Abstract
Semliki Forest virus (SFV) is an enveloped alphavirus whose membrane fusion is triggered by low pH and promoted by cholesterol and sphingolipid in the target membrane. Fusion is mediated by E1, a viral membrane protein containing the putative fusion peptide. Virus mutant studies indicate that SFV's cholesterol dependence is controlled by regions of E1 outside of the fusion peptide. Both E1 and E1*, a soluble ectodomain form of E1, interact with membranes in a reaction dependent on low pH, cholesterol, and sphingolipid and form highly stable homotrimers. Here we have used detergent extraction and gradient floatation experiments to demonstrate that E1* associated selectively with detergent-resistant membrane domains (DRMs or rafts). In contrast, reconstituted full-length E1 protein or influenza virus fusion peptide was not associated with DRMs. Methyl β-cyclodextrin quantitatively extracted both cholesterol and E1* from membranes in the absence of detergent, suggesting a strong association of E1* with sterol. Monoclonal antibody studies demonstrated that raft association was mediated by the proposed E1 fusion peptide. Thus, although other regions of E1 are implicated in the control of virus cholesterol dependence, once the SFV fusion peptide inserts in the target membrane it has a high affinity for membrane domains enriched in cholesterol and sphingolipid.
Enveloped viruses use cellular membranes during viral entry into the cell via membrane fusion and during virus exit as the source of the virus envelope proteins and lipid bilayer. Viruses have clearly evolved to take advantage of specific cell surface proteins as receptors during entry and/or fusion. Recent reports suggest that some viruses also make use of specific membrane lipids during budding, particularly the laterally segregating cholesterol- and sphingolipid-enriched membrane domains known as rafts (35, 40, 41, 51). These membrane domains are known by a variety of terms, including detergent-resistant membranes (DRMs), due to their relative resistance to solubilization by Triton X-100 (TX-100) in the cold (6). Within cells, DRMs are involved in a variety of important cellular processes, including membrane sorting, signal transduction, and apical targeting (for review, see references 4, 5, 33, and 46). The property of detergent resistance is a function of the lipid composition of the domains (1, 5) and is widely used as an operational definition and an experimental tool.
The enveloped alphavirus Semliki Forest virus (SFV) infects cells via endocytic uptake in clathrin-coated vesicles and low-pH-dependent fusion within the endosome and buds from the plasma membrane (for review, see references 18 and 47). Alphaviruses are icosahedrally symmetrical viruses containing a nucleocapsid composed of the capsid protein and the positive-sense RNA. The nucleocapsid is surrounded by a lipid bilayer containing 80 trimers (E1/E2/E3)3 of the E1 and E2 transmembrane (TM) polypeptides, each of about 50 kDa, and a peripheral E3 polypeptide of about 10 kDa. Virus fusion is mediated by the E1 protein, which contains a highly conserved hydrophobic internal region proposed to be the virus fusion peptide (11). During fusion, E1 interacts with the target bilayer and undergoes specific low-pH-triggered conformational changes that result in exposure of previously masked epitopes and formation of a highly stable E1 homotrimer that appears to be required for fusion (22). A proteolytically truncated ectodomain form of E1, E1*, has been used to follow E1's membrane interactions and conformational changes in the absence of virus fusion (12, 13, 20, 25). The SFV E1 ectodomain has recently been crystallized and characterized structurally (27). The native E1 structure is remarkably similar to that of the fusion protein of the flavivirus tick-borne encephalitis (TBE) virus (39), with the protein lying down on the surface of the virus and the putative fusion peptide located at the tip of the molecule.
An interesting feature of the alphavirus membrane fusion reaction is its requirement for specific lipids in the target membrane (19, 53). Fusion is greatly promoted by the presence of cholesterol and sphingolipid in target liposomes, and fusion and infection are strongly inhibited by depleting cells of cholesterol. Cholesterol and sphingolipids act to promote the low-pH-dependent conformational changes in either E1 or E1*, including the protein's membrane interaction, acid epitope exposure, and homotrimer formation. srf-3 (sterol requirement in function), an SFV mutant with a decreased requirement for cholesterol in fusion, is less dependent on cholesterol for the conformational changes in E1 (8). The decrease in srf-3's cholesterol requirement is due to a single point mutation on the E1 protein, P226→S (50), which lies outside of the putative fusion peptide.
