Abstract
The role of the three conserved cysteine residues on Azotobacter vinelandii IscU in accepting sulfane sulfur and forming a covalent complex with IscS has been evaluated using electrospray-ionization mass spectrometry studies of variants involving individual cysteine-to-alanine substitutions. The results reveal that IscS can transfer sulfur to each of the three alanine-substituted forms of IscU to yield persulfide or polysulfide species, and formation of a heterodisulfide covalent complex between IscS and Cys37 on IscU. It is concluded that S transfer from IscS to IscU does not involve a specific cysteine on IscU or the formation of an IscS–IscU heterodisulfide complex.
Keywords: Iron-sulfur cluster assembly, IscS, IscU, Mass spectrometry, Sulfur transfer
Abbreviations: isc, iron sulfur cluster; nif, nitrogen fixation; suf, sulfur utilization factor; DTT, dithiothreitol; ESI-MS, electrospray-ionization mass spectrometry
1. Introduction
Iron–sulfur (Fe–S) clusters are one of the most common prosthetic groups in biology with functions including electron transfer, coupling proton and electron transfer, substrate binding and activation, regulation of enzyme activity and gene expression, sensing of reactive species, radical generation, disulfide cleavage, sulfur donation, and storage of iron or electrons, for a recent comprehensive review see [1]. Fe–S clusters are also believed to constitute one of the most ancient types of prosthetic groups. In accord with this hypothesis, the fundamental process of Fe–S cluster biosynthesis appears to be remarkably conserved in almost all organisms [2–4]. In prokaryotes, three distinct operons have emerged, the nif [5], isc [6], and suf [7] operons, that encode proteins essential for Fe–S cluster biosynthesis. All three operons have either U-type or A-type proteins that are proposed to function as scaffolds for the assembly of [2Fe–2S] and/or [4Fe–4S] clusters. Cluster-loaded forms of both U- and A-type scaffolds have been used for the in vitro maturation of a variety of apo Fe–S proteins via intact cluster transfer [8–18]. All Fe–S cluster biosynthetic systems described so far also contain a gene encoding a pyridoxal phosphate-dependent cysteine desulfurase enzyme, NifS [19,20], IscS [6]and SufS (or SufSE) [21–24], responsible for the mobilization of sulfur by the catalytic conversion of l-cysteine to l-alanine and sulfane sulfur (S0). The objectives of this study were to investigate the mechanism of S transfer from Azotobacter vinelandii IscS to IscU, a ubiquitous Fe–S cluster scaffolding protein containing three rigorously conserved cysteine residues.
Previous electrospray-ionization mass spectrometry (ESI-MS) studies of chromatographically separated fractions of a reaction mixture involving IscS, IscU and l-cysteine concluded that S0 sequestered by IscS from substrate l-cysteine is directly transferred from a cysteine persulfide on the active-site cysteine of IscS to form persulfides or polysulfides on IscU [25]. Furthermore, a covalently bound disulfide or polysulfide complex (~58 kDa) involving IscS (~44 kDa) and IscU (~14 kDa) was observed [25]. The current study examines the role of each rigorously conserved cysteine residue on IscU by investigating the effect of individual cysteine-to-alanine substitution on sulfur transfer and disulfide complex formation between IscS and IscU. The results show that none of the individual Cys-to-Ala substitutions prevent S0 transfer from IscS to IscU, indicating that the transfer mechanism is not critically dependent on a specific cysteine residue on IscU. In addition, they demonstrate the capacity for formation of a specific heterodisulfide between the active-site cysteine located within IscS and the Cys37 residue of IscU. However, formation of a heterodisulfide IscU–IscS complex is not required to affect S transfer from IscS to IscU.
2. Materials and methods
2.1. Plasmid constructions
Plasmids used for the high-level heterologous expression of A. vinelandii wild-type and substituted forms of IscU in Escherichia coli included pDB956 (wild-type), pDB1214 (IscUC37A), pDB1235 (IscUC63A), and pDB1229 (IscUC106A). Plasmid pDB956 was described previously [9] and it contains IscU whose expression is controlled by the T7 promoter within the pT7-7 expression vector. Other recombinant plasmids are identical to pDB956 with the exception of the replacement of UGC (cys) codons for residues 37, 63, and 106 by GCG, GCC, and GCG (alaa) codons, respectively. The plasmid used for heterologous expression of IscS in E. coli (pDB943) was described previously [6]. The E. coli strain BL21(DE3) was used as the host for heterologous expression.
