Abstract
The function of the transcriptional enhancer, Eμ3′, of the IgH locus of the channel catfish, Ictalurus punctatus, involves the interaction of E-protein and Oct family transcription factors. The E-proteins [class I basic helix–loop–helix (bHLH) family] are encoded in mammals by three genes: E2A (of which E12/E47 are alternatively spliced products), HEB, and E2-2. An E2A homologue has been identified in a catfish B-cell cDNA library and contains regions homologous to the bHLH and activation domains of mammalian and other vertebrate E2A proteins. E2A message is widely expressed, being readily detected in catfish B cells, T cells, kidney, spleen, brain, and muscle. Its expression is lower than that previously observed for TF12/CFEB, the catfish homologue of HEB. E2A strongly activated transcription of a μE5 motif-dependent construct in catfish B cells, and also activated transcription from the core region of the catfish IgH enhancer (Eμ3′) in a manner dependent on the presence of the μE5 site. Catfish E2A, expressed in vitro, bound the μE5 motif present in the core region of Eμ3′. These results document the conservation of structure and function in vertebrate E2A and suggest a potential role of E2A in driving expression of the IgH locus at the phylogenetic level of a teleost fish.
Keywords: Catfish, E2A, Transcription factor, Gene regulation, Immunoglobulin
Introduction
The class I family of basic helix–loop–helix (bHLH) transcription factors is widely expressed and plays key regulatory roles in a variety of developmental processes. The class I bHLH family of genes includes E2A (which encodes by alternative RNA processing two proteins, E12 and E47, Murre et al. 1989a), TF12/HEB, and E2-2. E12 and E47 are characterized by their broad tissue distribution, their involvement in many tissue-specific developmental processes, and their ability to interact with other classes of bHLH factors. The E-proteins form homo- or heterodimers with other E-proteins, but also have important interactions with the tissue-restricted class II bHLH proteins such as MyoD (Quong et al. 1999, 2002; Murre et al. 1989b) and with the inhibitory class V Id factors (Yokota 2001), which lack the DNA-binding domain and function as dominant-negative inhibitors of E-proteins (Nagata and Todokoro 1994). The activity of the class I E-proteins is regulated by two main mechanisms: first, the relative concentrations of E-proteins, class II bHLH factors, and the Id proteins, and second, covalent modification. For example, homodimers of E47 are unable to bind DNA when phosphorylated (Sloan et al. 1996), and disulfide bond formation is important in the stable homodimerization of E-proteins (Markus and Benezra 1999).
E2A transcription factors regulate the transcription of many B-lineage genes such as λ5, early B-cell factor, TdT, recombination-activating gene 1 (Bain et al. 1997; Schlissel et al. 1991; Choi et al. 1996; Kee and Murre 1998), and play major roles in controlling processes such as immunoglobulin (Ig) gene rearrangement (Romanow et al. 2000; Goebel et al. 2001) and the expression of activation-induced cytidine deaminase to mediate Ig class switch recombination and somatic hypermutation (Sayegh et al. 2003). E-proteins bind a series of E-box motifs of consensus sequence CANNTG, which were originally detected in Eμ, the IgH μ gene-associated enhancer (Ephrussi et al. 1985). Two of the E-protein binding motifs, μE2 and μE5, are important regulatory sites in many immune system genes (Perez-Mutul et al. 1988; Nelson et al. 1990). The picture that has emerged of the role of E2A-encoded proteins in the control of transcription is a complex one. E47 was first identified as a protein that binds Ig gene regulatory elements (Murre et al. 1989a). In B lymphocytes, the active DNA-binding complex consists of E47 homodimers (Sloan et al. 1996; Chu and Kohtz 2001), but in non-B cells, phosphorylated E47 is unable to bind DNA and is thus active as a heterodimer with tissue-restricted class II bHLH proteins, such as MyoD or NeuroD (Lassar et al. 1991; Lee et al. 1995). Although E12 can bind to DNA as heterodimers with tissue-specific bHLH proteins in non-B cells, E12 binds poorly as a homodimer due to the presence of an inhibitory domain (Sun and Baltimore 1991; Vitola et al. 1996). Formation of a stable intermolecular disulfide bond appears to be a prerequisite for the formation of functional E12 homodimers in B lymphocytes (Markus and Benezra 1999; Benezra 1994).
Although the vertebrate IgH locus shows strong conservation of the structural, antibody-encoding elements (Litman et al. 2004), it has undergone striking changes in the nature of its transcriptional control (Litman et al. 2004; Cioffi et al. 2001; Magor et al. 1994, 1999). Thus, although the transcriptional control region, Eμ3′, of the IgH locus of the channel catfish, Ictalurus punctatus, has strong B-cell-specific activity when tested in both mammals and fish, it differs from the mammalian Eμ enhancer in its location in the locus and in the nature of the transcription factors through which it drives expression (Cioffi et al. 2001). The mammalian Eμ enhancer contains a single copy of motifs that bind transcription factors of the Ets, Oct, E-protein, and bHLH-Zip families (Erman et al. 1998; Nikolajczyk et al. 1996, 1997; Rao et al. 1997). However, the “core” region of this enhancer consists of (in mouse) a μE3 site that binds TFE3 or (in humans) a core-binding factor site (Rao et al. 1997) that is flanked by two Ets factor-binding sites (μA and μB). In contrast, the core of the catfish Eμ3′ enhancer contains two variant (but highly functional) Oct motifs and a single consensus μE5 site (Cioffi et al. 2001). The function of the Eμ3′ enhancer has been shown to depend on the interaction of factors bound to the Oct and μE5 sites (Cioffi et al. 2001). The major isoforms of catfish Oct2 (octamer-binding transcription factor) have been cloned and their function characterized (Ross et al. 1998, 1999; Cioffi et al. 2002). However, by analogy with the situation in mammals, catfish can be assumed to possess a diverse array of E-proteins. The catfish homologue of TF12/HEB has recently been cloned and characterized (Hikima et al. 2004), but nothing is known of catfish homologues of other E-proteins. Whereas it is known that teleost fish (e.g., zebrafish and carp) (Wulbeck et al. 1994; Nihei et al. 1999) express messages with sequence homology to E12, there has been essentially no experimental investigation of the expression and function of E2A factors in fish. We report here the results of a study to identify E2A in catfish, define the expression of this transcription factor, and investigate its potential role in driving transcription of the IgH locus.