A variety of evidence thus indicates that SFV requires both cholesterol and sphingolipid for fusion. Although this dual requirement is reminiscent of the involvement of these two lipids in the formation of cellular raft domains, no evidence has been presented for raft involvement in SFV fusion. For example, srf-3 has a decreased cholesterol requirement but is unchanged in its sphingolipid requirement, suggesting their independent control (8). Moreover, galactosylceramide, a sphingolipid that does not interact with cholesterol in model monolayer studies, is nonetheless fully active in SFV fusion (36). Given the efficient insertion of the E1 ectodomain into target liposomes at low pH, however, we used this system to examine the association of membrane-bound E1 with the DRM fraction. Our results indicated that the E1 fusion peptide is strongly associated with cholesterol- and sphingolipid-dependent membrane rafts and that it is specifically released following reduction in the cholesterol content of the membrane.
(The data in Fig. 8 are from a thesis to be submitted by D. L. Gibbons in partial fulfillment of the requirements for the Doctor of Philosophy degree from the Sue Golding Graduate Division of Medical Sciences, Albert Einstein College of Medicine, Yeshiva University, Bronx, N.Y.)
FIG. 8.
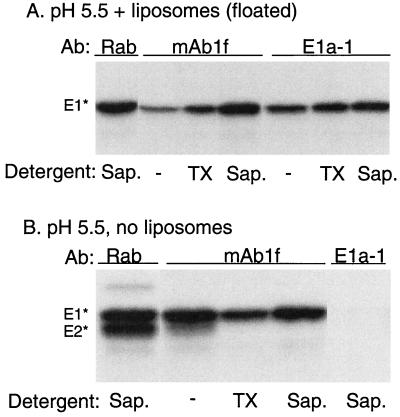
E1* associates with rafts via its fusion peptide. Radiolabeled ectodomains were mixed with 1 mM complete liposomes (A) or buffer (B), and the mixtures were acid treated at pH 5.5 for 3 min at 37°C. The sample for panel A was then floated on a sucrose gradient as described for Fig. 2, and the top four fractions containing liposomes and associated protein were collected and pooled. Aliquots of the panel A and B samples were then, where indicated, extracted for 30 min with either 1% TX-100 on ice or 1% saponin at room temperature. The samples were then immunoprecipitated in the absence of any additional detergent using a polyclonal rabbit antiserum against the SFV envelope proteins (Rab), MAb 1f against E1 residues 85 to 95, or MAb E1a-1 against the acid conformation of E1. A representative example of three experiments is shown. −, no detergent.
MATERIALS AND METHODS
Virus and ectodomain preparation.
The SFV used in these experiments was a well-characterized plaque-purified stock (12) propagated in BHK-21 cells. Virus was biosynthetically labeled with [35S]methionine and purified by banding on sucrose gradients. The soluble ectodomain forms of E1 and E2 were prepared by protease digestion of a mixture of radiolabeled and unlabeled SFV and purified by chromatography on concanavalin A, all as previously described (25). The bromelain fragment of the influenza virus hemagglutinin (BHA) was a kind gift of Judy White and Jennifer Gruenke and was prepared by bromelain digestion of influenza virus and purified by chromatography on ricin as described previously (10).
Antibodies and immunoprecipitation analysis.
