Abstract
Genetically identical cells and organisms exhibit remarkable diversity even when they have identical histories of environmental exposure. Noise, or variation, in the process of gene expression may contribute to this phenotypic variability. Recent studies suggest that this noise has multiple sources, including the stochastic or inherently random nature of the biochemical reactions of gene expression. In this review, we summarize noise terminology and comment on recent investigations into the sources, consequences, and control of noise in gene expression.
Any individual in a population of living organisms or cells is unique. Much of population variability is due to genetic differences, but environment and history also contribute to variability in cellular phenotype. Indeed, identical twin humans or cloned cats differ in appearance and behavior (Fig. 1). However, even cells or organisms with the same genes, in the same environment, with the same history, display variations in form and behavior that can be subtle or dramatic. Investigations have focused on the possibility that such variability is inevitable in biological systems because of the random nature of chemical reactions within a cell (1). When large numbers of molecules are present, chemical reactions may proceed in a predictable manner. However, when only a few molecules of a specific type exist in a cell, stochastic effects can become prominent.
Fig. 1.

Examples of possible stochastic influences on phenotype. (A) The fingerprints of identical twins are readily distinguished on close examination. Reprinted from (37) with permission from Elsevier. (B) Cc, the first cloned cat (left) and Rainbow, Cc's genetic mother (right), display different coat patterns and personalities (38). Photo credit, College of Veterinary Medicine and Biomedical Sciences, Texas A&M University.
Gene expression, as defined by the set of reactions that control the abundance of gene products, influences most aspects of cellular behavior, and its variation is often invoked to explain phenotypic differences in a population of cells. Because DNA, RNA, and proteins can be present and active at a few copies per cell, the abundance of gene products is theoretically sensitive to stochastic fluctuations. Four potential sources of variation in gene expression must be considered: (i) as described above, the inherent stochasticity of biochemical processes that are dependent on infrequent molecular events involving small numbers of molecules; (ii) variation in gene expression owing to differences in the internal states of a population of cells, either from predictable processes such as cell cycle progression or from a random process such as partitioning of mitochondria during cell division; (iii) subtle environmental differences, such as morphogen gradients in multicellular development; and (iv) ongoing genetic mutation, either random or directed. We use the term “noise” in gene expression to refer to the measured level of variation in gene expression among cells, regardless of source, within a supposedly identical population.
Measurement Techniques and Definitions
Recent investigations have employed green fluorescent protein (GFP) variants, which allow the quantification of protein levels in living cells by flow cytometry or fluorescence microscopy. The coefficient of variation, or noise η, is defined as the ratio of the standard deviation to the mean of the population. Other metrics of variability can be useful as well (SOM Text).
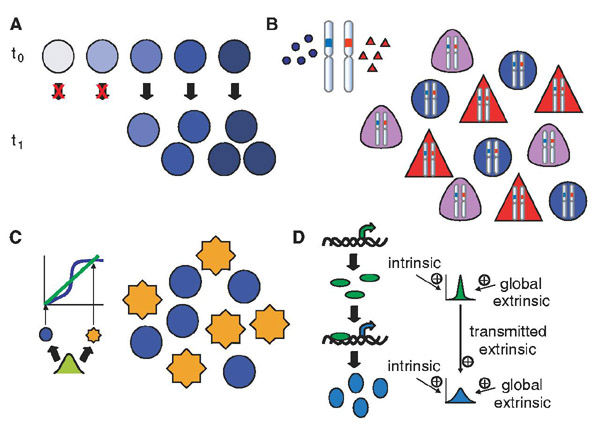
Once genetic mutation and local microenvironments are eliminated as sources of noise, an elegant experimental method can assist in differentiating among the remaining sources (2). This method involves quantifying expression of two equivalent, independent gene reporters placed in the same cell, which then allows noise sources to be partitioned into two categories: intrinsic, meaning noise sources that create differences between the two reporters within the same cell (Fig. 2A), and extrinsic, referring to sources that affect the two reporters equally in any given cell but create differences between two cells (Fig. 2B). Stochastic events during the process of gene expression, from the level of promoter-binding to mRNA translation to protein degradation, will manifest as intrinsic noise. Differences between cells, either in local environment or in the concentration or activity of any factor that affects gene expression, will result in extrinsic noise. Extrinsic noise should be further subdivided into two categories (3, 4): global noise, or fluctuations in the rates of the basic reactions that affect expression of all genes (Fig. 2C), and gene-or pathway-specific extrinsic noise (Fig. 2D), such as fluctuations in the abundance of a particular transcription factor or stochastic events in a specific signal transduction pathway. If a factor that causes extrinsic noise is experimentally manipulable, it is possible to eliminate such extrinsic noise by reduction of variability in that factor; for example, cell cycle synchronization will reduce extrinsic noise due to differences in cell cycle stage in a population (Fig. 2E).