Materials and methods
Molecular cloning of catfish E2A homologues
Construction of a cDNA library from the catfish B-cell line, 1B10 (Miller et al. 1994) and its screening with a probe for the bHLH regions of catfish E-proteins was as previously described (Hikima et al. 2004). Full-length cDNA sequences of catfish E2A were obtained using 5′- and 3′-RACE (SMART RACE Kit, Clontech, Palo Alto, CA). Total RNA of the 1B10 cell line was isolated using Trizol (Invitrogen Life Technologies, San Diego, CA), reverse transcribed for cDNA, and the 5′ upstream region of E2A was amplified by 5′-RACE PCR using the following reverse primer: 5′-TTA CTG TTG TGG TCT GAA GGG TAG ATC G-3′ (G-2166, positions 1,503–1,530 in the sequence of catfish E2A, accession no. AY770493). The 5′-RACE PCR was conducted using the proof-reading Taq polymerase, Ex-Taq (Takara Bio Inc., Shiga, Japan). PCR cycling conditions were carried out with a denaturing step of 95°C for 3 min followed by 30 cycles of 94°C for 15 s and 68°C for 5 min, and 5 min at 72°C for the final extension. DNA sequencing was performed by the Biomolecular Resource Laboratory of the Medical University of South Carolina. Sequence analysis for homology, multiple-alignment, and secondary structure prediction was performed using the DNAStar (Madison, WI) or GENETIX-Mac version 10.3 (SDC Software Development, Tokyo, Japan) suites of programs.
Phylogenetic analysis
Inferred amino acid sequences of catfish E2A (accession no. AY770493), CFEB1 and 2 (AY528668 and AY528669), human E12 (AAA52331), human ITF1 (S10099), mouse E2A (AAH18260), mouse E47 (AAK18618), hamster E2A (P98180), rat E2A (P21677), Xenopus E12 (S23391), zebrafish E12 (I50518), human HEB (M80627), mouse ALF1A (C45020), mouse ALF1B (S19958), mouse TF12 (NP_035674), rat TF12 (NP_037308), chicken TF12 (P30985), human TF4 (NP_003190), mouse TF4 (NP_038713), mouse MITF2A (AAC52414), mouse SEF2 (CAA62868), and dog TF4, ITF2 (P15881) were aligned using the MegaAlign program (DNAStar) with PAM 250 residue weight table, gap penalty of 10, and gap-length penalty of 10. The alignment was used to generate most parsimonious phylogenetic trees (branch swapping, tree bisection reconnection, 1,000 bootstrap replicates) in the PAUP program version 4.0 beta (Swofford 2002). The Drosophila bHLH protein Da (NP_477189) was used as the outgroup.
S1 nuclease protection assay
S1 nuclease protection assay was carried out with total RNA from catfish head kidney, trunk kidney, spleen, brain, and muscle and from the B-cell (1B10) and T-cell (G14D) lines, as described in (Hikima et al. 2004). A 112-bp oligonucleotide (G-2188) that would detect catfish E2A had the following sequence: 5′-TGA AGC CCT CTG GTG CTC CTG CAG GCT GTG GGG TGC CCG AGG CGT GGG ACG TTG GTA ATA GGC TGC TGC TGC TGG GAG GGA GGC ACG CAG GGT CAT CAT GGT TGA CAA TGG A-3′. A 49-bp oligonucleotide probe for catfish β-actin (G-1034) had the following sequence: 5′-GGG TCA CAC CAT CAC CAG AGT CCA TCA CGA TAC CAG TGG GCA TCA ACT C-3′.
DNA constructs
The series of luciferase-expressing constructs containing (1) the minimal c-fos promoter (pGL3/Δ56), (2) the minimal c-fos promoter with the core region of the catfish enhancer (pGL3/Δ56/R♯2), (3) the minimal c-fos promoter with the core region of the catfish enhancer with the μE5 site mutated (pGL3/Δ56/R♯2-ΔμE5), or (4) the minimal c-fos promoter with a trimer of μE5 motifs (pGL3/Δ56/μE5×3) that was used in this study has previously been described (Hikima et al. 2004). The sequence of the μE5 motif used in pGL3/Δ56/μE5×3 (TGCAGGTGTG) is the native sequence of the consensus μE5 site in the Eμ3′ enhancer (Magor et al. 1994). The full-length coding region of catfish E2A was directionally cloned (HindIII and NotI) into the expression vector pRc/CMV (Invitrogen Life Technologies). The sequences to be cloned were PCR-amplified using the following primers containing the cloning site (underlined) and a Kozak consensus sequence in the sense primer (italics). The E2A sense primer was 5′-TTT AAGCTTGGC ACC ATG AAC GAT CAG CAG GGC CAC AGA ATG G-3′ (G-2325), and the E2A antisense primer was 5′-ATA AGA TTG CGG CCG CTC ATA TAT GCC CAA CAG AAC TGT GTC CA-3′ (G-2326). PCR was performed using Ex-Taq polymerase (Takara Bio Inc.) for 30 cycles with the following profile: 15 s at 94°C and 5 min at 68°C. Plasmids for transfection were purified using Nucleo-bond AX mega prep kit (Clontech) and dissolved in dH2O.
Cell lines, DNA transfection, and luciferase reporter assay
The catfish B lymphoblastoid cell line 1G8, T-cell line G14D, and the mouse plasmacytoma cell line J558L were used for transfections as described previously (Cioffi et al. 2001; Hikima et al. 2004). Equimolar amounts of construct were transfected. Each group contained the same amount of reporter, 2.44 pM [corresponding to 8 μg for the empty reporter vector pGL3/Δ56 (Hikima et al. 2004), 8.1 μg for pGL3/Δ56/μE5×3, and 8.19 μg for pGL3/Δ56/R♯2], the same amount of expression vector, 1.64 pM (corresponding to 6 μg for the empty expression vector pRc/CMV, 8.3 μg for pRc/CMV/CFEB1, and 8.17 μg for pRc/CMV/E2A), and the same amount of the Renilla luciferase construct pRL/CMV, 0.371 pM (1.0 μg). Optimal electroporation conditions for transfection were as previously described (Cioffi et al. 2001; Hikima et al. 2004). Transfected cells were harvested 36–40 h after electroporation and the luciferase activity was measured using the Dual luciferase reporter assay system (Promega) and a TD-20/20 luminometer (Turner Designs, Sunnyvale, CA). Normalization for transfection activity was performed using the activity of the Renilla luciferase, and values were calculated as mean±SD.
Antiserum production
In order to produce the peptide antiserum against catfish E2A, the peptide sequence, YPRDSAGYPGSKPGC, corresponding to amino acids 181–195 of catfish E2A, was synthesized and conjugated to KLH (Sigma-Genosys, The Woodlands, TX). Two rabbits (Sigma Genosys) were immunized with the synthesized peptide, and the IgG fraction of the antiserum was prepared by affinity chromatography on Protein A (Invitrogen Life Technologies). The antibody to CFEB has been described previously (Hikima et al. 2004).
Constructs for in vitro transcription and translation
For in vitro transcription and translation of epitope-tagged catfish E2A, the full-length coding sequence was cloned into the EcoRI–BglII sites (with S-tag) of the pCITE-4b(+) vector (Novagen, Cambridge, MA). The DNA fragment to be cloned was amplified using Ex-Taq polymerase (Takara Bio Inc.) by 30 cycles, with the following profile: 15 s at 94°C and 5 min at 68°C. For the pCITE-4b(+) vector with S-tag, the forward and reverse primers were as follows (restriction sites underlined): 5′-TGA ATT CGA TGA ACG ATC AGC AGG GCC ACA GAA TG-3′ (G-2376) and 5′-TTT AGA TCT TCA TAT ATG CCC AAC AGA ACT GTG TCC A-3′ (G-2377).