2.2. Protein purification
Purification of IscU, substituted forms of IscU, and IscS was performed as previously described [6,9]. Protein homogeneity was determined to be >95% by densitometry measurements of SDS–PAGE gels.
2.3. ESI-MS sample preparation
Samples of IscU, IscUC37A, IscUC63A, and IscUC106A for ESI-MS analysis were prepared under an argon atmosphere in a Vacuum Atmospheres glove box (O2 < 5 ppm) at room temperature in 50 mM Tris/HCl buffer, pH 7.8, without β-mercaptoethanol or dithiothreitol (DTT). Before use, all IscU samples were pre-incubated with 10 mM DTT for 30 min and DTT was removed using a 50-mL G25 desalting column controlled by a Pharmacia FPLC located within an anoxic glove box. Samples of 0.1 mM IscU and IscU substituted forms were incubated with 2 mM l-cysteine and 0.1 mM IscS for 20 min before quenching the reaction in liquid nitrogen. Protein concentrations were assessed using a Bio-Rad modified DC protein assay with bovine serum albumen (Roche) as the standard.
2.4. ESI-MS measurements
Samples for ESI-MS were chromatographically separated using a BioBasic-4 column (1 mm × 150 mm with a 5 μm particle size and 300 Å pore size) from Keystone Scientific Inc before acquiring the mass spectrum. The HPLC arrangement used an Applied Biosystems (ABI) 140B solvent delivery system where solvent A was water with 0.1% trifluoroacetic acid and solvent B was acetonitrile. Sample volumes used 2.0–3.0 μL injections at 100% A. Solvent B was ramped to 40% in 3 min; then to 70% over 32 min, finally solvent B reached 100% over the last 5 min. The HPLC flow rate was 30 μL per minute and the UV absorption at 214 nm was measured using an ABI 759A detector. After flowing through the detector, the effluent was split so that 18 μL per minute went into the PE Sciex API I plus single quadrapole mass spectrometer equipped with an electrospray ionization source and the remaining 12 μL of eluant went to waste. The mass spectrometer scanned from 800 to 1500 m/z with a 2.0 ms dwell time and a 0.2 m/z step size. The reported masses are accurate to ±0.01%.
3. Results
3.1. IscS to IscU sulfur transfer
Sulfur transfer from IscS to wild-type and substituted forms of IscU in the presence of excess l-cysteine (1:1:20 IscU:IscS: l-cysteine) was assessed by ESI-MS analysis of fractions separated from the reaction mixture using reverse-phase HPLC, after 20 mins of anaerobic incubation at room temperature. The reconstructed mass spectra of IscU, IscUC37A, IscUC63A, and IscUC106A after DTT treatment, but prior to incubation with IscS and excess l-cysteine, are shown in the left panel of Fig. 1. The mass spectra confirm the respective Cys-to-Ala substitutions because the ion mass of each of alanine-substituted forms is 32-Da less than wild-type IscU. The previously published ESI-MS studies of wild-type IscU indicated a mass of 13744 Da corresponding to the predicted mass based on the primary sequence, i.e., 13875 Da, minus 131 Da due to post-translational cleavage of the N-terminal methionine residue [25]. The parent ion mass (13712 Da) for all three IscU-substituted forms is also in excellent agreement with the predicted mass based on the predicted amino acid sequence minus the N-terminal methionine residue (13712 Da). The mass spectra of DTT incubated samples of IscU and substituted forms of IscU are all very similar with a dominant parent ion peak and successively weaker peaks at +31 and +62 Da, corresponding to the addition of one and two S atoms, respectively. While the same three peaks are also observed in IscUC106A, the dominant form contains one additional S atom.
Fig. 1.

Reconstructed ESI-quadrupole mass spectra of monomeric A. vinelandii IscU, IscUC37A, IscUC63A, and IscUC106A before and after incubation with IscS and l-cysteine. Left panel: IscU and IscU variants (0.1 mM) after incubation with excess DTT. Right panel: IscU and IscU variants (0.1 mM) after incubation with 0.1 mM IscS and 2.0 mM l-cysteine for 20 min.