Fig. 2.
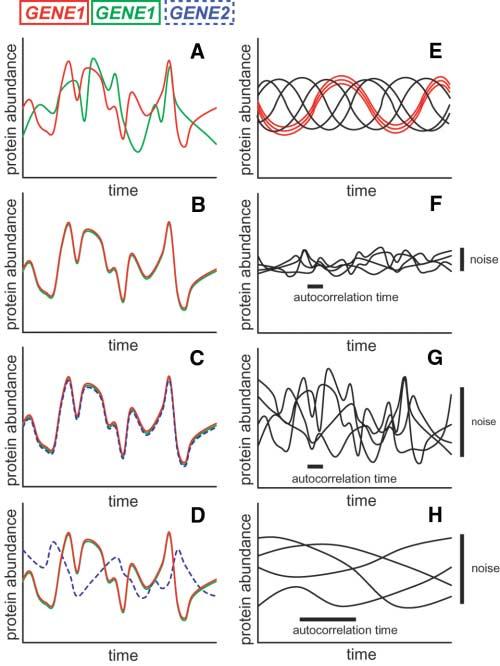
Noise definitions and characteristics. (A) Intrinsic noise results in differences between two reporters of the same gene in a single cell. (B) Extrinsic noise affects two reporters of the same gene equally in a single cell but causes differences from cell to cell or in a single cell over time. (C) Global noise affects two distinct genes equally but results in differences from cell to cell or in a single cell over time. (D) Gene- or pathway-specific extrinsic noise affects two reporters of the same gene equally but causes differences from a reporter of a second distinct gene in a single cell. (E to H) Noise in a population; each line represents a different cell. (E) Manipulable extrinsic noise: In a synchronized population of cells, cell cycle progression results in predictable changes in protein abundance over time (red lines); when the cells grow asynchronously, the population displays variability (black lines). (F) Noise of low magnitude and short autocorrelation time. (G) Noise of high magnitude and short autocorrelation time. (H) Noise of high magnitude and long autocorrelation time.
Experimental Investigation of Noise Sources
The division of noise into “extrinsic” and “intrinsic” categories has proven practical experimentally. Elowitz et al. pioneered the two-reporter method in studies of noise in gene expression in Escherichia coli, which quantified levels of cyan and yellow fluorescent proteins expressed from identical promoters on the same prokaryotic chromosome (5). These studies demonstrated that the stochastic nature of gene expression gives rise to noise in protein levels in a clonal population of E. coli and that the relative contributions of extrinsic and intrinsic components to the total noise vary with expression level. Ozbudak et al. also quantified noise in gene expression in the prokaryote Bacillus subtilis (6). By comparing the noise observed in reporters with altered efficiency of transcription and translation, they concluded that prokaryotic transcription is the dominant source of noise in protein levels, as predicted by basic models of stochastic gene expression (7-10).
We employed the two-reporter system to measure gene expression noise in cells of the diploid eukaryote Saccharomyces cerevisiae (3). These studies revealed that intrinsic noise in reporter protein levels is detectable and, for one gene, results from slow interconversion between inactive and active promoter states due to stochastic chromatin-remodeling events. However, extrinsic noise is the predominant form of noise for all gene promoters measured in these experiments. Simultaneous measurement of two independent, unrelated gene promoters indicated that much of this extrinsic noise is global in nature, presumably due to fluctuations in some factor that affects expression of all genes and not due to fluctuations in extrinsic factors that affect a particular gene. Blake et al. quantified noise in S. cerevisiae using a single-reporter method (11). Their results for the GAL1 gene are consistent with the extrinsic noise profile of GAL1 measured by the two-reporter method, which suggests that this noise is not the result of stochastic chromatin remodeling or transcription. Rather, most of the single-reporter noise is likely due to extrinsic factors such as global noise or noise in GAL signaling.