Electrophoretic mobility shift assay
The probe containing the μE5 consensus sequence (underlined) in the context of the native surrounding sequence in the Eμ3′ enhancer (Magor et al. 1994) was created by annealing the following oligonucleotides: forward (G-2616), 5′-TTC CTG TGC AGG TGT GTT TCA-3′ (21 mer), and reverse (G-2617), 5′-TGA AAC ACA CCT GCA CAG GA-3′ (20 mer). The 5′ overhang of the annealed probe was provided to permit labeling by a Klenow-catalyzed fill-in reaction. The corresponding probe with a scrambled μE5 site (underlined) was created by annealing the following oligonucleotides: forward (G-2618), 5′-TTC CTG ACG TGTGG GTT TTC A-3′, and reverse (G-2619), 5′-TGA AAA CCC ACA CGT CAG GA-3′. After annealing, the double-stranded DNA was purified by electrophoresis on 8% nondenaturing polyacrylamide gel (BioRad, Hercules, CA) followed by electroelution. The annealed probes were radiolabeled by fill-in with Klenow fragment (Fisher, Suwanee, GA) using [α-32P]dATP (New England Nuclear, Boston, MA) and purified using, sequentially, two Micro-spin columns, G-50 and G-25 (Amersham Pharmacia Biotech, Piscataway, NJ). In vitro synthesized E2A proteins were produced by transcription and translation using the TNT quick coupled transcription/translation systems (Promega). The electrophoretic mobility shift assay (EMSA) reaction mixtures containing a 3-μl aliquot of 5× gel shift binding buffer (Promega), 6 μl of TNT products (estimated amount of recombinant protein: 18–36 ng; see Anon 1993), and 2 μg of purified IgG in a total volume of 13–14 μl were incubated at room temperature for 15 min, and then 1 μl of 32P-labeled probe (105 cpm/μl, specific activity 5×104 cpm/ng), unlabeled competitor, or scrambled competitor (each 100 times the concentration of the labeled probe) were added. After 30-min incubation, DNA–protein complexes were analyzed on 4% nondenaturing polyacrylamide gels in 0.5× TBE buffer (45 mM Tris–HCl, 45 mM boric acid, 1 mM EDTA). Gels were dried, exposed to a phosphoimager screen, and analyzed using a Typhoon phosphoimager and the Imagequant program (Amersham Biosciences).
Western blot analysis of the synthesized proteins was performed as follows. After 50 μl of TNT proteins were purified with S-protein-agarose, purified proteins were fractionated on a 7% SDS-PAGE gel and electrotransferred to a nitrocellulose filter. After transfer, the filter was incubated for 2 h at room temperature in 5% skim milk. Primary antibody incubation was carried out at room temperature for 1 h in TBS-T (10 mM Tris–HCl, 150 mM NaCl, 0.05% Tween 20) containing 400 ng/ml anti-E2A-peptide IgG. After primary antibody binding, the filter was washed three times with TBS-T. Secondary antibody incubation was carried out at room temperature for 1 h in TBS-T containing anti-rabbit IgG conjugated to horseradish peroxidase (Bio-Rad) diluted 1:3,000. The filter was then washed four times with TBS-T, and proteins were detected using the ECL Western blotting detection reagent (Amersham Biosciences).
Results
Molecular cloning of an E2A homologue in catfish
The catfish B lymphoblastoid cell (1B10) cDNA library was screened with a probe for the bHLH domain of catfish E-proteins, and a total of 149 positive clones were identified (Hikima et al. 2004). Of these clones, only 2 were identifiable as putative homologues of E2A, as determined by DNA sequence analysis and PCR screening. The E2A clones were completely sequenced, and neither contained full-length open reading frames. The sequence of E2A mRNA was extended and completed by 5′-RACE and shown to be 4,574 bases in length, encoding 667 amino acid residues. The catfish sequence could easily be aligned to human E12 and showed regions homologous to the functional domains defined in E12. The most obvious difference between the catfish E2A and human E12 structures is the presence of a longer LH-AD2 domain in catfish (Fig. 1a). The sequence identities between catfish E2A and E12 of other vertebrates (human, mouse, chicken, and zebrafish) were striking, particularly in the bHLH and the putative activation domains (AD1 and HL-AD2, Fig. 1a). The zebrafish E12 was closest in overall sequence to the catfish E2A, although the percent identities of the AD1 and AD2 domains of catfish E2A were higher in comparisons with mouse or human E12 than with zebrafish E12 (Fig. 1a). Although other vertebrate E-protein structures also aligned well with the catfish E2A, the highest identities in the HLH domains were shown to E12 (Fig. 1a).
Fig. 1.
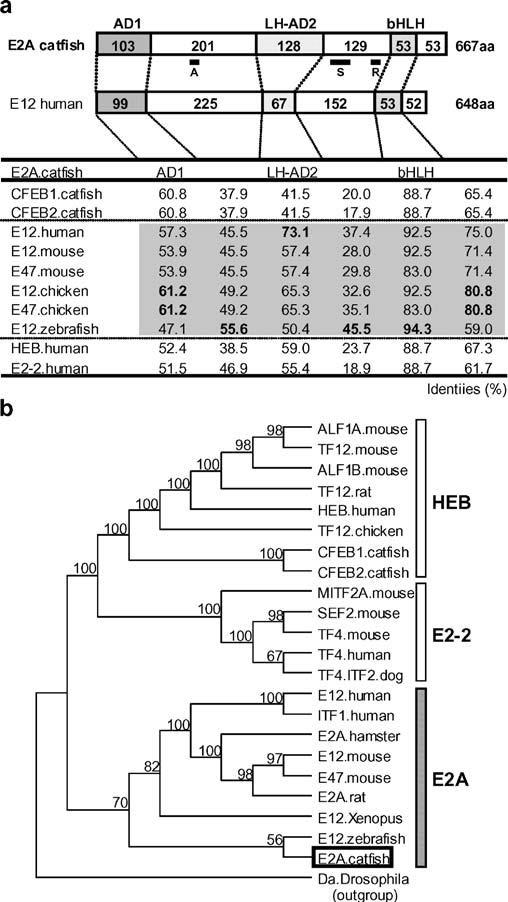
The structure and evolutionary relationships of catfish E2A. a Schematic depiction of the domain structure of catfish E2A in comparison with human E12, shown above a table of percent identities. The lengths (amino acid sequence) of the activation domain 1 (AD1), the loop-helix activation domain 2 (LH-AD2), the basic helix–loop–helix (bHLH) domain, and the intervening regions are shown. The table lists the percent identities of the inferred amino acid sequences of the domains (AD1, LH-AD2, bHLH, and their intervening regions) of the catfish E2A as compared with those published for other species. Top, A, S, and R indicate the position of the peptide sequence used for antibody production, the probe sequence used in the S1 nuclease protection assay, and the Rep domain, respectively. b Phylogenetic tree of vertebrate E-proteins. The class I bHLH daughterless protein (Da) of Drosophila was used as the outgroup. Catfish E2A, is indicated by an open box, and bootstrap values (>50%) in support of each node are shown
A more precise analysis of phylogenetic relationships, using parsimony-based methods (Fig. 1b), confirmed that separate HEB, E2-2, and E2A lineages can clearly be identified within vertebrate E-proteins (Hikima et al. 2004). The catfish E2A was firmly assigned to the E2A branch and showed the closest relationship with zebrafish E12.