A rabbit antiserum to purified envelope proteins was prepared as described previously (21). Monoclonal antibody (MAb) E1a-1 was a previously described mouse MAb against the acid conformation of the SFV E1 protein (21), which was mapped by an antibody-resistant virus mutant to an E1 region in the vicinity of G157 (2). MAb 1f, a mouse MAb against the SFV E1 protein, was a kind gift from Holland Cheng and Lena Hammar and was mapped to residues 85 to 95 by Pepscan (Pepscan Systems) analysis (Lena Hammar, personal communication). Immunoprecipitation was performed by incubation of the samples with buffer, detergent, or methyl β-cyclodextrin (MβCD) as indicated, followed by incubation of the samples with the indicated antibody and retrieval with zysorbin. Samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and fluorography (12) and were quantified by PhosphorImager analysis with ImageQuant, v. 1.2, software (Molecular Dynamics, Sunnyvale, Calif.).
Liposomes and E1 protein analysis.
Liposomes were prepared by extrusion (8), with a molar ratio for complete liposomes of 1:1:1:3 (phosphatidylcholine [PC; from egg yolk]-phosphatidylethanolamine [PE; from egg PC by transphosphatidylation]-sphingomyelin [from bovine brain]-cholesterol) plus trace amounts of [3H]cholesterol or with the same lipids minus either sphingomyelin or cholesterol as indicated. All phospholipids were purchased from Avanti Polar Lipids (Alabaster, Ala.), cholesterol was from Steraloids (Wilton, N.H.), and [3H]cholesterol was from Amersham (Arlington Heights, Ill.). For assays, SFV ectodomains, BHA, or SFV was mixed with liposomes at a final concentration of 0.8 to 1 mM in morpholineethanesulfonic acid (MES)-saline buffer (20 mM MES, 130 mM NaCl), adjusted to the indicated pH by the addition of precalibrated amounts of 0.5 N acetic acid, incubated at 37°C for the indicated time, and then adjusted to neutral pH prior to further analysis. Bovine brain sphingomyelin contains predominantly 18:0 acyl chains that have transition temperatures above 37°C (information supplied by Avanti Polar Lipids), and thus, rafts should be present in the liposomes at the 37°C incubation temperature. Low-pH treatment of BHA was performed at pH 5.0, in keeping with the fusion threshold of influenza virus (10).
Ectodomain-membrane association was assayed by pretreating the samples with the indicated detergent or with MβCD as described below. The samples (usually 150 μl) were then adjusted to 40% sucrose by the addition of a 60% sucrose stock and transferred to a TLS55 tube. Samples were overlaid with 1.4 ml of 25% (wt/vol) sucrose and 200 μl of 5% (wt/vol) sucrose (all ice-cold in 50 mM Tris [pH 7.4]-100 mM NaCl). Similar results for the TX-100 samples were obtained when 0.1% TX-100 was included in the sucrose solutions (data not shown). Gradients were centrifuged for 3 h at 54,000 rpm at 4°C in the TLS55 rotor and fractionated by hand into seven 300-μl fractions. Aliquots were analyzed by scintillation counting for the distribution of [3H]cholesterol, concentrated by acid precipitation, and quantified by SDS-PAGE analysis as described above for the distribution of ectodomains. The distribution of BHA was determined by Western blot analysis using a 1:15,000 dilution of C-HA1, a polyclonal antiserum provided by Jennifer Gruenke and Judy White, and visualized by peroxidase-conjugated secondary antibody and enhanced chemiluminescence. The E1* homotrimer was assayed by solubilization in SDS gel buffer for 3 min at 30°C without reducing agents (12); other samples were routinely reduced to enable the separation of E1*, E2*, and BHA1.
MβCD treatment.
Liposome-associated E1* or BHA was treated for 30 min at 37°C with 20 mM MβCD (Sigma Chemical Co., St. Louis, Mo.), assuming an average molecular weight of 1,311. Samples were then analyzed for membrane association or homotrimer formation as described above. Alternatively, samples were treated for 30 min at 37°C with 20 mM MβCD that was precomplexed with saturating levels of cholesterol (9) (ratio of ∼1 mol of cholesterol to 5.6 mol of MβCD) (Sigma).
Reconstitution of the SFV envelope proteins by detergent dialysis.