Recent measurements of gene expression in single E. coli cells over long time periods have provided insights into the relative amplitude and time scales of intrinsic and extrinsic noise (12). Extrinsic noise is the primary source of variability in gene expression, similar to the observation in budding yeast. The authors calculated autocorrelation times for noise, or the time scale over which the protein production rate fluctuates in any given cell (Fig. 2, F to H). The autocorrelation time for intrinsic noise is ≤10 min, consistent with the hypothesis that rapid fluctuations in mRNA numbers are the source of intrinsic noise. The autocorrelation time for global noise factors in protein production rate is ∼40 min, similar to the observed cell cycle length, which suggests that whatever factors result in global noise persist on average for about one cell cycle.
In Drosophila melanogaster, Ahmad and Henikoff studied the behavior of a GFP reporter subject to position-effect variegation (13). The reporter gene displays an expression pattern suggestive of repeated rounds of stochastic activation and inactivation of gene expression, resulting in patches of cells expressing the reporter. Fluctuations in the chromatin state of the reporter gene uncover transcription-factor binding sites, and recruitment of chromatin-modulating activities after transcription-factor binding slows the rate of heterochromatin reformation. In another eukaryotic system, each allele of the gene encoding the cytokine IL-4 is expressed in a probabilistic manner in response to signaling through the T cell receptor (TCR) in mouse T helper 2 lymphocytes (14, 15). The probability of expression for each allele is independent of its parental origin and increased with the strength of TCR stimulation, leading to biallelic expression at higher levels of stimulation. Furthermore, the pattern of allelic expression is different over multiple rounds of activation for some clonal populations, which suggests that the stochastic gene activation observed is a reversible process.
Other mammalian genes may show similar patterns of both monoallelic and biallelic ongoing stochastic expression. In a survey of allelic imbalances in gene expression in heterozygous human cells, 18% of the more than 120 genes assayed displayed consistent biases in expression patterns toward one allele (16). Such imbalanced expression may be due to slow, reversible stochastic fluctuations in gene expression, or it may be due to stochastic events in processes other than gene expression, nonrandom epigenetic factors, or polymorphism in regulatory sequences.
Consequences of Noise in Gene Expression
Both the magnitude and the frequency of the noise affect the consequence. Small changes in protein abundance may have dramatic effects on fitness if they persist long enough, whereas large fluctuations in abundance may not have any effect if they occur too frequently to affect a cellular process (12). The observation that the time scale for intrinsic noise fluctuations is much shorter than that for extrinsic noise suggests that extrinsic noise may affect cellular phenotypes more strongly than intrinsic noise, at least in E. coli (12).
Small differences in protein abundance may confer a fitness advantage or disadvantage (Fig. 3A). Intrinsic noise can produce fluctuations in the relative expression of two alleles of the same gene in a heterozygote, potentially resulting in cells that express no allele, either individual allele, or both alleles. If the two alleles are functionally divergent, the population of cells could acquire heterogeneity (Fig. 3B). Such fluctuations may contribute to the still-debated phenomenon of hybrid vigor. Alternatively, intrinsic noise in the case of haploinsufficiency may result in increased levels of noise or complete loss of function in a subset of cells. Such a mechanism has been proposed in the case of the human tumor suppressor gene NF1 (17) and prostate neoplasia formation in the mouse (18).
Fig. 3.
Consequences of noise. (A) Small differences in gene product abundance affect reproductive fitness. (B) In a heterozygous diploid population, cells display the phenotypes associated with each homozygote as well as the heterozygote. (C) Noise allows simultaneous achievement of multiple steady-state phenotypes in a population. (D) Noise can be transmitted from one gene, in this case a transcription factor, to a downstream target. The intrinsic and global extrinsic noise of the transcription factor can cause extrinsic noise in the downstream gene.
A brief period of intrinsic noise followed by feedback may allow for stochastic choice in stable monoallelic expression. This model has been proposed to explain the process of odorant receptor choice in olfactory neurons (19, 20). Each murine olfactory neuron expresses a single allele of one odorant receptor gene out of a choice of ∼1500 odorant receptor genes. A functional odorant receptor is required to prevent expression of other odorant receptors, which suggests that receptor choice occurs through stochastic activation of a single promoter followed by inhibitory signaling to the inactive odorant receptor promoters. The resulting heterogeneous population of olfactory neurons enables sensitive differentiation of odorant molecules; the stochastic nature of gene expression may create a functional sense of smell.
