8,12 and RGD21-5), and the C-terminal region (RGD13-19,23,34 and RGD21-15,16) did not. Several of these latter RGD viruses also failed to transduce human cells (RGD13-12,19,23,24 and RGD21-5,15,16), whereas for others (RGD13-4,5,8 and RGD21-2,3) the defect was mouse cell specific. Second, most RGD21 viruses transduced NIH 3T3 cells with efficiencies comparable to those of the equivalent RGD13 viruses, and none of the RGD21 viruses transduced NIH 3T3 cells with greater efficiency than the equivalent RGD13 virus.
FIG. 1.
Transduction of NIH 3T3 cells and A375 human melanoma cells by RGD13 viruses. NIH 3T3 and A375 human melanoma cells were infected with an RGD virus and then selected with G418 for 2 weeks, fixed, and stained with Giemsa, and the colonies were counted. The amphotropic virus, Amph, was generated by expressing the amphotropic envelope, pCAA, and the ecotropic virus, Eco, was generated by expressing the wild-type ecotropic envelope, pCEE. Note the log scale. The pCAA expression vector was generated by removing the amphotropic envelope gene from the full-length infectious clone(17) and insertion into pCEE after removal of the ecotropic envelope. The packaging construct, LAPNL, was generated by removal of the VSV-G envelope from LGRNL (30) and insertion of the secreted alkaline phosphatase gene (SEAP; Tropix). Pseudotyped virus producer cell lines were generated by cotransfection of Anjou 65 cells with LAPNL and a plasmid expressing a chimeric envelope derivative using Dotap (Boehringer), followed by selection in zeocin (200 μg/ml) for 2 weeks. RGD13 required cotranfection of a zeocin expression plasmid (Invitrogen).
FIG. 2.
Transduction of NIH 3T3 cells and A375 human melanoma cells by RGD21 viruses (as described for Fig. 1).
The RGD viruses were next tested for transduction of A375 human melanoma cells, which have been previously used to study integrin receptor binding (2, 8, 19). As expected, viruses bearing unmodified MLV envelope failed to transduce this human cell line. Significantly, however, many of the RGD viruses were able to transduce A375 human melanoma cells (Fig. 1 and 2). Transduction occurred when the RGD peptide was inserted at the N terminus (RGD13-1-3 and RGD21-1), within the N-terminal region (RGD13-4-6 and RGD21-2,3), within the VRA region (RGD13-7,8,10,11 and RGD21-4,6,7), and upstream of the PRR (RGD13-14,15 and RGD21-8,9). RGD viruses with insertions in the PRR and C-terminal region failed to transduce human cells. Several of the RGD viruses that transduced human cells failed to transduce NIH 3T3 cells (RGD13-4,5,8 and RGD21-2,3), indicating that viral tropism had been switched. In all cases tested, RGD viruses that transduced A375 human melanoma cells also transduced other human and nonhuman cell lines that contained integrin receptors (data not shown).
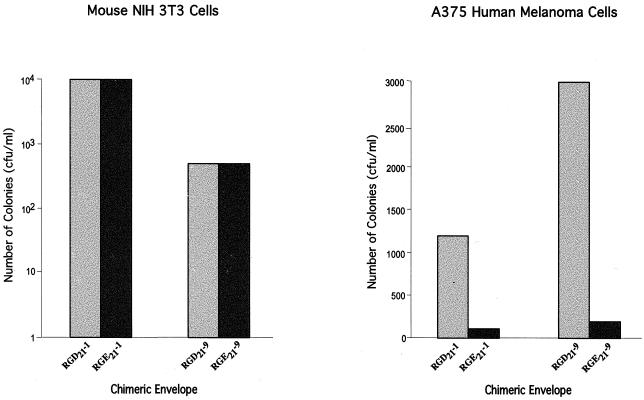
To examine the basis and specificity of human cell transduction, two experimental approaches were undertaken. First, the RGD21 ligand was replaced with the equivalent RGE21 sequence. Pseudotyped virus expressing an RGE21 chimeric envelope derivative transduced NIH 3T3 host cells with efficiencies comparable to the equivalent RGD21 derivative; in contrast, transduction of A375 human melanoma cells was significantly reduced (Fig. 3). Second, we analyzed the effect of antibodies to integrin receptors on the transduction of RGD viruses. NIH 3T3 and A375 human melanoma cells were pretreated with integrin receptor antibodies, and transduction was performed with two of the RGD21 viruses. Transduction of human but not mouse cells was substantially reduced (Fig. 4).
FIG. 3.
Requirement of the RGD sequence for transduction of human cells. NIH 3T3 (A) and A375 human melanoma (B) cells were infected with an RGD21 or RGE21 virus, and transduction was analyzed as described in the legend to Fig. 1.
FIG. 4.