Reconstitution of the SFV envelope proteins by detergent dialysis was performed by a method similar to one that was previously published (15). Briefly, 6 μmol of the complete lipid mix described above plus four times this mass of octylglucoside in acetone solution (Fluka Chemical Co.) were dried down on a rotary evaporator, washed twice with chloroform and twice with t-butanol, lyophilized for >90 min, and dissolved in 1 ml of a solution containing 25 mM octylglucoside, 100 μg of purified SFV, and 107 cpm of [35S]methionine-labeled SFV. The clear solution of lipid and virus proteins was then dialyzed against 1 liter of MES-saline buffer with two changes for a total of 39 h. The reconstituted proteoliposomes were then assayed by gradient floatation with or without prior detergent extraction. Similar liposomes were prepared without SFV proteins and used for ectodomain assays.
RESULTS
Lipid requirements and raft association of the E1 ectodomain.
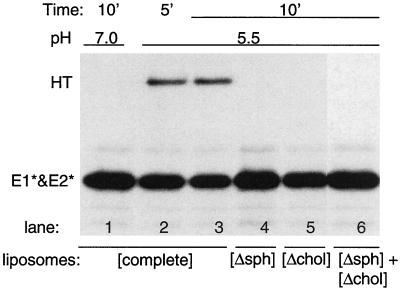
We wished to explore the dual role of cholesterol and sphingolipid in the membrane interaction of the SFV E1 fusion protein. In order to follow membrane interaction in the absence of the E1 TM domain and membrane fusion, we used the previously described E1 ectodomain, E1*, a proteolytically truncated form of E1 active in membrane binding, homotrimer formation, and acid epitope exposure (12, 13, 25). The ectodomain preparation used for these studies contains both E1* and the E2 ectodomain, E2*, but these polypeptides are monomeric and removal of E2* has no effect on the activity of the E1 ectodomain (25). We first tested the ability of various lipid mixtures to support low-pH-triggered E1* homotrimer formation and acid epitope exposure. As previously observed, complete liposomes containing both lipids supported efficient homotrimer formation at low pH (Fig. 1, lanes 2 and 3), while liposomes lacking either cholesterol or sphingolipid were inactive (lanes 4 and 5). Interestingly, mixtures of sphingolipid- and cholesterol-deficient liposomes were also inactive, even though the reaction contained optimal amounts of both lipids (lane 6). Identical results were obtained when E1* was assayed for exposure of the acid-specific MAb E1a-1 epitope (data not shown). Solubilization of complete liposomes with a wide variety of detergents prior to assay resulted in the loss of their ability to support E1* trimerization and epitope exposure (data not shown). Thus, E1* activity was only observed when the protein was treated at low pH in the presence of a lipid bilayer containing both cholesterol and sphingolipid.
FIG. 1.
Lipid requirements for formation of the E1* homotrimer. [35S]methionine-labeled ectodomains were mixed with various types of liposomes at a final lipid concentration of 1 mM. The liposomes were either “complete liposomes” containing PE, PC, cholesterol, and sphingomyelin ([complete]) or were deficient in either sphingomyelin ([Δsph]) or cholesterol ([Δchol]). One sample ([Δsph] + [Δchol]) contained both deficient liposome types, each at a lipid concentration of 1 mM. All samples were preincubated for 5 min at 37°C and then treated at the indicated pH at 37°C for the indicated time. The samples were then solubilized in SDS gel buffer at 30°C in the absence of reduction and alkylation, conditions that preserve the E1* homotrimer (HT) but do not separate E1* and E2*, and analyzed by SDS-PAGE. A representative example of two experiments is shown.