Many reports have been made of differentiation of a population of unicellular organisms into two distinct states of gene expression, for example, for the lysis-lysogeny decision in lambda phage-infected E. coli (21, 22) as well as the lac operon in E. coli (23). The stochastic factor that generates the two states of expression has not been experimentally confirmed in any such case, but noise in gene expression remains a plausible culprit (9, 10, 21, 24). Noise in gene expression in the context of positive feedback may be sufficient to create switching between the two stable states. The use of stochasticity to populate multiple steady states may play an important role in differentiation in multicellular organisms (Fig. 3C) or in survival in fluctuating environments for unicellular organisms.
Genes are organized into regulatory circuits where the expression of one gene can influence the expression of another. A consequence of this organization is that noise in the expression of one gene may propagate to affect noise in the expression of a downstream gene (Fig. 3D). Recent work in E. coli has demonstrated that a synthetic cascade of three transcription factors produces more noise in output than a linear cascade of two transcription factors or than one transcription factor alone (25). Such transmission of noise has been analyzed further in other recent work, also in E. coli, where the authors examined the sources of noise in a synthetic transcriptional cascade (4). Intrinsic noise in the expression of a transcription factor causes extrinsic noise in a downstream target gene. Additionally, global noise affecting expression of the transcription factor propagates to the downstream target. Because the factor acts to repress transcription, global fluctuations in the repressor counteract the effects of global fluctuations in the expression of the downstream gene, which suggests that global fluctuations in a transcriptional activator will exacerbate the noise in the target gene.
Control of Noise in Gene Expression
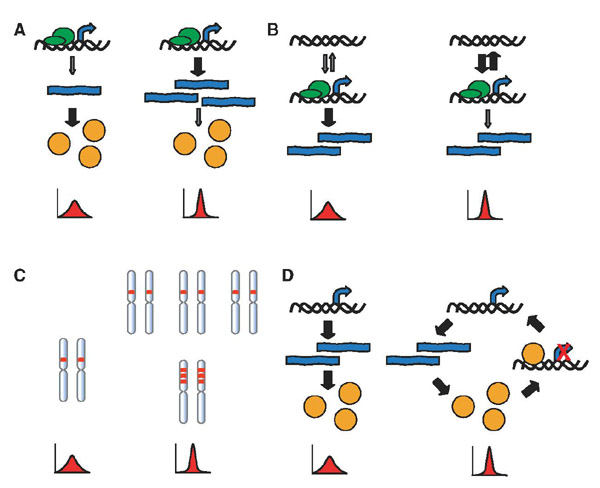
It is expected that control of noise in gene expression is under evolutionary pressure. Several models suggest how such control could generate or suppress the intrinsic noise of gene expression. A theoretical model (7-10), consistent with experimental evidence from B. subtilis (6), suggests that frequent transcription followed by inefficient translation results in lower intrinsic noise in protein levels than does infrequent transcription followed by efficient translation (Fig. 4A). Similarly, when promoter fluctuations contribute to intrinsic noise, as in S. cerevisiae, frequent promoter activation events followed by inefficient transcription will result in less noise in mRNA levels than infrequent promoter fluctuations followed by efficient transcription (Fig. 4B) (3, 11, 26). In these models, the control of noise comes at the energetic cost of producing few proteins from numerous mRNA or the cost of repeated rounds of promoter remodeling resulting in a few mRNA. It has been noted that key regulatory proteins in E. coli display low translation rates, which could lower noise in protein levels (6). Similarly, yeast genes that are essential or encode proteins involved in multi-subunit complexes tend to have higher rates of transcription and lower rates of translation (27).
Fig. 4.
Control of noise. (A) Infrequent transcription followed by efficient translation results in high intrinsic noise in protein levels (left); frequent transcription and inefficient translation results in low intrinsic noise (right). (B) Infrequent promoter transitions between inactive and active states followed by efficient transcription result in high intrinsic noise in mRNA levels (left); frequent promoter transitions followed by inefficient transcription result in low intrinsic noise (right). (C) Increases in gene copy number through polyploidy (top right) or gene duplication (bottom right) result in decreased intrinsic noise relative to a single gene copy (left). (D) Negative feedback, as when a transcription factor represses its own transcription (right), results in decreased noise relative to a linear pathway (left).
The control of gene copy number represents a second way to lower the intrinsic noise in gene expression (Fig. 4C), which is predicted to scale with the inverse square root of the gene copy number. Noise control can therefore be added to the reasons cited for the widespread presence of polyploidy. Also, noise control by increased copy number provides an evolutionary rationalization for exact gene duplications and the maintenance of identical copies of the same gene. For example, two copies of many ribosomal protein genes have been maintained in S. cerevisiae after an ancient genome duplication event (28).