Incubation of IscU and the substituted forms of IscU with IscS and l-cysteine results in the incorporation of covalently attached S in the form of persulfides or polysulfides as evidenced by increased intensities for peaks corresponding to multiple S atom additions relative to the parent ion (see right panel of Fig. 1). For wild-type IscU, the dominant forms have 2 or 3 S atoms incorporated, although species with 1–6 additional S atoms are observed. Compared to wild-type IscU, less S is incorporated in each of the single ala-substituted forms on incubation with IscS and l-cysteine. For IscUC37A, the dominant peak in the mass spectrum corresponds to the addition of two S atoms, although forms with 1–4 additional S atoms are observed. For IscUC63A, increases in forms with 1–4 additional S atoms are apparent compared to the parent ion. For IscUC106A, the dominant form changes from one with 1 additional S atom to one with 2 additional S atoms after incubation with IscS and l-cysteine. Quantitation of mass spectrometry data based on areas of individual peaks indicates IscS-mediated incorporation of covalently attached S in wild-type IscU and all three substituted forms, with the net addition corresponding to 2.2 S atoms for wild-type IscU, 1.9 S atoms for IscU C37A, 0.9 S atoms for IscUC63A, and 0.5 S atoms for IscUC106A.
3.2. IscS–IscU complex formation
Previous ESI-MS studies of IscU and IscS revealed the presence of a small fraction of a covalently bound disulfide or polysulfide complex between IscS and IscU monomers when incubated in the presence of excess l-cysteine [25]. Hence, one of the objectives of parallel ESI-MS studies using the ala-substituted variants was to address the possibility that a specific IscU cysteine residue is responsible for complex formation. The reconstructed mass spectra of IscU and the IscU ala-substituted forms in the region of the IscS–IscU complex (mass range 58–59 kDa) are shown in Fig. 2. Wild-type IscU, IscUC63A, and IscUC106A each form a covalently bound disulfide or polysulfide complex with IscS, having average masses of 58530, 58400 and 58460 Da, respectively. The predicted masses for these complexes based on the observed masses of IscS and IscU in the absence of polysulfides or persulfides (44647 Da for IscS, see below, and 13744 Da for wild-type IscU, and 13712 for IscUC63A and IscUC106A) are 58391 Da for the complex involving wild-type IscU and 58359 Da for the complexes involving IscUC63A and IscUC106A. The observed masses and breadth of the peaks indicate heterogeneous complexes involving disulfide or polysulfide covalent linkages between IscU and IscS and/or multiple polysulfides or persulfides on IscS and IscU (2–7 additional S atoms for the wild-type IscU–IscS complex and 1–4 additional S atoms for the complexes involving IscUC63A or IscUC106A and IscS). In contrast no covalent complex was observed to form between IscUC37A and IscS, see Fig. 2, suggesting that Cys37 is required for the formation of disulfide or polysulfide complexes between IscU and IscS.
Fig. 2.

Reconstructed ESI-quadrupole mass spectra of A. vinelandii IscU, IscUC37A, IscUC63A, and IscUC106A after incubation with IscS and l-cysteine in the mass range corresponding to a covalent IscS–IscU complex. Samples are the same as those used in the right panel of Fig. 1.
ESI-MS studies of IscU and IscS 1:1 reaction mixtures were also carried out with a 20-fold excess of S-methyl-l-cysteine in place l-cysteine. S-methyl-l-cysteine is an effective alternative substrate for cysteine desulfurases (R.S. Phillips, personal communication). Consequently, these experiments were designed to investigate the possibility that an S-methyl group could be transferred from IscS to IscU, thereby preventing the formation of polysulfides or persulfides either on or between the conserved cysteines. However, neither the wild-type nor any substituted forms of IscU showed any mass change when incubated in the presence of S-methyl- l-cysteine and IscS. The reconstructed mass spectra were identical to those shown in the left panel of Fig. 1 (data not shown).