Expression of E2A message in catfish tissues and cell lines
S1 nuclease protection assays were used to identify the expression patterns of E2A in catfish cell lines and tissues. The results indicated that E2A message was widely expressed, being readily detected in catfish B cells, T cells, kidney, spleen, brain, and muscle (Fig. 2). The expression of E2A was higher in T cells, brain, and muscle, than in B cells, head kidney, trunk kidney, and spleen (Fig. 2).
Fig. 2.
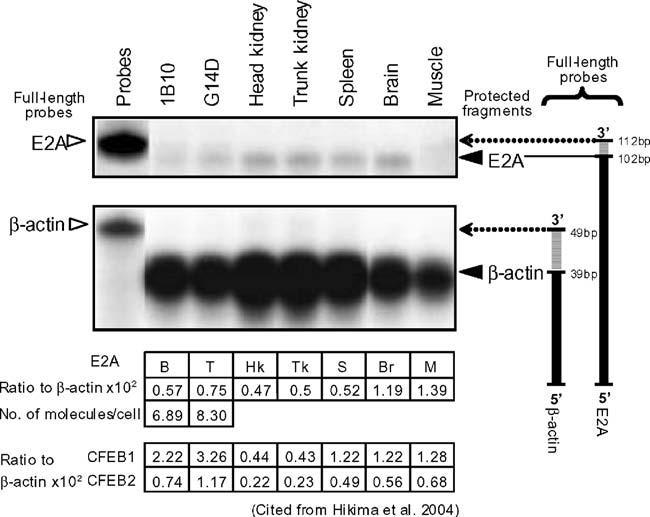
Wide expression of catfish E2A mRNA demonstrated by S1 nuclease protection. Radiolabeled oligonucleotide probes for catfish E2A and β-actin were hybridized to 50 μg of total RNA from the catfish B-cell line (1B10), T-cell line (G14D), and head kidney, trunk kidney, spleen, brain, and muscle tissue (lanes 2 and 8). All samples were digested with S1 nuclease, except lane 1, which is a control for probe integrity. After digestion, samples were separated by electrophoresis on a 10% acrylamide/urea gel and visualized by phosphoimaging. Full-length oligonucleotides for E2A and β-actin are indicated by open triangles (left) and protected fragments by solid triangles (right). A schematic showing the full-length and protected fragments is on the right of the figure. The relative intensity of the E2A signal (expressed as a ratio to β-actin expression) and calculated as the mean of duplicate experiments is shown in the table under the figure. The number of molecules of catfish E2A and β-actin message per cell was calculated for the B and T lymphoid cell lines. The data for CFEB1 and CFEB2 in each tissue are from Hikima et al. (2004)
Transcriptional activation by E2A
To assess the ability of catfish E2A to activate transcription, a simple construct containing a minimal promoter (TATA box and transcription start site) and three copies of the μE5 motif found in the Eμ3′ enhancer (Fig. 3a) was used. In initial experiments, this reporter construct was cotransfected into the catfish B-cell line (1G8) with constructs expressing E2A. Transfections with CFEB expression constructs were also conducted to allow direct comparison between the levels of transcription induced by E2A and CFEB. The results show that E2A is a very strong activator in catfish B cells: it was able to increase expression of the reporter by greater than 2,000-fold above control levels. The activity of E2A was 2.5-fold higher than that of CFEB1 (Fig. 3b). To evaluate whether the activity of E2A is B-cell specific, the ability of E2A to drive transcription was also tested in a catfish T cell line (G14D) and in a mouse plasmacytoma (J558L). The results showed that E2A also drives transcription in G14D and J558L, but to lower levels than seen in the 1G8 catfish B cells (increased transcription of approximately 850- and 120-fold, respectively, as compared with greater than 2,000-fold enhancement in the catfish B cells). The trend of higher transcriptional activity by E2A, as compared to CFEB, was seen in catfish and mouse B cells as well as in catfish T cells (Fig. 3b).
Fig. 3.
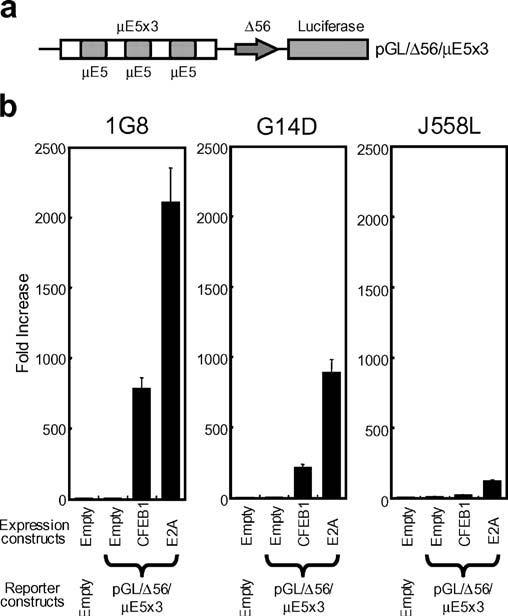
E2A drives transcription from a μE5-dependent reporter. a Schematic of the reporter construct (pGL/Δ56/μE5×3) that contains a minimal c-fos promoter (Δ56) with, upstream, a trimer of μE5 motifs. b Transcription driven from the reporter constructs by cotransfection of vectors expressing E2A (pRc/CMV/E2A) or CFEB1 (pRc/CMV/CFEB1) into the catfish B-cell line (1G8), T-cell line (G14D), or the mouse plasmacytoma (J558L). Expression is compared to the basal transcription assessed by cotransfection of an empty expression vector (pRc/CMV) with the reporter construct. Values are shown as mean±SD for six replicates
Next, the ability of the E2A to drive transcription from the physiologically relevant core of the catfish Eμ3′ enhancer (Region ♯2, Fig. 4a) was tested. Whereas E2A was active from the core of the enhancer, this activity was less than that seen with the artificial enhancer containing a trimer of μE5 motifs (compare Figs. 3 and 4). The activity of E2A, when tested from the core of the Eμ3′ enhancer was, in catfish B cells, less than that of CFEB. Interestingly, when the activity of E2A from the core (Region ♯2) of the Eμ3′ enhancer was tested in a catfish T-cell line (G14D) and in a mouse B-lineage cell (J558L) the results showed that although E2A was able to drive transcription at a low level from the Region ♯2-dependent construct in catfish T cells (G14D), it was inactive in the mouse B-lineage cells (J558L) (Fig. 4b).
Fig. 4.