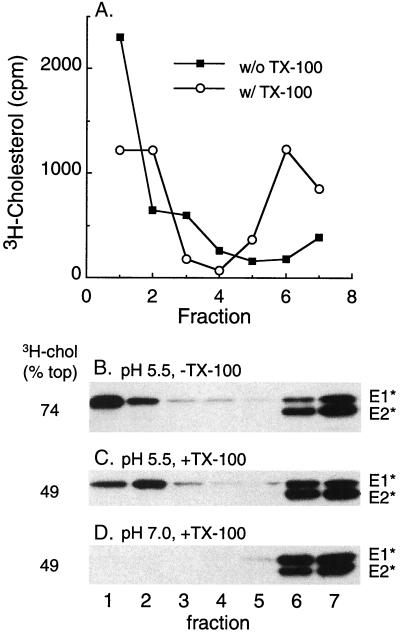
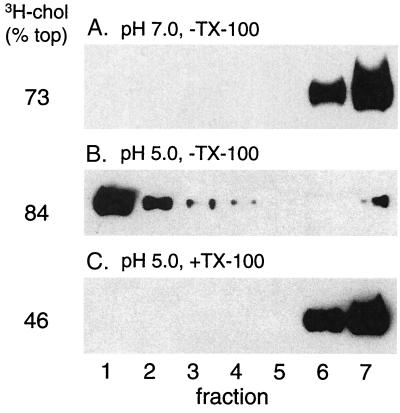
These requirements suggested a possible role for cholesterol- and sphingolipid-enriched raft domains in the activity and/or membrane interaction of E1*. To follow raft domains, the target liposomes were extracted with TX-100 on ice to solubilize the bulk membrane while preserving lipid rafts and were then analyzed by floatation on a sucrose step gradient. Gradient analysis of complete liposomes containing trace amounts of [3H]cholesterol showed that in the absence of detergent extraction, most of the radiolabeled cholesterol was recovered in the top three gradient fractions, with an average of 74% for six experiments (Fig. 2A). Following detergent extraction, a substantial amount of the radiolabeled cholesterol still floated to the top of the gradient (average, 49% for six determinations), demonstrating the presence of a DRM fraction or rafts. We then tested whether E1* was associated with such raft domains after its insertion into the target membrane. Ectodomains were mixed with complete liposomes, treated at pH 5.5 to trigger membrane insertion, and analyzed on sucrose floatation gradients with or without prior extraction in TX-100 on ice. As previously reported, E1* bound to liposomes at low pH and floated to the top of the gradient (Fig. 2B). Such membrane-bound E1* was resistant to carbonate extraction, a standard technique used to demonstrate the intrinsic association of a membrane protein with the lipid bilayer (data not shown). Following extraction with TX-100 on ice, most of this E1* was still found in the top three gradient fractions (Fig. 2C). Floatation with DRMs required prior low-pH-dependent membrane insertion, since in the absence of low-pH treatment no E1* was found in the top of the gradient with or without TX-100 extraction (Fig. 2D and data not shown).
FIG. 2.
Membrane-inserted E1* is associated with DRM domains. (A) Sucrose gradient floatation pattern of [3H]cholesterol-labeled liposomes before and after extraction with TX-100. Complete liposomes containing trace amounts of [3H]cholesterol were extracted on ice for 10 min with 1% TX-100 where indicated and were loaded on the bottom of a sucrose step gradient. The gradients were centrifuged to float the liposomes to the top of the gradient and were fractionated, and the radioactive cholesterol was quantified. Fraction 1 is the top of the gradient. (B to D) [35S]methionine-labeled ectodomains were mixed with complete liposomes at a final lipid concentration of 1 mM, treated at the indicated pH for 10 min at 37°C as for Fig. 1, and then adjusted to neutral pH. The samples were extracted with Triton X-100 on ice where indicated and separated by sucrose gradient floatation as for panel A. The fractions were analyzed by SDS-PAGE and scintillation counting to quantify the position of the viral ectodomains and the [3H]cholesterol, respectively. The percentage of the total [3H]cholesterol recovered in the top three fractions is shown on the left side of the figure and is an average of that obtained for six experiments.