Complex formation between IscU and IscS when incubated in the presence of S-methyl-l-cysteine was also evaluated. The results of these experiments are shown in the left panel of Fig. 3, which compares the reconstructed mass spectra of the IscS–IscU covalent complex formed in the presence of l-cysteine or S-methyl-l-cysteine. Similar results were obtained using the IscUC63A and IscUC106A, but no complex formation was observed with IscUC37A when incubated in the presence of l-cysteine or S-methyl-l-cysteine (data not shown). In the presence of S-methyl-l-cysteine, the complex is more homogeneous due to the lack of polysulfide or persulfides, with a well-defined major peak corresponding to a mass of 58 398 Da, see right panel of Fig. 3. Moreover, on the basis of the reconstructed mass spectra of IscS in reaction mixtures containing l-cysteine and S-methyl-l-cysteine, see right panel of Fig. 3, the mass of the complex formed in the presence of S-methyl-l-cysteine can only be interpreted in terms of IscU and IscS covalently attached via a disulfide rather than a polysulfide. The broad mass peak centered near 44730 Da that is observed for IscS in the presence of l-cysteine is a consequence of persulfides or polysulfides formation on IscS, as evidenced by a substantial sharpening and a shift to lower mass, 44666 Da, on addition of DTT [25]. In the presence of S-methyl-l-cysteine, the mass spectrum of IscS is much better resolved, see right panel of Fig. 3, indicating that IscS has a mass of 44652 Da in the absence of persulfides or polysulfides. The observed mass of the IscU–IscS complex in the presence of S-methyl-l-cysteine (58398 Da) corresponds to the sum of the masses of IscU and IscS without additional S atoms (13744 Da + 44652 Da = 58396 Da), within experimental error. An analogous IscU–IscS covalent complex without additional sulfur atoms was observed in the mass spectrum of 1:1 IscU:IscS mixture in the absence of substrate (data not shown). Hence, the mass spectrometry results indicate that covalent complex formation between IscU and IscS involves a disulfide between Cys37 of IscU and the active-site cysteine on IscS, Cys325, and that the formation of this heterodisulfide does not require the presence of the l-cysteine substrate.
Fig. 3.
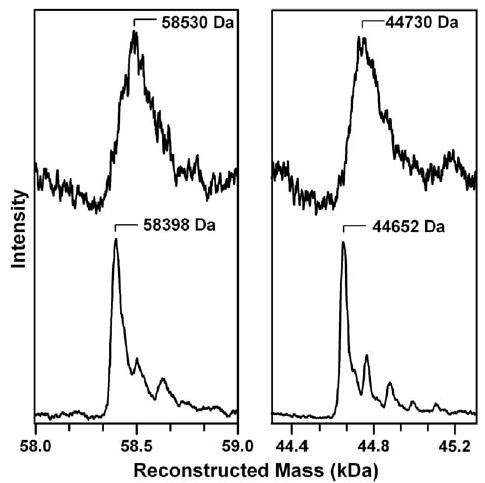
Comparison of the reconstructed ESI-quadrupole mass spectra of the IscU–IscS complex (left panel) and IscS (right panel) in a reaction mixture containing 0.1 mM IscU, 0.1 mM IscS and 2.0 mM l-cysteine (upper spectra) or 2.0 mM S-methyl-l-cysteine (lower spectra).
4. Discussion
Gel-filtration and chemical cross-linking studies have demonstrated the formation of a noncovalent α2β2 complex between homodimeric A. vinelandii IscU and IscS [3,9]. However, this complex is distinct from the covalent complex between monomeric IscS and IscU that was originally identified as a heterodisulfide or heteropolysulfide IscU–IscS complex observed by ESI-MS studies when IscU is incubated with stoichiometric IscS and excess l-cysteine [25]. The mass spectrometry studies of IscU and substituted forms of IscU reported in this work demonstrate that the covalent complex between IscS and IscU does not require l-cysteine and involves a disulfide rather than a polysulfide. Moreover, the loss of the capacity to form a complex with IscUC37A, but not with IscUC63A or IscUC106A, identifies Cys37 as the residue involved in the formation of the heterodisulfide complex with IscS. These results concur with recent SDS/PAGE studies of E. coli IscU and IscS [26] in finding a heterodisulfide complex between IscS and IscU, but disagree with respect to the specific cysteine on IscU that participates in the heterodisulfide, Cys37 in A. vinelandii IscU versus Cys63 in E. coli IscU. This discrepancy is particularly puzzling as the same cysteine variants were used in both studies to identify the IscU cysteine involved with heterodisulfide formation with IscS. The origin of this discrepancy is unclear at present, but it should noted that the studies involving E. coli IscU and IscS involved His-tagged IscU and were not conducted under anaerobic conditions [26]. Hence it is possible that the C-terminal His-tag on IscU or the presence of O2 perturb the structure of IscU such that a different activesite cysteine is used in heterodisulfide formation.