Antibodies to integrin receptors block transduction of human cells. NIH 3T3 (A) and A375 human melanoma (B) cells were pretreated with polyclonal antibodies to β1, β3, and αν integrin receptors (Santa Cruz Biotechnology), infected with RGD21-1, RGD21-4, or RGD21-9 viruses, and transduction was analyzed as described in the legend to Fig. 1. The three antibodies were diluted 1:100 in Dulbecco modified Eagle medium and incubated with cells for 4 h. Cells were then incubated with pseudotyped virus for 6 h.
We have developed a strategy for altering the host range of ecotropic retrovirus vectors by using chimeric envelope derivatives bearing short peptide ligands. In many instances, pseudotyped virus expressing a chimeric envelope derivative transduced human cells without removal of the N-terminal region or coexpression of wild-type envelope. Ligand insertions at multiple locations within the N-terminal half of envelope enabled transduction of human cells. In contrast, transduction of human cells did not occur with RGD peptide insertions in the PRR or C-terminal region of envelope, although many of these viruses could transduce mouse cells. Some of the RGD viruses bearing insertions at the N terminus or VRA region (RGD13-4,5,8 and RGD21-2,3) transduced human but not mouse cells. Collectively, these observations indicate that the position of the inserted ligand can dictate tropism.
Transduction efficiencies differed among the RGD viruses, indicating that the precise location of the ligand within envelope is important and can be optimized. In general, RGD13 and RGD21 ligands transduced NIH 3T3 cells with comparable efficiencies, suggesting that envelope can accommodate ligands of different sizes, perhaps longer than those examined in this study. Longer ligands may be more disruptive but may also have increased affinity for the target receptor. The accompanying study confirms and extends these conclusions and demonstrates the generality of the experimental approach.
Acknowledgments
We thank A. J. Mackrell, D. Ott, and J. K. Yee for plasmids and W. S. Pear for the Anjou 65 cell line.
This work was supported in part by an NIH grant to M.R.G.M.R.G. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Albritton, L. M., L. Tseng, D. Scadden, and J. M. Cunningham. 1989. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell 57:659-666. [DOI] [PubMed] [Google Scholar]
- 2.Allman, R., P. Cowburn, and M. Mason. 2000. In vitro and in vivo effects of a cyclic peptide with affinity for the ανβ3 integrin in human melanoma cells. Eur. J. Cancer 36:410-422. [DOI] [PubMed] [Google Scholar]
- 3.Aumailley, M., M. Gerl, A. Sonnenberg, R. Deutzmann, and R. Timpl. 1990. Identification of the Arg-Gly-Asp sequence in laminin A chain as a latent cell-binding site being exposed in fragment P1. FEBS Lett. 262:82-86. [DOI] [PubMed] [Google Scholar]
- 4.Chu, T. H., and R. Dornburg. 1997. Toward highly efficient cell-type-specific gene transfer with retroviral vectors displaying single-chain antibodies. J. Virol. 71:720-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cosset, F. L., F. J. Morling, Y. Takeuchi, R. A. Weiss, M. K. Collins, and S. J. Russell. 1995. Retroviral retargeting by envelopes expressing an N-terminal binding domain. J. Virol. 69:6314-6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D'Souza, S. E., T. A. Haas, R. S. Piotrowicz, V. Byers-Ward, D. E. McGrath, H. R. Soule, C. Cierniewski, E. F. Plow, and J. Smith.W. 1994. Ligand and cation binding are dual functions of a discrete segment of the integrin beta 3 subunit: cation displacement is involved in ligand binding. Cell 79:659-667. [DOI] [PubMed] [Google Scholar]
- 7.Etienne-Julan, M., P. Roux, S. Carillo, P. Jeanteur, and M. Piechaczyk. 1992. The efficiency of cell targeting by recombinant retroviruses depends on the nature of the receptor and the composition of the artificial cell-virus linker. J. Gen. Virol. 73:3251-3255. [DOI] [PubMed] [Google Scholar]
- 8.Gehlsen, K. R., G. E Davis., and P. Sriramarao. 1992. Integrin expression in human melanoma cells with differing invasive and metastatic properties. Clin. Exp. Metastasis 10:111-120. [DOI] [PubMed] [Google Scholar]
- 9.Goodman, S. L., M. Aumailley, and H. von der Mark. 1991. Multiple cell surface receptors for the short arms of laminin: α1β1 integrin and RGD-dependent proteins mediate cell attachment only to domains III in murine tumor laminin. J. Cell Biol. 113:931-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenspoon, N., R. Hershkoviz, R. Alon, D. Varon, G. B. Shenkman, Marx, S. Federman, G. Kapustina, and O. Lider. 1993. Structural analysis of integrin recognition and the inhibition of integrin-mediated cell functions by novel nonpeptidic surrogates of the Arg-Gly-Asp sequence. Biochemistry 32:1001-1008. [DOI] [PubMed] [Google Scholar]