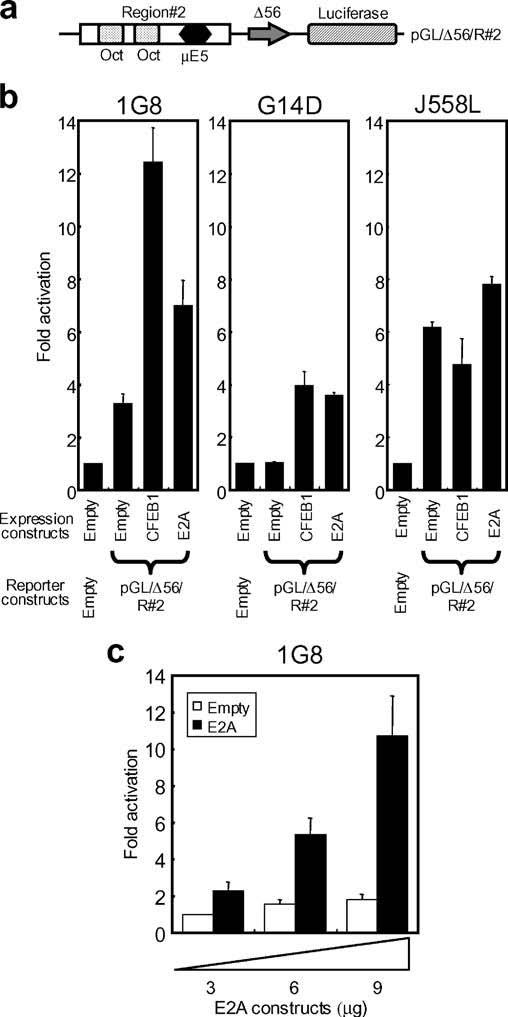
E2A drives transcription from the core region of the Eμ3′ enhancer. a Schematic of the reporter construct (pGL/Δ56/R♯2) that contains the core of the Eμ3′ enhancer (Region♯2) upstream of the minimal c-fos promoter. b Activation of expression from the pGL/Δ56/R♯2 reporter plasmid by E2A and CFEB1. Six micrograms of the E2A or CFEB1 expression vectors were cotransfected with the reporter construct into the catfish B-cell line (1G8), T-cell line (G14D), or the mouse plasmacytoma (J558L). c Three, 6, or 9 μg of the E2A expression vector were cotransfected with the reporter construct into 1G8, G14D, or J558L. Expression is compared to basal transcription assessed by cotransfection of an empty expression vector (pRc/CMV) with the reporter construct. All values are shown as mean±SD for six replicate experiments
The core of the Eμ3′ enhancer contains a single μE5 site, along with two variant octamer motifs. In order to test if the activity of E2A from the core enhancer was μE5 dependent, a Region ♯2-containing construct was used in which the μE5 motif had been scrambled (Fig. 5a). This construct was no longer responsive to E2A (Fig. 5b), indicating that E2A drives transcription, from a physiologically relevant enhancer, in a μE5-dependent manner (Fig. 5b).
Fig. 5.
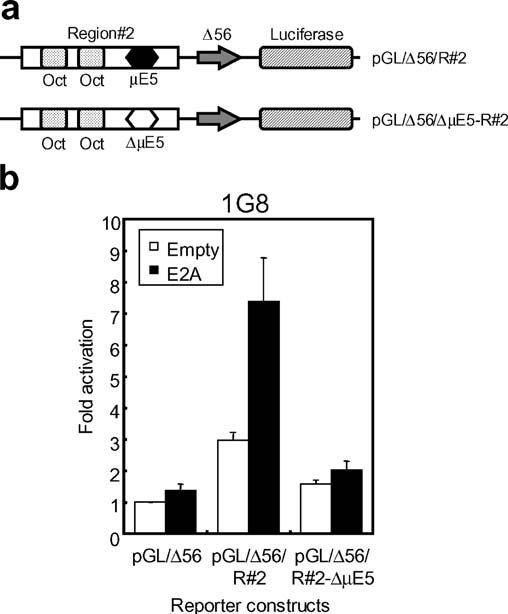
E2A activation from the Eμ3′ enhancer is dependent on the μE5 motif. a Schematic of the reporter constructs that contained the core (Region♯2) of the Eμ3′ enhancer (pGL/Δ56/R♯2), or mutated region♯2 (pGL/Δ56/ΔμE5-R♯2). The sequences of the μE5 site in region ♯2 (black hexagon) and in the mutated region ♯2 (white hexagon) are described in Hikima et al. (2004). b The μE5 motif is essential for E2A activation from the Eμ3′ enhancer. Expression constructs (E2A or CFEB1) and reporter constructs driven by region♯2 or region♯2 in which the μE5 site was mutated were cotransfected into the catfish B-cell line (1G8). Activity is shown as mean±SD for six replicate experiments, relative to the transcription driven by the empty expression plasmid, pRc/CMV
Catfish E2A binds the μE5 motif
The ability of E2A to drive transcription from μE5-dependent constructs (Fig. 3) does not formally demonstrate their ability to bind a μE5 site. To assess its binding properties, E2A was expressed as a recombinant protein by in vitro transcription and translation and tested for the ability to bind the μE5 motif by EMSA (Fig. 6a). The μE5 motif sequence used in the artificial (trimer) enhancer and in this probe was the same as that found in the native Eμ3′ enhancer. The results clearly showed that E2A was capable of binding to the μE5 motif in a manner that was specifically inhibited by an excess of unlabeled competitor. That the mobility shift was due to the E2A protein was confirmed using antibody to E2A and to CFEB. The antibody to E2A generated a super-shifted band (Fig. 6a, lane 5), but the antibody to CFEB generated a complex that did not migrate out of the well (Fig. 6a, lane 4). This latter result also demonstrates that the sequence similarities between CFEB and E2A are sufficient that antigenic cross-reactivity occurs. The SDS-PAGE analysis of the 35S-labeled E2A demonstrates that the recombinant protein expressed by the TNT system was of the predicted molecular size (Fig. 6b), and Fig. 6c shows that the antipeptide E2A antibody recognizes recombinant E2A in a Western blot analysis.
Fig. 6.