The SDS/PAGE studies of E. coli IscU and IscS interpreted the heterodisulfide complex as an integral part of the mechanism of S-transfer from IscS to IscU [26]. However, the mass spectrometry results reported here for A. vinelandii IscS and IscU argue against a role for the IscS–IscU heterodisulfide complex in S transfer from IscS to IscU. This is best illustrated by IscUC37A which does not form a heterodisulfide complex, but retains the capacity to accept ~2 S atoms from IscS in the presence of l-cysteine. Indeed, the heterodisulfide is likely to provide an impediment to S transfer based on the observation of increased S transfer for the IscUC37A substituted-form compared to the IscUC63A and IscUC106A IscU substituted forms.
One of the major objectives of this study was to establish if S transfer from IscS involves a specific cysteine on IscU. The ESIMS results on the Cys-to-Ala substituted forms clearly demonstrate that this is not the case, as S transfer from IscS to IscU occurs with each of the three IscU variants investigated. Hence, the in vitro transfer mechanism is not critically dependent on a specific Cys on IscU. Rather the results indicate that only two of the three conserved cysteines on IscU are needed for S transfer from IscS and that any pair of cysteines is functional in mediating S transfer. The simplest interpretation is that any one of the three conserved cysteines on IscU can accept a sulfane S from IscS to form a cysteine persulfide.
Finally it is worthwhile to consider the ESI-MS results reported herein in the context of the recent NMR solution structure for Haemophilis influenzae IscU [27] and X-ray crystal structure of the E. coli SufE sulfurtransferase [28]. The NMR structure of H. influenzae IscU with Zn bound at the Fe–S cluster assembly site reveals a protein with compact globular core and flexibility in the N- and C-terminal regions. The Zn is ligated by the three conserved cysteine residues (Cys37, Cys63, and Cys106) and one histidine residue (His105) in a solvent exposed site near the end of three conformationally dynamic loops at one end of the IscU structure. In accord with the ESI-MS evidence for a IscS–IscU heterodisulfide involving Cys37 on IscU, Cys63 and Cys106 are partially solvent exposed and Cys37 is fully solvent exposed in the IscU NMR structure [27]. However, the proximity of the three active-site cysteine residues in Zn-bound IscU, coupled with the conformational flexibility of the three loops which each contain one of the active-site cysteine residues, is consistent with ability of the cysteine persulfide on the flexible side arm of IscS [29] to transfer S0 to any one of the three conserved cysteines to form a cysteine persulfide on IscU.
SufE has not been shown to function as a scaffold for the assembly of Fe–S clusters, but does function as a sulfurtransferase to mobilize sulfane S generated on the SufS cysteine desulfurase, by transferring the active-site cysteine presulfide on SufS to a more accessible active-site cysteine persulfide on SufE [23,24]. Interestingly, SufE shows strong structural homology to IscU [27,28], and the cysteine that constitutes the S acceptor site on SufE coincides with the location of the cysteine residues involved in Fe–S cluster assembly on IscU. Cys37 occupies the equivalent topographical position to the active-site cysteine, Cys51, on SufE. Hence, SufE provides indirect evidence that S transfer from the cysteine persulfide on IscS to a single cysteine on IscU can occur and does not require the presence of additional cysteine residues on IscU or the formation of a heterodisulfide between IscS and IscU.
Acknowledgments
This work was supported by grants from the National Institutes of Health (GM62542 to M.K.J.) and the National Science Foundation (MCB-021138 to D.R.D.). We thank Dr. Robert S. Phillips (University of Georgia) for informing us that S-methyl-l-cysteine is an effective alternative substrate for cysteine desulfurases.
Footnotes
Edited by Hans Eklund
References
- 1.Johnson, M.K. and Smith, A.D. (2005) Iron–sulfur proteins in: Encyclopedia of Inorganic Chemistry (King, R.B., Ed.), 2nd edn, John Wiley & Sons, Chichester.