We quantified the efficiency of association of E1* with membranes and DRMs (Table 1) and found that, on average, 56% of the input E1* showed cofloatation with complete liposomes. Following TX-100 extraction, cofloatation was 47%. Thus, >80% of the membrane-inserted E1* was found in the DRM after TX-100 treatment, a high efficiency of DRM association compared to those of influenza virus hemagglutinin (HA) or placental alkaline phosphatase, two typical raft-associated TM proteins (6, 42). Even though about 50% of the cholesterol was present outside of DRM domains, the membrane-inserted E1* was selectively associated with the detergent-resistant membrane fraction. This E1* efficiency was obtained with complete liposomes having a 1:1 molar ratio of cholesterol to phospholipid, the optimal ratio for E1* insertion. Using complete liposomes with a lower cholesterol-to-phospholipid ratio (1:2), the levels of both detergent-resistant [3H]cholesterol and low-pH-induced E1* membrane binding were decreased, but the same high proportion of the membrane-bound E1* was resistant to cold TX-100 extraction (data not shown).
TABLE 1.
Lipid dependence of DRM formation and E1* interactiona
| Liposome compositionb (no. of expts) | [3H]cholesterol (% in the top 3 fractions)
|
E1* (% in the top 3 fractions)
|
||
|---|---|---|---|---|
| Without TX-100 | With TX-100 | Without TX-100 | With TX-100 | |
| Complete (3) | 74 | 50 | 56 | 47 |
| Δchol (1) | 70 | 12 | 0 | 0 |
| Δsph (1) | 85 | 32 | 13 | 0 |
| Δsph + DPPC (1) | 80 | 45 | 3 | 1 |
| Δchol + epicholesterol (2) | 70 | 30 | 1 | 0 |
| Δchol + androstanol (2) | 82 | 26 | 38 | 0 |
| Δchol + coprostanol (2) | 88 | 32 | 11 | 0 |
A mixture of the indicated liposomes (1 mM lipid) and radiolabeled ectodomains was treated at pH 5.5 for 10 min at 37°C and extracted as indicated with TX-100 on ice, and the membrane association was measured by sucrose gradient floatation, scintillation counting ([3H]cholesterol), and SDS-PAGE quantitation (E1*).
Δchol and Δsph refer to cholesterol- and sphingolipid-deficient liposomes, respectively, and replacement of these lipids with DPPC or various sterols is as indicated.
A series of experiments was performed to analyze the membrane and DRM interactions of radiolabeled SFV particles (data not shown). As previously observed (23), SFV showed very efficient cofloatation with complete liposomes after low-pH treatment. In contrast to the E1 ectodomain, extraction with TX-100 in the cold released almost all of the virus envelope proteins from the liposomes. This result is in keeping with the very rapid fusion of virus with complete liposomes, resulting in two populations of E1, a small population in which the fusion peptide is inserted in the target bilayer and a much larger population that only interacts with the liposomes via the TM domain following fusion. This result is also in agreement with experiments using hydrophobic photolabeling to detect membrane insertion, which suggest that the viral E1 not directly involved in mediating fusion is inactive in target membrane insertion (data not shown). Thus, the E1 ectodomain was preferable for use in our present studies since it produced one protein population uniformly and efficiently inserted into the membrane in the absence of virus fusion.
The lipid requirements for DRM formation and E1* membrane insertion were compared by varying the lipid composition of the target liposomes and monitoring the floatation of the [3H]cholesterol marker and E1* with or without cold TX-100 extraction (Table 1). E1* membrane association was dependent on both cholesterol and sphingolipid (also see reference 25), a condition that also promoted the formation of a DRM fraction. Prior studies indicated that dipalmitoylphosphatidylcholine (DPPC) could substitute for sphingomyelin to produce a liquid-ordered DRM domain (1). However, even though DPPC produced DRMs in our liposome system, it did not promote the initial interaction of E1* with the membrane (Table 1), in keeping with the specific requirement for sphingolipid in SFV fusion (36, 53). Although epicholesterol (3α-OH) had been reported to promote DRM formation (54), in our system epicholesterol was substantially less efficient for DRM formation than cholesterol (Table 1). Epicholesterol was also inactive in E1*-membrane interaction and in virus fusion activity, in agreement with the requirement for the sterol 3β-OH group for these activities (Table 1) (23). In contrast, although the sterols androstanol and coprostanol did not efficiently support DRM formation (Table 1) (54), they permit some virus fusion activity (23), and androstanol in particular allowed considerable E1*-membrane interaction (Table 1). The membrane-bound E1* was efficiently solubilized by cold TX-100 extraction of the androstanol liposomes. Thus, the data with DPPC demonstrate that a liquid-ordered domain per se is not sufficient to mediate E1*-membrane interaction, while the cholesterol and androstanol results indicate that the detergent resistance properties of membrane-inserted E1* strongly correlate with those of the sterol present in the target membrane. This results in strong detergent resistance for cholesterol plus E1* and a lack of detergent resistance for androstanol plus E1*.