- 11.Han, X., N. Kasahara, and Y. W. Kan. 1995. Ligand-directed retroviral targeting of human breast cancer cells. Proc. Natl. Acad. Sci. USA 92:9747-9751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hart, S. L., A. M. Knight, R. P. Harbottle, A. Mistry, H. D. Hunger, D. F. Cutler, R. Williamson, and C. Coutelle. 1994. Cell binding and internalization by filamentous phage displaying a cyclic Arg-Gly-Asp-containing peptide. J. Biol. Chem. 269:12468-12474. [PubMed] [Google Scholar]
- 13.Kasahara, N., A. M. Dozy, and Y. W. Kan. 1994. Tissue-specific targeting of retroviral vectors through ligand-receptor interactions. Science 266:1373-1376. [DOI] [PubMed] [Google Scholar]
- 14.MacKrell, A. J., N. W. Soong, C. M. Curtis, and W. F. Anderson. 1996. Identification of a subdomain in the Moloney murine leukemia virus envelope protein involved in receptor binding. J. Virol. 70:1768-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McClure, M. O., M. A. Sommerfelt, M. Marsh, and R. A. Weiss. 1990. The pH independence of mammalian retrovirus infection. J. Gen. Virol. 71:767-773. [DOI] [PubMed] [Google Scholar]
- 16.Nilson, B. H., F. J. Morling, F. L. Cosset, and S. J. Russell. 1996. Targeting of retroviral vectors through protease-substrate interactions. Gene Ther. 3:280-286. [PubMed] [Google Scholar]
- 17.Ott, D., R. Friedrich, and A. Rein. 1990. Sequence analysis of amphotropic and 10A1 murine leukemia viruses: close relationship to mink cell focus-inducing viruses. J. Virol. 64:757-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfaff, M., M. Aumailley, U. Specks, J. Knolle, H. G. Zerwes, and R. Timpl. 1993. Integrin and Arg-Gly-Asp dependence of cell adhesion to the native and unfolded triple helix of collagen type VI. Exp. Cell Res. 206:167-176. [DOI] [PubMed] [Google Scholar]
- 20.Pierschbacher, M. D., and E. Ruoslahti. 1987. Influence of stereochemistry of the sequence Arg-Gly-Asp-Xaa on binding specificity in cell adhesion. J. Biol. Chem. 262:17294-17298. [PubMed] [Google Scholar]
- 21.Roux, P., P. Jeanteur, and M. Piechaczyk. 1989. A versatile and potentially general approach to the targeting of specific cell types by retroviruses: application to the infection of human cells by means of major histocompatibility complex class I and class II antigens by mouse ecotropic murine leukemia virus-derived viruses. Proc. Natl. Acad. Sci. USA 86:9079-9083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Russell, S. J., R. E. Hawkins, and G. Winter. 1993. Retroviral vectors displaying functional antibody fragments. Nucleic Acids Res. 21:1081-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Solowska, J., J. L. Guan, E. E. Marcantonio, J. E. Trevithick, C. A. Buck, and R. O. Hynes. 1989. Expression of normal and mutant avian integrin subunits in rodent cells. J. Cell Biol. 109:853-861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Somia, N. V., M. Zoppe, and I. M. Verma. 1995. Generation of targeted retroviral vectors by using single-chain variable fragment: an approach to in vivo gene delivery. Proc. Natl. Acad. Sci. USA 92:7570-7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valsesia-Wittmann, S., A. Drynda, G. Deleage, M. Aumailley, J. M. Heard, O. Danos, G. Verdier, and F. L. Cosset. 1994. Modifications in the binding domain of avian retrovirus envelope protein to redirect the host range of retroviral vectors. J. Virol. 68:4609-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valsesia-Wittmann, S., F. J. Morling, B. H. Nilson, Y. Takeuchi, S. J. Russell, and F. L. Cosset. 1996. Improvement of retroviral retargeting by using amino acid spacers between an additional binding domain and the N terminus of Moloney murine leukemia virus SU. J. Virol. 70:2059-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valsesia-Wittmann, S., F. J. Morling, T. Hatziioannou, S. J. Russell, and F. L. Cosset. 1997. Receptor co-operation in retrovirus entry: recruitment of an auxiliary entry mechanism after retargeted binding. EMBO J. 16:1214-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu, B. W., J. Lu, T. K. Gallaher, W. F. Anderson, and P. M. Cannon. 2000. Identification of regions in the Moloney murine leukemia virus SU protein that tolerate the insertion of an integrin-binding peptide. Virology 269:7-17. [DOI] [PubMed] [Google Scholar]
- 29.Yamada, T., M. Matsushima, K. Inaka, T. Ohkubo, A. Uyeda, T. Maeda, K. Titani, K. Sekiguchi, and M. Kikuchi. 1993. Structural and functional analyses of the Arg-Gly-Asp sequence introduced into human lysozyme. J. Biol. Chem. 268:10588-10592. [PubMed] [Google Scholar]
- 30.Yee, J. K., T. Friedmann, and J. C. Burns. 1994. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 43:99-112. [DOI] [PubMed] [Google Scholar]
- 31.Zhao, Y., L. Zhu, S. Lee, L. Li, E. Chang, N. W. Soong, D. Douer, and W. F. Anderson. 1999. Identification of the block in targeted retroviral-mediated gene transfer. Proc. Natl. Acad. Sci. USA 96:4005-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]