Becskei and Serrano demonstrated the reduction of noise by means of negative feedback in a simple model in which a transcription factor negatively regulates its own synthesis (Fig. 4D) (29). More subtle forms of feedback may exist in which the rates of earlier gene expression steps are affected by later events. For example, the association of histone methylase activity with the elongating RNA polymerase complex during transcription results in the stable methylation of histones, which may affect the subsequent activation of the promoter (30).
Noise in gene expression may fundamentally limit the accuracy of cellular processes, such as the circadian oscillator. In a synthetic oscillator based on three transcription factors in E. coli, the transmission of noise may have resulted in the loss of coordination among cells (31). Mihalcescu et al. demonstrated in the unicellular cyanobacterium Synechococcus elongatus that circadian oscillations persist for weeks with a stable oscillatory period without extracellular entraining cues or intercellular communication (32), which suggests that the circadian network is strongly resistant to biochemical noise. Recent work has asserted that this oscillator relies on posttranslational molecular events (33). Perhaps a core posttranslational oscillator, which can rely on large numbers of molecules and avoid the small-number stochasticity of gene expression, is required for robust oscillations.
Cellular control mechanisms may exist to enable the switch between globally noisy or globally “quiet” states of gene expression. Queitsch et al. demonstrated that reduction of heat-shock protein 90 (Hsp90) chaperone activity in Arabidopsis thaliana increases morphological diversity in inbred lines, in addition to revealing otherwise silent genetic variation among different lines (34). Hsp90 chaperone activity is hypothesized to reduce the effect of stochastic molecular events that might otherwise result in developmental variability.
There may exist buffering agents that reduce either the magnitude of noise in gene expression or the impact of such noise on cellular or organismal phenotype. These buffering agents may be regulated, especially in times of stress, to produce a phenotypically diverse population. Waddington's theories of canalization and genetic assimilation propose that wasteful phenotypic variability in a population is suppressed when the population is well adapted to its environment (35). However, if environmental conditions shift, phenotypic noise becomes advantageous because a noisy population will produce some members that are better adapted to the new environment. Recent work supports the idea that it is advantageous to increase variability in times of stress and decrease variability when organisms are well adapted to the environment (36). Regulation of global noise factors could provide a molecular basis for such evolutionary flexibility.
Concluding Remarks
Many questions remain concerning the generation of noise in gene expression and its consequences for cellular behavior. The presence of stochasticity in gene expression has been confirmed to result in noise in protein abundance, but other sources of noise may result in phenotypic variability. Beyond the identification of true examples of phenotypic consequence, much work must be done to understand how cellular processes behave robustly in the presence of underlying stochasticity. Such work often requires a nontraditional collaboration between mathematicians, physicists, and in vivo experimentalists. Many biologists are beginning to focus on the limitations and benefits that stochasticity creates for biological systems, and we expect that future investigations will reveal results both unexpected and unpredictable.
Footnotes
Supporting Online Material www.sciencemag.org/cgi/content/full/309/5743/2010/DC1 SOM Text References
References and Notes
- 1.van Kampen NG. Stochastic Processes in Physics and Chemistry. North-Holland, Amsterdam: 1992. [Google Scholar]
- 2.Swain PS, Elowitz MB, Siggia ED. Proc. Natl. Acad. Sci. U.S.A. 2002;99:12795. doi: 10.1073/pnas.162041399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raser JM, O'Shea EK. Science. 2004;304:1811. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pedraza JM, van Oudenaarden A. Science. 2005;307:1965. doi: 10.1126/science.1109090. [DOI] [PubMed] [Google Scholar]