- 2.Mansy SS, Cowan JA. Iron–sulfur cluster biosynthesis: toward an understanding of cellular machinery and molecular mechanism. Acc Chem Res. 2004;37:719–725. doi: 10.1021/ar0301781. [DOI] [PubMed] [Google Scholar]
- 3.Johnson D, Dean DR, Smith AD, Johnson MK. Structure, function and formation of biological iron–sulfur clusters. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 4.Lill R, Mühlenho U. Iron–sulfur-protein biogenesis in eukaryotes. Trends Biochem Sci. 2005;30:133–141. doi: 10.1016/j.tibs.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 5.Dean DR, Bolin JT, Zheng L. Nitrogenase metalloclusters: structures, organization, and synthesis. J Bacteriol. 1993;175:6737–6744. doi: 10.1128/jb.175.21.6737-6744.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng L, Cash VL, Flint DH, Dean DR. Assembly of iron–sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J Biol Chem. 1998;273:13264–13272. doi: 10.1074/jbc.273.21.13264. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi Y, Tokumoto U. A third bacterial system for the assembly of iron–sulfur clusters with homologs in archaea and plastids. J Biol Chem. 2002;277:28380–28383. doi: 10.1074/jbc.C200365200. [DOI] [PubMed] [Google Scholar]
- 8.Yuvaniyama P, Agar JN, Cash VL, Johnson MK, Dean DR. NifS-directed assembly of a transient [2Fe–2S] cluster within the NifU protein. Proc Natl Acad Sci USA. 2000;97:599–604. doi: 10.1073/pnas.97.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agar JN, Zheng L, Cash VL, Dean DR, Johnson MK. Role of the IscU protein in iron–sulfur cluster biosynthesis: IscS-mediated assembly of a [Fe2S2] cluster in IscU. J Am Chem Soc. 2000;122:2136–2137. [Google Scholar]
- 10.Agar JN, Krebs B, Frazzon J, Huynh BH, Dean DR, Johnson MK. IscU as a scaffold for iron–sulfur cluster biosynthesis: sequential assembly of [2Fe–2S] and [4Fe–4S] clusters in IscU. Biochemistry. 2000;39:7856–7862. doi: 10.1021/bi000931n. [DOI] [PubMed] [Google Scholar]
- 11.Krebs C, Agar JN, Smith AD, Frazzon J, Dean DR, Huynh BH, Johnson MK. IscA, an alternative scaffold for Fe–S cluster biosynthesis. Biochemistry. 2001;40:14069–14080. doi: 10.1021/bi015656z. [DOI] [PubMed] [Google Scholar]
- 12.Ollagnier-de-Choudens S, Mattioli T, Takahashi Y, Fontecave M. Iron–sulfur cluster assembly. Characterization of IscA and evidence for a specific functional complex with ferredoxin. J Biol Chem. 2001;276:22604–22607. doi: 10.1074/jbc.M102902200. [DOI] [PubMed] [Google Scholar]
- 13.Ollagnier-de-Choudens S, Nachin L, Sanakis Y, Loiseau L, Barras F, Fontecave M. SufA from Erwinia chrysanthemi. Characterization of a scaffold protein required for iron–sulfur cluster assembly. J Biol Chem. 2003;278:17993–18001. doi: 10.1074/jbc.M300285200. [DOI] [PubMed] [Google Scholar]
- 14.Ollagnier-de-Choudens S, Sanakis Y, Fontecave M. SufA/IscA: reactivity studies of a class of scaffold proteins involved with [Fe–S] cluster assembly. J Biol Inorg Chem. 2004;9:828–838. doi: 10.1007/s00775-004-0581-9. [DOI] [PubMed] [Google Scholar]
- 15.Wollenberg M, Berndt C, Bill E, Schwenn JD, Seidler A. A dimer of the FeS cluster biosynthesis protein IscA from cyanobacteria binds a [2Fe2S] cluster between two protomers and transfers it to [2Fe2S] and [4Fe4S] apo proteins. Eur J Biochem. 2003;270:1662–1671. doi: 10.1046/j.1432-1033.2003.03522.x. [DOI] [PubMed] [Google Scholar]
- 16.Wu G, Mansy SS, Wu SP, Surerus KK, Foster MW, Cowan JA. Characterization of an iron–sulfur cluster assembly protein (ISU1) from Schizosaccharomyces pombe. Biochemistry. 2002;41:5024–5032. doi: 10.1021/bi016073s. [DOI] [PubMed] [Google Scholar]