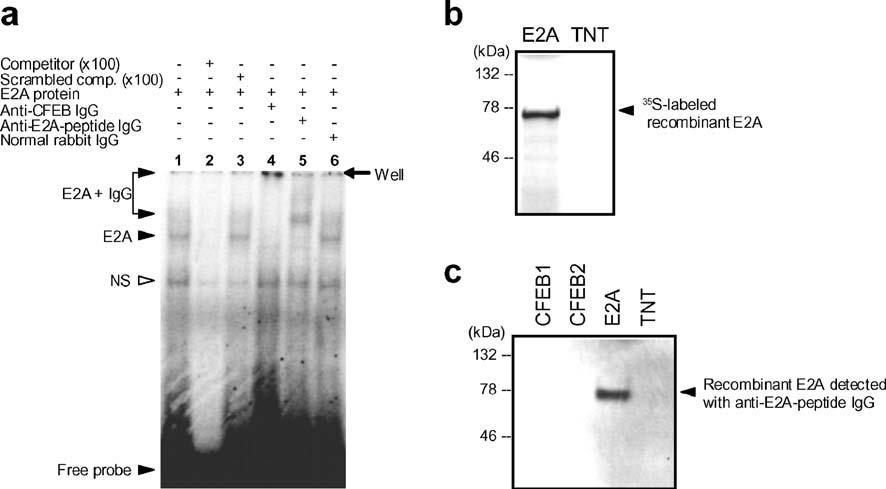
E2A binds the μE5 motif. a E2A protein (S-tagged) was expressed by in vitro transcription and translation and assessed for its ability to bind the μE5 motif in electrophoretic mobility shift assays. The presence in the reaction mix of the E2A protein, unlabeled competitor, scrambled competitor, and IgG from normal rabbit serum, rabbit anti-CFEB serum (Hikima et al. 2004), or rabbit anti-E2A-peptide serum, are indicated above the figure. Left, the shifted and supershifted bands are indicated by arrows. The figure shows results developed by phosphoimaging as described in Materials and methods. b The presence of the 35S-labeled recombinant E2A protein was demonstrated by SDS-PAGE and phosphoimaging. c Western blot analysis showing recognition of the E2A protein with anti-E2A-peptide IgG
Discussion
The results presented here demonstrate that, at the phylogenetic level of a teleost fish, the E-protein family includes a functional homologue of E2A. In many respects, such as its overall structure, its binding to the consensus μE5 motif and its powerful transcriptional activation, catfish E2A confirms the strong evolutionary conservation of this molecule within the vertebrates. However, in other respects, such as its level of expression relative to the CFEB transcription factor (Fig. 2), catfish E2A demonstrates that, in comparing evolutionary homologues across the vertebrates, considerable differences must be expected that will affect our understanding of transcriptional control in diverse vertebrate species.
In terms of its overall structure, the catfish E2A homologue was highly conserved when compared to other vertebrate E-proteins. Two transactivation domains (AD1 and LH-AD2), and the bHLH domain that is involved in dimerization and DNA-binding (Massari et al. 1996; Inukai et al. 1998; Atchley and Fitch 1997) were readily identified. Although both sequence alignments and phylogenetic trees clearly support the conclusion that the molecule studied here is a catfish homologue of E2A, it is not possible to conclude definitively whether it is a homologue of mammalian E12 or E47 (the two alternatively spliced isoforms of E2A expressed in mammals). Although the catfish E2A shows a close relationship in the phylogenetic analyses to a molecule identified as zebrafish E12 (Fig. 1), it is clear that a comprehensive analysis (at the genomic, transcriptomic, and proteomic levels) of E2A in the zebrafish and catfish is lacking. It is likely that an accurate understanding of the relationship between fish and mammalian E2A will be possible only when many more data are available.
Information on the expression of E2A reveals a major difference between the catfish and mammals. Whereas E2A is the predominant E-protein expressed in mammals, especially in lymphoid tissues, this is not the case with catfish. It is clear that CFEB (the catfish homologue of TF12/HEB) is the major E-protein expressed, as measured by transcript levels. Whether measured relative to actin or as the number of messages per cell, E2A is expressed in catfish B- and T-cell lines at levels three- to four-fold lower than CFEB1 (Hikima et al. 2004). Similarly, in head kidney, trunk kidney, spleen, brain, and muscle the expression of E2A message was consistently lower than that of CFEB1 (compare the differences of expression level between E2A and CFEB1 in Fig. 2). Whereas catfish CFEB1 was expressed at higher levels in B- and T-cell lines than in any tissue, the converse was true for E2A, where expression was lower in the T- and B-cell lines than in some tissues. These results indicate, however, that the catfish E2A gene is expressed in those tissues where mammalian E12 has important functions, i.e., for T cell development, myogenesis, and neurogenesis (Quong et al. 2002; Massari and Murre 2000).
E2A drove transcription over 100-fold more strongly from the artificial enhancer (containing three μE5 motifs) than from the core enhancer that contains one μE5 and two octamer motifs (compare Figs. 3 and 4). The binding of E2A molecules to three adjacent sites will increase the likelihood of intermolecular interactions that stabilize binding and/or increase transcriptional activity in a synergistic manner. Differences were observed between catfish and mouse cells in both the background levels of transcription from the reporter constructs used, as well as the degree of transcriptional activation driven by ectopically expressed catfish E2A and CFEB (Figs. 3 and 4). Differences in background expression of the reporter construct may reflect the preexisting levels of transcription factors in the cells. That J558L is a more terminally differentiated B-lineage cell (a plasma cell) than is 1G8 (a B lymphoblastoid cell) may be reflected in higher levels of transcription factors that drive Ig gene expression. The differences in response of catfish and mouse cells to the transfected catfish E-proteins could reflect the optimization (or coevolution) of the components of the active transcription complex that drives IgH transcription in different vertebrate lineages. The transcriptional activity of E2A was higher than that of CFEB1 (by approximately three-fold) when measured using an artificial enhancer containing a trimer of μE5 motifs. On the other hand, when the physiologically relevant core region from the catfish IgH enhancer (Eμ3′) was used, E2A showed, in catfish B cells, weaker activation of transcription than CFEB1 (Figs. 3 and 4). This result supports the suggestion that CFEB may be the dominant E-protein driving transcription of the IgH locus in catfish B cells, by virtue of both its higher expression and its higher activity. It also raises the possibility that CFEB may interact more efficiently than E2A with the Oct transcription factors that bind to the core enhancer. There is strong evidence that E2A proteins (both E12 and E47) are of major importance in mammalian B cell development and function (Kee et al. 2000; Bain and Murre 1998; Bain et al. 1997; Greenbaum et al. 2004); for example, the induction of E2A is required to promote IgH class switch recombination during B-cell activation (Quong et al. 1999). Our observations on the relative levels of expression and functional activities of CFEB and E2A in catfish B cells suggest that the situation observed in mammals, where E2A is the dominant E-protein in the development and function of B cells, may not be the case at the phylogenetic level of a teleost fish.
The basis of some functional differences in E2A proteins may lie in the region of the Rep domain. The amino acid sequence of the region around the inhibitory domain of mammalian E12, which prevents homodimerization, was compared with the sequences of E2A proteins of other vertebrates. Interestingly, sequence insertions are present in this region of the E2A of lower vertebrates (fish and frogs), but absent from the E2A of birds and mammals (Fig. 7). The Rep domain, important in E-protein homodimerization, is also involved in repressing transactivation by the AD1 and AD2 domains (Markus et al. 2002). Furthermore, since several acidic amino acids are observed in the inserted sequences, phosphorylation of these residues may influence homodimerization (Sloan et al. 1996; Mitsui et al. 1993). These results suggest that in the details of their structure and function, as well as in their levels of expression relative to other E-proteins, E2A molecules characterized in lower vertebrates may not possess the properties of the classical E12/E47 proteins described in mammals.
Fig. 7.