- 5.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Science. 2002;297:1183. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 6.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Nat. Genet. 2002;31:69. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- 7.McAdams HH, Arkin A. Proc. Natl. Acad. Sci. U.S.A. 1997;94:814. doi: 10.1073/pnas.94.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kierzek AM, Zaim J, Zielenkiewicz P. J. Biol. Chem. 2001;276:8165. doi: 10.1074/jbc.M006264200. [DOI] [PubMed] [Google Scholar]
- 9.Kepler TB, Elston TC. Biophys. J. 2001;81:3116. doi: 10.1016/S0006-3495(01)75949-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thattai M, van Oudenaarden A. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8614. doi: 10.1073/pnas.151588598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blake WJ, Kaern M, Cantor CR, Collins JJ. Nature. 2003;422:633. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- 12.Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Science. 2005;307:1962. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- 13.Ahmad K, Henikoff S. Cell. 2001;104:839. doi: 10.1016/s0092-8674(01)00281-1. [DOI] [PubMed] [Google Scholar]
- 14.Bix M, Locksley RM. Science. 1998;281:1352. doi: 10.1126/science.281.5381.1352. [DOI] [PubMed] [Google Scholar]
- 15.Riviere I, Sunshine MJ, Littman DR. Immunity. 1998;9:217. doi: 10.1016/s1074-7613(00)80604-4. [DOI] [PubMed] [Google Scholar]
- 16.Pastinen T, et al. Physiol. Genomics. 2004;16:184. doi: 10.1152/physiolgenomics.00163.2003. [DOI] [PubMed] [Google Scholar]
- 17.Kemkemer R, Schrank S, Vogel W, Gruler H, Kaufmann D. Proc. Natl. Acad. Sci. U.S.A. 2002;99:13783. doi: 10.1073/pnas.212386999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magee JA, Abdulkadir SA, Milbrandt J. Cancer Cell. 2003;3:273. doi: 10.1016/s1535-6108(03)00047-3. [DOI] [PubMed] [Google Scholar]
- 19.Chess A, Simon I, Cedar H, Axel R. Cell. 1994;78:823. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- 20.Serizawa S, Miyamichi K, Sakano H. Trends Genet. 2004;20:648. doi: 10.1016/j.tig.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 21.Arkin A, Ross J, McAdams HH. Genetics. 1998;149:1633. doi: 10.1093/genetics/149.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isaacs FJ, Hasty J, Cantor CR, Collins JJ. Proc. Natl. Acad. Sci. U.S.A. 2003;100:7714. doi: 10.1073/pnas.1332628100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ozbudak EM, Thattai M, Lim HN, Shraiman BI, van Oudenaarden A. Nature. 2004;427:737. doi: 10.1038/nature02298. [DOI] [PubMed] [Google Scholar]
- 24.Hasty J, Pradines J, Dolnik M, Collins JJ. Proc. Natl. Acad. Sci. U.S.A. 2000;97:2075. doi: 10.1073/pnas.040411297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hooshangi S, Thiberge S, Weiss R. Proc. Natl. Acad. Sci. U.S.A. 2005;102:3581. doi: 10.1073/pnas.0408507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ko MS. J. Theor. Biol. 1991;153:181. doi: 10.1016/s0022-5193(05)80421-7. [DOI] [PubMed] [Google Scholar]
- 27.Fraser HB, Hirsh AE, Giaever G, Kumm J, Eisen MB. PLoS Biol. 2004;2:e137. doi: 10.1371/journal.pbio.0020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kellis M, Birren BW, Lander ES. Nature. 2004;428:617. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 29.Becskei A, Serrano L. Nature. 2000;405:590. doi: 10.1038/35014651. [DOI] [PubMed] [Google Scholar]
- 30.Ng HH, Robert F, Young RA, Struhl K. Mol. Cell. 2003;11:709. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 31.Elowitz MB, Leibler S. Nature. 2000;403:335. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 32.Mihalcescu I, Hsing W, Leibler S. Nature. 2004;430:81. doi: 10.1038/nature02533. [DOI] [PubMed] [Google Scholar]
- 33.Tomita J, Nakajima M, Kondo T, Iwasaki H. Science. 2005;307:251. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- 34.Queitsch C, Sangster TA, Lindquist S. Nature. 2002;417:618. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- 35.Waddington CH. Nature. 1959;183:1654. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- 36.Pigliucci M, Murren CJ. Evolution Int. J. Org. Evolution. 2003;57:1455. doi: 10.1111/j.0014-3820.2003.tb00354.x. [DOI] [PubMed] [Google Scholar]
- 37.Jain AK, et al. Pattern Recognition. 2002;35:2653. [Google Scholar]
- 38.Shin T, et al. Nature. 2002;415:859. doi: 10.1038/nature723. [DOI] [PubMed] [Google Scholar]
- 39.We regret not acknowledging numerous contributors to this topic. We thank A. Arkin, M. Elowitz, and A. van Oudenaarden for critical commentary. Supported by the Howard Hughes Medical Institute, NIH grant GM51377, the David and Lucile Packard Foundation (E.K.O.), the Burroughs Wellcome/UCSF Interfaces of Science Fellowship program, and the UCSF Medical Scientist Training Program (J.M.R.). E.K.O. is a member of the science board and occasional consultant for Chiron Corporation