- 17.Wu SP, Wu G, Surerus KK, Cowan JA. Iron–sulfur cluster biosynthesis: Kinetic analysis of [2Fe–2S] cluster transfer from holo ISU to apo Fd: role of redox chemistry and a conserved aspartate. Biochemistry. 2002;41:8876–8885. doi: 10.1021/bi0256781. [DOI] [PubMed] [Google Scholar]
- 18.Wu SP, Cowan JA. Iron–sulfur cluster biosynthesis. A comparative kinetic analysis of native and cys-substituted ISA-mediated [2Fe–2S]2+ cluster transfer to an apoferredoxin target. Biochemistry. 2003;42:5784–5791. doi: 10.1021/bi026939+. [DOI] [PubMed] [Google Scholar]
- 19.Zheng L, White RH, Cash VL, Jack RF, Dean DR. Cysteine desulfurase activity indicates a role for NIFS in metallocluster biosynthesis. Proc Natl Acad Sci USA. 1993;90:2754–2758. doi: 10.1073/pnas.90.7.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng L, White RH, Cash VL, Dean DR. A mechanism for the desulfurization of l-cysteine catalyzed by the nifS gene product. Biochemistry. 1994;33:4714–4720. doi: 10.1021/bi00181a031. [DOI] [PubMed] [Google Scholar]
- 21.Patzer SL, Hantke K. SufS is a NifS-like protein, and SufD is necessary for stability of the [2Fe–2S] FhuF protein in Escherichia coli. J Bacteriol. 1999;181:3307–3309. doi: 10.1128/jb.181.10.3307-3309.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loiseau L, Ollagnier-de-Choudens S, Nachin L, Fontecave M, Barras F. Biogenesis of Fe–S cluster by the bacterial Suf system. SufS and SufE form a new type of cysteine desulfurase. J Biol Chem. 2003;278:38352–38359. doi: 10.1074/jbc.M305953200. [DOI] [PubMed] [Google Scholar]
- 23.Ollagnier-de-Choudens S, Lascoux D, Loiseau L, Barras F, Forest E, Fontecave M. Mechanistic studies of the SufS–SufE cysteine desulfurase: evidence for S transfer from SufS to SufE. FEBS Lett. 2003;555:263–267. doi: 10.1016/s0014-5793(03)01244-4. [DOI] [PubMed] [Google Scholar]
- 24.Outten FW, Wood MJ, Muñoz FM, Storz G. The SufE protein and the SufBCD complex enhance SufS cysteine desulfurase activity as part of a sulfur transfer pathway for Fe–S cluster assembly in E. coli. J Biol Chem. 2003;278:45713–45719. doi: 10.1074/jbc.M308004200. [DOI] [PubMed] [Google Scholar]
- 25.Smith AD, Agar JN, Johnson KA, Frazzon J, Amster IJ, Dean DR, Johnson MK. Sulfur transfer from IscS to IscU: the first step in iron–sulfur cluster biosynthesis. J Am Chem Soc. 2001;123:11103–11104. doi: 10.1021/ja016757n. [DOI] [PubMed] [Google Scholar]
- 26.Kato S, Mihara H, Kurihara T, Takahashi Y, Tokumoto U, Yoshimura T, Esaki N. Cys-328 of IscS and Cys-63 of IscU are the sites of disulfide bridge formation in a covalently bound IscS/IscU complex: implications for the mechanism of iron–sulfur cluster assembly. Proc Natl Acad Sci USA. 2002;99:5948–5952. doi: 10.1073/pnas.082123599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramelot TA, Cort JR, Goldsmith-Fischman S, Kornhaber GJ, Xiao R, Shastry R, Acton TB, Honig B, Montelione GT, Kennedy MA. Solution NMR structure of the iron–sulfur cluster assembly protein U (IscU) with zinc bound at the active site. J Mol Biol. 2004;344:567–583. doi: 10.1016/j.jmb.2004.08.038. [DOI] [PubMed] [Google Scholar]
- 28.Goldsmith-Fischman S, Kuzin A, Edstrom WC, Benach J, Shastry R, Xiao R, Acton TB, Honig B, Montelione GT, Hunt JF. The SufE sulfur-acceptor protein contains a conserved core structure that mediates interdomain interactions in a variety of redox protein complexes. J Mol Biol. 2004;344:549–565. doi: 10.1016/j.jmb.2004.08.074. [DOI] [PubMed] [Google Scholar]
- 29.Mihara H, Esaki N. Bacterial cysteine desulfurases: their function and mechanisms. Appl Microbiol Biotechnol. 2002;60:12–23. doi: 10.1007/s00253-002-1107-4. [DOI] [PubMed] [Google Scholar]