Specificity of DRM association for the SFV E1 ectodomain.
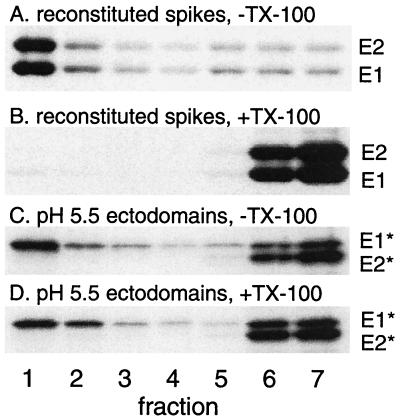
The association of E1* with the DRM fraction could be caused by intrinsic properties of the cholesterol-rich target membrane’s interaction with any membrane-associated protein. To test this, we solubilized full-length viral E1 and E2 and reconstituted them into complete liposomes by detergent dialysis at neutral pH. Both proteins were reconstituted, as determined by cofloatation with liposomes (Fig. 3A) and resistance to carbonate extraction (data not shown), but could be completely solubilized by extraction with TX-100 in the cold (Fig. 3B), in agreement with the known properties of the E1 and E2 proteins in the virus and in infected cells (31, 41). Complete liposomes were prepared in parallel by detergent dialysis and were active for low-pH-triggered E1* binding, conformational changes, and DRM association (Fig. 3C and D and data not shown).
FIG. 3.
The TM domains of reconstituted viral E1 and E2 do not associate with DRMs. The DRM association of the E1 ectodomain and of the full-length SFV E1 and E2 proteins was compared. Radiolabeled full-length SFV E1 and E2 were reconstituted into complete liposomes at neutral pH via detergent dialysis (A and B). Alternatively, radiolabeled ectodomains were mixed with complete liposomes previously prepared by detergent dialysis and treated at pH 5.5 for 10 min at 37°C to trigger insertion (C and D). Samples were then extracted with TX-100 on ice where indicated, separated by sucrose gradient floatation, and analyzed as described for Fig. 2. A representative example of two experiments is shown.
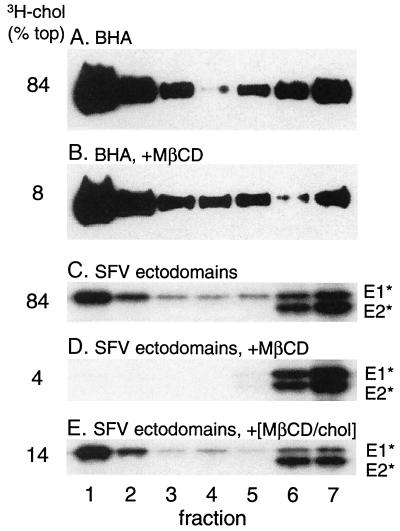
It was also possible that the DRM association of E1* was a general feature of fusion peptides or was somehow triggered by low-pH insertion. We therefore tested BHA, the ectodomain of influenza virus HA, a low-pH-dependent fusion protein without a strong requirement for specific target membrane lipids (10). BHA binding to complete liposomes was efficiently triggered by low-pH treatment (Fig. 4A versus B), but the membrane-associated BHA was completely solubilized by TX-100 in the cold (Fig. 4C). Both the [3H]cholesterol marker and parallel E1* experiments demonstrated the presence of DRM in the same liposome preparations (Fig. 4C and data not shown). Taken together, these data indicate that DRM association is a specific property of the E1 ectodomain following its low-pH-triggered insertion into the target membrane.