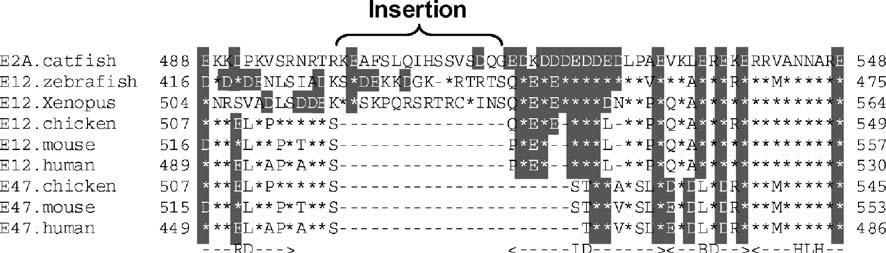
The inhibition domain region of fish and amphibian E2A contains insertions. An alignment is shown of the amino acid sequences of the inhibition domain region of vertebrate E2A/E12/E47 sequences from catfish, zebrafish, chicken, mouse, and human. Asterisks denote residues identical with catfish E2A, and dashes indicate gaps. Acidic residues are indicated as shaded boxes. RD Rep domain, ID inhibitory domain, BD basic domain, HLH helix–loop–helix domain
Acknowledgements
This work was supported by awards from the National Science Foundation (MCB9807531) and the National Institutes of Health (R01-GM62317 and R01-AI-19530). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact. Publication ♯13 from the Marine Biomedicine and Environmental Sciences Center.
Footnotes
The nucleotide sequences reported in this paper have been submitted to GenBank with accession number YA770493
Electronic Supplementary Material Supplementary material is available for this article at http://dx.doi.org/10.1007/s00251-005-0793-3.
References
- Anon . Promega protein guide: tips and techniques. Promega Corporation; WI: 1993. In vitro translation of proteins; p. 36. [Google Scholar]
- Atchley WR, Fitch WM. A natural classification of the basic helix–loop–helix class of transcription factors. Proc Natl Acad Sci U S A. 1997;94:5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain G, Murre C. The role of E-proteins in B- and T-lymphocyte development. Semin Immunol. 1998;10:143–153. doi: 10.1006/smim.1998.0116. [DOI] [PubMed] [Google Scholar]
- Bain G, Robanus Maandag EC, te Riele HP, Feeney AJ, Sheehy A, Schlissel M, Shinton SA, Hardy RR, Murre C. Both E12 and E47 allow commitment to the B cell lineage. Immunity. 1997;6:145–154. doi: 10.1016/s1074-7613(00)80421-5. [DOI] [PubMed] [Google Scholar]
- Benezra R. An intermolecular disulfide bond stabilizes E2A homodimers and required for DNA binding at physiological temperatures. Cell. 1994;79:1057–1067. doi: 10.1016/0092-8674(94)90036-1. [DOI] [PubMed] [Google Scholar]
- Choi JK, Shen CP, Radomska HS, Eckhardt LA, Kadesch T. E47 activates the Ig-heavy chain and TdT loci in non-B cells. EMBO J. 1996;15:5014–5021. [PMC free article] [PubMed] [Google Scholar]
- Chu C, Kohtz DS. Identification of the E2A gene products as regulatory targets of the G1 cycle-dependent kinase. J Biol Chem. 2001;276:8524–8534. doi: 10.1074/jbc.M008371200. [DOI] [PubMed] [Google Scholar]
- Cioffi CC, Middleton DL, Wilson MR, Miller NW, Clem LW, Warr GW. An IgH enhancer that drives transcription through basic helix–loop–helix and Oct transcription factor binding motifs. Functional analysis of the Eμ3′ enhancer of the catfish. J Biol Chem. 2001;276:27825–27830. doi: 10.1074/jbc.M100110200. [DOI] [PubMed] [Google Scholar]
- Cioffi CC, Pollenz RS, Middleton DL, Wilson MR, Miller NW, Clem LW, Warr GW, Ross DA. Oct2 transcription factor of a teleost fish: activation domains and function from an enhancer. Arch Biochem Biophys. 2002;404:55–61. doi: 10.1016/s0003-9861(02)00227-8. [DOI] [PubMed] [Google Scholar]
- Ephrussi A, Church GM, Tonegawa S, Gilbert W. B lineage-specific interactions of an immunoglobulin enhancer with cellular factors in vivo. Science. 1985;227:134–140. doi: 10.1126/science.3917574. [DOI] [PubMed] [Google Scholar]
- Erman B, Cortes M, Nikolajczyk BS, Speck NA, Sen R. ETS-core binding factor: a common composite motif in antigen receptor gene enhancers. Mol Cell Biol. 1998;18:1322–1330. doi: 10.1128/mcb.18.3.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel P, Janney N, Valenzuela JR, Romanow WJ, Murre C, Feeney AJ. Localized gene-specific induction of accessibility to V(D)J recombination induced by E2A and early B cell factor in nonlymphoid cells. J Exp Med. 2001;194:645–656. doi: 10.1084/jem.194.5.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum S, Lazorchak AS, Zhuang Y. Differential functions for the transcription factor E2A in positive and negative gene regulation in pre-B lymphocytes. J Biol Chem. 2004;279:45028–45035. doi: 10.1074/jbc.M400061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikima J, Cioffi CC, Middleton DL, Wilson MR, Miller NW, Clem LW, Warr GW. Evolution of transcriptional control of the IgH locus: characterization, expression, and function of TF12/HEB homologs of the catfish. J Immunol. 2004;173:5476–5484. doi: 10.4049/jimmunol.173.9.5476. [DOI] [PubMed] [Google Scholar]
- Inukai T, Inaba T, Ikushima S, Look AT. The AD1 and AD2 transactivation domains of E2A are essential for the antiapoptotic activity of the chimeric oncoprotein E2A-HLF. Mol Cell Biol. 1998;18:6035–6043. doi: 10.1128/mcb.18.10.6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee BL, Murre C. Induction of early B cell factor (EBF) and multiple B lineage genes by the basic helix–loop–helix transcription factor E12. J Exp Med. 1998;188:699–713. doi: 10.1084/jem.188.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee BL, Quong MW, Murre C. E2A proteins: essential regulators at multiple stages of B-cell development. Immunol Rev. 2000;175:138–149. [PubMed] [Google Scholar]
- Lassar AB, Davis RL, Wright WE, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell. 1991;66:305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix–loop–helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- Litman GW, Flajnik MF, Warr GW. Diverse forms of immunoglobulin genes in lower vertebrates. In: Honjo T, Alt FW, Neuberger M, editors. Molecular biology of B cells. Elsevier Academic Press; San Diego: 2004. pp. 417–432. [Google Scholar]
- Magor BG, Wilson MR, Miller NW, Clem LW, Middleton DL, Warr GW. An Ig heavy chain enhancer of the channel catfish Ictalurus punctatus: evolutionary conservation of function but not structure. J Immunol. 1994;153:5556–5563. [PubMed] [Google Scholar]
- Magor BG, Ross DA, Pilstorm L, Warr GW. Transcriptional enhancers and the evolution of the IgH locus. Immunol Today. 1999;20:13–17. doi: 10.1016/s0167-5699(98)01380-2. [DOI] [PubMed] [Google Scholar]
