Abstract
We have developed an analysis toolbox called NUTMEG (Neurodynamic Utility Toolbox for Magnetoencephalography) for reconstructing the spatiotemporal dynamics of neural activations and overlaying them onto structural MR images. The toolbox runs under MATLAB in conjunction with SPM2 and can be used with the Linux/UNIX, Mac OS X, and even Windows platforms. Currently, evoked magnetic field data from 4-D Neuroimaging, CTF, and KIT systems can be imported to the toolbox for analysis. NUTMEG uses an eigenspace vector beamforming algorithm to generate a tomographic reconstruction of spatiotemporal magnetic source activity over selected time intervals and spatial regions. The MEG coordinate frame is coregistered with an anatomical MR image using fiducial locations and, optionally, headshape information. This allows the reconstruction to be superimposed onto an MRI to provide a convenient visual correspondence to neuroanatomy. Navigating through the MR volume automatically updates the displayed time series of activation for the selected voxel. Animations can also be generated to view the evolution of neural activity over time. Since NUTMEG displays activations using SPM2's engine, certain SPM functions such as brain rendering and spatial normalization may be applied as well. Finally, as a MATLAB package, the end user can easily add customized functions. Source code is available at http://bil.ucsf.edu/ and distributed under a BSD-style license.
Keywords: Beamformer, Magnetoencephalography, MEG, Inverse method, Functional neuroimaging
INTRODUCTION
Magnetoencephalography (MEG) and electroencephalography (EEG) noninvasively measure the magnetic and electric fields generated by neuronal currents, respectively, and can detect such activity with fine temporal resolution. The potential to localize the sources of these signals more accurately has emerged with the advent of dense MEG and EEG sensor arrays. Traditional multiple-dipole methods assume that a small set of current dipoles can adequately represent the distribution of an unknown source. While these methods are ideal for focal sources when the number of sources is known a priori or can be deduced from the spatiotemporal distribution of the data, problems arise if the sources have a large spatial extent or are otherwise not well characterized by current dipoles [Jeffs, 1987]. This is often the case, for example, in cognitive studies or studies involving sensorimotor integration. In addition, finding the dipole model which best accounts for the actual distribution of sources may be difficult because of problems in estimating the correct model order and in finding the optimal solution of the nonlinear optimization problem associated with multiple-dipole source models.
Studies of the inverse problem have traditionally focused on improving the spatial resolution of MEG and EEG, with the accepted limit being on the order of 2-5 mm [Leahy, 1998]. However, a second problem in MEG/EEG involves estimating the time course of source activations. A spatial filtering technique called beamforming provides an efficient solution, without requiring a priori assumptions about the location or number of neural sources. Using such a class of algorithms, MEG source reconstructions can be viewed as a “virtual depth electrode” measurement technique. We have developed the Neurodynamic Utility Toolbox for Magnetoencephalography (NUTMEG) to implement a data-dependent spatial filter for MEG spatiotemporal source reconstructions and to coregister the results of these reconstructions onto anatomical MRI volumes. NUTMEG provides an easy-to-use software suite to generate and visualize neural source reconstructions from MEG sensor arrays.
METHODS & RESULTS
NUTMEG implements an adaptive spatial filter called an eigenspace vector beamformer [Sekihara, 2001]. We currently employ the spherically symmetric volume conductor model in our computations [Sarvas, 1987]. Lead field calculations with this forward model have been vectorized and, using a spatial resolution of 5 mm, can be generated within a few seconds even for large reconstruction volumes. The vector beamformer separates sources into three orthogonal directions (although the radial component has no contribution with a spherical head model). As with all beamformers, the sensor covariance plays an important role; the eigenspace modification separates this covariance matrix into signal and noise subspace components using singular value decomposition [Sekihara, 2001] (see Fig. 1).
Figure 1.
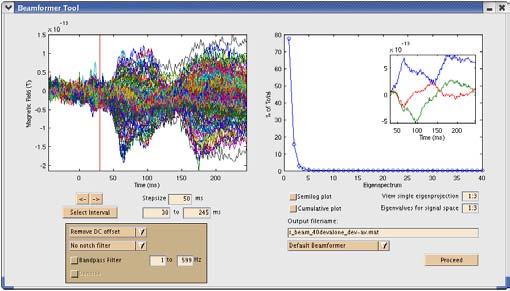
Interactive tool showing MEG sensor data (left) and singular values of the sensor covariance matrix (right) with associated eigenprojections (right, inset). The time interval and signal subspace desired for beamforming reconstruction can be selected here.
For visualization, the MEG coordinate system is coregistered onto an anatomical MRI using fiducial information to calculate an appropriate affine transformation matrix. Optionally, information obtained from a headshape digitizer may be used to further refine this transformation. The reconstruction volume can be selected by simply drawing outlines on the orthogonal cross-sections of the MRI (see Fig. 2). Final results can be navigated in both space and time through a graphical interface (see Fig. 3).
Figure 2.