- Markus M, Benezra R. Two isoform of protein disulfide isomerase alter the dimerization status of E2A proteins by a redox mechanism. J Biol Chem. 1999;274:1040–1049. doi: 10.1074/jbc.274.2.1040. [DOI] [PubMed] [Google Scholar]
- Markus M, Du Z, Benezra R. Enhancer-specific modulation of E protein activity. J Biol Chem. 2002;277:6469–6477. doi: 10.1074/jbc.M110659200. [DOI] [PubMed] [Google Scholar]
- Massari ME, Murre C. Helix–loop–helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:426–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari ME, Jennings PA, Murre C. The AD1 transactivation domain of E2A contains a highly conserved helix which is required for its activity in both Saccharomyces cerevisiae and mammalian cells. Mol Cell Biol. 1996;16:121–129. doi: 10.1128/mcb.16.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller NW, Rycyzyn MA, Wilson MR, Warr GW, Naftel JP, Clem LW. Development and characterization of channel catfish long term B cell lines. J Immunol. 1994;152:2180–2189. [PubMed] [Google Scholar]
- Mitsui K, Shirakata M, Paterson BM. Phosphorylation inhibits the DNA-binding activity of MyoD homodimers but not MyoD-E12 heterodimers. J Biol Chem. 1993;268:24415–24420. [PubMed] [Google Scholar]
- Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989a;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN, Cabrera CV, Buskin JN, Hauschka SD, Lassar AB, Weintraub H, Baltimore D. Interactions between heterologous helix–loop–helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989b;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Nagata Y, Todokoro K. Activation of helix–loop–helix proteins Id1, Id2 and Id3 during neural differentiation. Biochem Biophys Res Commun. 1994;199:1355–1362. doi: 10.1006/bbrc.1994.1380. [DOI] [PubMed] [Google Scholar]
- Nelson B, Kadesch T, Sen R. Complex regulation of the immunoglobulin μ heavy chain gene enhancer: μB, a new determinant of enhancer function. Mol Cell Biol. 1990;10:3145–3154. doi: 10.1128/mcb.10.6.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nihei Y, Kobiyama A, Hirayama Y, Kikuchi K, Watabe S. The mRNA expression patterns of E12 transcription factor in various developmental stages and adult tissues of carp. Fish Sci. 1999;65:600–605. [Google Scholar]
- Nikolajczyk BS, Nelsen B, Sen R. Precise alignment of sites required for μ enhancer activation in B cells. Mol Cell Biol. 1996;16:4544–4554. doi: 10.1128/mcb.16.8.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolajczyk BS, Cortes M, Feinman R, Sen R. Combinatorial determinants of tissue-specific transcription in B cells and macrophages. Mol Cell Biol. 1997;17:3527–3535. doi: 10.1128/mcb.17.7.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Mutul J, Macchi M, Wasylyk B. Mutational analysis of the contribution of sequence motifs within the IgH enhancer to tissue specific transcriptional activation. Nucleic Acids Res. 1988;16:6085–6096. doi: 10.1093/nar/16.13.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quong MW, Harris DP, Swain SL, Murre C. E2A activity is induced during B-cell activation to promote immunoglobulin class switch recombination. EMBO J. 1999;18:6307–6318. doi: 10.1093/emboj/18.22.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quong MW, Romanow WJ, Murre C. E-protein function in lymphocyte development. Annu Rev Immunol. 2002;20:301–322. doi: 10.1146/annurev.immunol.20.092501.162048. [DOI] [PubMed] [Google Scholar]
- Rao E, Dang W, Tian G, Sen R. A three-protein–DNA complex on a B cell-specific domain of the immunoglobulin μ heavy chain gene enhancer. J Biol Chem. 1997;272:6722–6732. doi: 10.1074/jbc.272.10.6722. [DOI] [PubMed] [Google Scholar]
- Romanow WJ, Langerak AW, Goebel P, Wolvers-Tettero IL, van Dongen JJ, Feeney AJ, Murre C. E2A and EBF act in synergy with the V(D)J recombinase to generate a diverse immunoglobulin repertoire in nonlymphoid cells. Mol Cell. 2000;5:343–353. doi: 10.1016/s1097-2765(00)80429-3. [DOI] [PubMed] [Google Scholar]
- Ross DA, Magor BG, Middleton DL, Wilson MR, Norman WM, Clem LW, Warr GW. Characterization of Oct2 from the channel catfish: functional preference for a variant octamer motif. J Immunol. 1998;160:3874–3882. [PubMed] [Google Scholar]
- Ross DA, Lyles M, Ledford BE, Magor BG, Wilson MR, Norman WM, Clem LW, Middleton DL, Warr GW. Catfish Oct2 binding affinity and functional preference for octamer motifs, and interaction with OBF-1. Dev Comp Immunol. 1999;23:199–211. doi: 10.1016/s0145-305x(99)00007-5. [DOI] [PubMed] [Google Scholar]
- Sayegh CE, Quong MW, Agata Y, Murre C. E-proteins directly regulate expression of activation-induced deaminase in mature B cells. Nat Immunol. 2003;4:586–593. doi: 10.1038/ni923. [DOI] [PubMed] [Google Scholar]
- Schlissel M, Voronova A, Baltimore D. Helix–loop–helix transcription factor E47 activates germ-line immunoglobulin heavy-chain gene transcription and rearrangement in a pre-T-cell line. Genes Dev. 1991;5:1367–1376. doi: 10.1101/gad.5.8.1367. [DOI] [PubMed] [Google Scholar]
- Sloan SR, Shen C-P, McCarrick-Walmsley R, Kadesch T. Phosphorylation of E47 as a potential determinant of B-cell-specific activity. Mol Cell Biol. 1996;16:6900–6908. doi: 10.1128/mcb.16.12.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XH, Baltimore D. An inhibitory domain of E12 transcription factor prevents DNA binding in E12 homodimers but not in E12 heterodimers. Cell. 1991;64:459–470. doi: 10.1016/0092-8674(91)90653-g. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic analysis using parsimony (* and other methods) version 4. Sinauer Associates; Sunderland: 2002. [Google Scholar]
- Vitola SJ, Wang A, Sun XH. Substitution of basic amino acids in the basic region stabilizes DNA binding by E12 homodimers. Nucleic Acids Res. 1996;24:1921–1927. doi: 10.1093/nar/24.10.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulbeck C, Fromental-Ramain C, Campos-Ortega JA. The HLH domain of a zebrafish HE12 homologue can partially substitute for functions of the HLH domain of Drosophila DAUGHTERLESS. Mech Dev. 1994;46:73–85. doi: 10.1016/0925-4773(94)90077-9. [DOI] [PubMed] [Google Scholar]
- Yokota Y. Id and development. Oncogene. 2001;20:8290–8298. doi: 10.1038/sj.onc.1205090. [DOI] [PubMed] [Google Scholar]