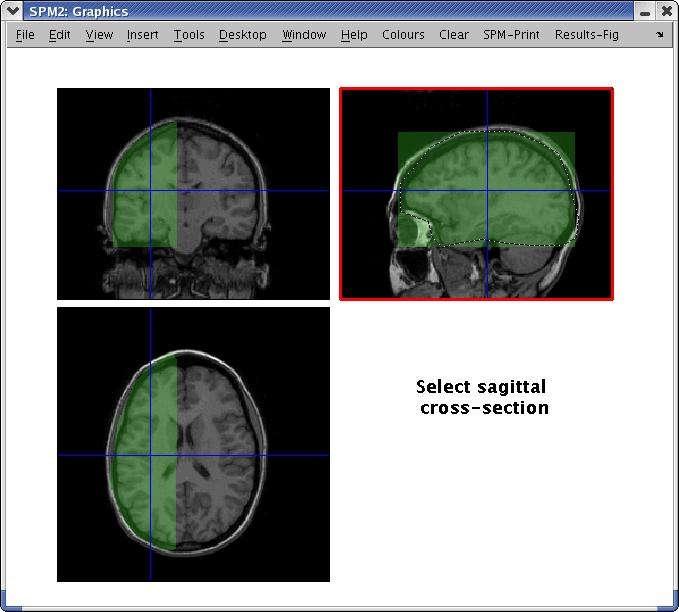
Interface for selecting volume of interest (VOI) for beamformer reconstruction. VOIs may be defined by drawing outlines on each of the three orthogonal cross-sections.
Figure 3.
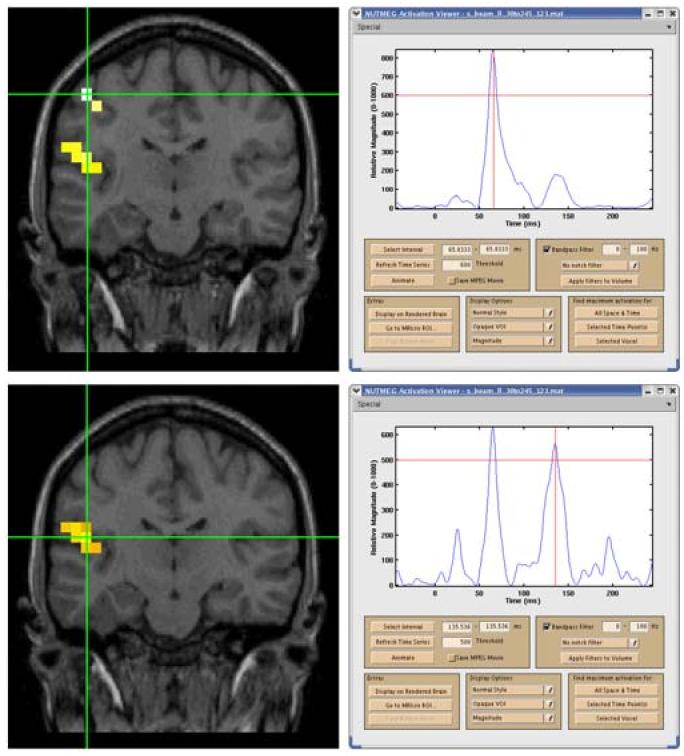
Beamformer reconstruction of evoked tactile stimulation of right D2 showing time series and localization for S1 at 66 ms (top) and S2 at 136 ms (bottom); note S2 also shows activity at 66 ms (slice has been tilted to include both S1 and S2). The peak at 20 ms is due to stimulus artifact.
NUTMEG is written for MATLAB (MathWorks, Natick, MA, USA), and requires SPM2 (http://www.fil.ion.ucl.ac.uk/spm). The interface with SPM2 allows activations to be overlaid onto standard orthogonal MRI slices or a rendered 3-D brain volume; at present, SPM2's analysis engine is not used. Activations may also be spatially normalized and displayed on an MNI template brain. All development and testing has been with dual 2GHz Pentium Xeon systems with 2GB of RAM running Red Hat Linux 9, though NUTMEG should be compatible with any platform running MATLAB 6.5 or 7. NUTMEG is open source and freely available (http://bil.ucsf.edu) for noncommercial use under a BSD-style license [Open Source Initiative, 2004]. Currently, MEG data may be readily imported from systems manufactured by CTF Systems, 4D Neuroimaging (BTi), and KIT/Yokogawa; shortly, it will also support data from Elekta Neuromag systems.
To demonstrate some of the features and capabilities of NUTMEG, we report results on a phantom and human study collected with a CTF Omega-275 system (Port Coquitlam, BC, Canada) at UCSF.
The phantom consisted of a spherical saline-filled volume conductor containing a current source set to oscillate at 7 Hz and 0.3 Vp-p, yielding a peak dipole moment of 17 nAm. The SNR was approximately 34 dB, calculated by dividing the signal subspace eigenvalue by the sum of the noise subspace eigenvalues. The theoretical location of the phantom dipole was (36,0,30) mm. Using a 2 mm reconstruction grid, NUTMEG localized the source in a voxel centered at (36.0,2.0,30.0) mm. Standard dipole source localization placed the source of the current phantom at (35.6,1.7,28.9) mm, which is located within the same voxel. These results are within the expected error range of 3 mm from the theoretical location.
The second data set was a somatosensory evoked field collected from a normal human adult subject; a piezoelectric device was used to stimulate a finger on the right hand (RD2) with a 25 ms 40 Hz vibration. NUTMEG localized two focal areas of activation; a peak at 66 ms showed a coactivation of areas corresponding to S1 and S2, while a broad peak near 136 ms activated primarily S2 (see Fig. 3).
CONCLUSION
We have developed a toolbox that uses an eigenspace vector beamformer to reconstruct the spatiotemporal dynamics of neural sources from MEG sensor arrays. This toolbox allows a user unfamiliar with the details of beamforming to reconstruct spatiotemporal activations from MEG sensor data. Results from this toolbox can facilitate comparisons of results with other functional neuroimaging modalities such as fMRI and PET.
Methods for assessing the statistical significance of beamformer activation maps are under investigation (e.g., [Sekihara, 2004]) and will soon be implemented. The vector beamformer currently used by NUTMEG is robust to weakly coherent sources and other forms of low-rank interferences [Sekihara, 2002] [Zumer, 2004]. However, all beamformers poorly resolve multiple strongly coherent sources. We are developing several methods to suppress activity from such interfering sources [Sahani, 2004] [Dalal, 2004]. Future directions for NUTMEG include incorporating support for more sophisticated head models as well as additional source localization methods.
ACKNOWLEDGEMENTS
We would like to acknowledge Alex Wade, Tom Holroyd, Fred Carver, and Darren Weber for significant contributions to source code as well as Dave McGonigle, Anne Findlay, and Tony Norcia for valuable feedback. Supported by NIH grants F31 DC006762 and R01 DC004855.
REFERENCES
- Dalal SS, Zumer JM, Sekihara K, Nagarajan SS. Modified beamformers for coherent source region suppression [abstract]. In: Halgren E, Ahlfors SP, Hamalainen MS, Cohen D, editors. Proceedings of the 14th International Conference on Biomagnetism; Boston: Biomag; Boston, USA. 2004 Aug 8-12.2004. p. 488. [Google Scholar]
- Jeffs B, Leahy R, Singh M. An evaluation of methods for neuromagnetic image reconstruction. IEEE Trans Biomed Eng. 1987;34:713–23. doi: 10.1109/tbme.1987.325996. [DOI] [PubMed] [Google Scholar]
- Leahy R, Mosher JC, Spencer ME, Huang MX, Lewine JD. A study of dipole localization accuracy for MEG and EEG using a human skull phantom. Electroencephalogr Clin Neurophysiol. 1998;107:159–73. doi: 10.1016/s0013-4694(98)00057-1. [DOI] [PubMed] [Google Scholar]
- The Open Source Initiative http://www.opensource.org/licenses/bsd-license.php. The BSD License. 2004 Available from: URL:
- Sahani M, Nagarajan SS. Reconstructing MEG sources with unknown correlations. In: Thrun S, Saul L, Scholkopf, editors. Advances in neural information processing systems 16. Cambridge: MIT Press; Cambridge, MA: 2004. in press. [Google Scholar]
- Sarvas J. Basic mathematical and electromagnetic concepts of the biomagnetic inverse problem. Phys Med Biol. 1987;32:11–22. doi: 10.1088/0031-9155/32/1/004. [DOI] [PubMed] [Google Scholar]
- Sekihara K, Nagarajan SS, Poeppel D, Marantz A, Miyashita Y. Reconstructing spatio-temporal activities of neural sources using an MEG vector beamformer technique. IEEE Trans Biomed Eng. 2001;48:760–71. doi: 10.1109/10.930901. [DOI] [PubMed] [Google Scholar]
- Sekihara K, Nagarajan SS, Poeppel D, Marantz A. Performance of an MEG adaptive-beamformer technique in the presence of correlated neural activties. IEEE Trans Biomed Eng. 2002;12:1534–46. doi: 10.1109/tbme.2002.805485. [DOI] [PubMed] [Google Scholar]
- Sekihara K, Sahani M, Nagarajan SS. Non-parametric statistical thresholding for MEG spatial-filter source reconstruction images. In: Halgren E, Ahlfors SP, Hamalainen MS, Cohen D, editors. Proceedings of the 14th International Conference on Biomagnetism; Boston: Biomag; Boston, USA. 2004 Aug 8-12.2004. pp. 595–6. [Google Scholar]
- Zumer JM, McGonigle DJ, Disbrow EA, Dalal SS, Zhu Z, Nagarajan SS. Magnetic source imaging of somatosensory frequency mismatch responses [abstract] Society for Neuroscience. 2004 Oct; Session #643.6. [Google Scholar]