FIG. 4.
The influenza HA fusion peptide does not associate with DRMs. BHA was mixed with complete liposomes (1 μg of BHA plus 1 mM lipid), treated for 5 min at the indicated pH, returned to neutral pH, and extracted as indicated with TX-100 on ice. The samples were separated by sucrose gradient floatation and analyzed as described for Fig. 2, except that BHA was detected by immunoblotting (see Materials and Methods). A representative example of two experiments is shown. A pH 7 sample was also extracted with TX-100 and showed identical results to those shown in panel A (data not shown).
Solubilization of E1* by cholesterol extraction from DRM.
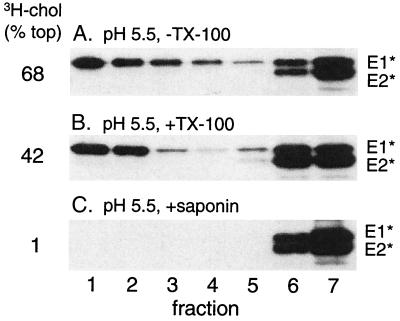
The membrane-associated form of E1* is resistant to solubilization by TX-100 in the cold, as is the [3H]cholesterol marker for DRM. Raft lipids can frequently be solubilized by TX-100 treatment at higher temperatures or by octylglucoside (6). Extraction of complete liposomes under these conditions rather inefficiently solubilized both [3H]cholesterol and E1*, shifting their positions to the middle of the gradient in the case of octylglucoside (data not shown), in keeping with reports on the solubilization of liposomes containing cholesterol and sphingolipids (1). Saponin is a nonionic detergent that interacts with cholesterol (37). It completely solubilized both the [3H]cholesterol and the DRM-associated E1* so that they were retained in the load volume at the bottom of the gradient (Fig. 5C). This detergent treatment, as expected, also completely solubilized other membrane markers such as reconstituted SFV E1 and E2 proteins or BHA (data not shown).
FIG. 5.
Extraction of membrane [3H]cholesterol and E1* with saponin. Radiolabeled ectodomains were mixed with complete liposomes, treated at pH 5.5 for 10 min at 37°C, extracted for 20 min on ice as indicated with either 1% TX-100 or 1% saponin, separated by sucrose gradient floatation, and analyzed as described for Fig. 2. A representative example of two experiments is shown.
Cyclodextrins are cyclic oligosaccharides with a polar surface and a hydrophobic cavity that in the case of β-cyclodextrins has a relatively high affinity for cholesterol versus other lipids (see references 9 and 24 and references therein). β-Cyclodextrins are commonly used as cholesterol acceptors to remove cholesterol from membranes and thus assay for acute effects of cholesterol depletion. Such cyclodextrin-mediated cholesterol depletion has been shown to decrease the DRM fraction, as assayed by subsequent TX-100 solubilization in the cold (24, 42). Conversely, cyclodextrin-cholesterol complexes have also been used to increase the cholesterol content of cell membranes (9). We treated either E1* or BHA at low pH in the presence of complete liposomes containing trace amounts of [3H]cholesterol. These membrane preparations were then treated as indicated with MβCD and assayed by sucrose gradient floatation. BHA showed efficient membrane binding following low-pH treatment (Fig. 6A) and binding was unchanged by exposure to MβCD (Fig. 6B), even though most of the cholesterol was removed from the membrane (8% floating in Fig. 6B versus 84% in Fig. 6A). This is in keeping with an intact but cholesterol-depleted bilayer following MβCD treatment. In addition, similar MβCD treatment did not release reconstituted E1 and E2 proteins from liposomes (data not shown). In contrast, the E1 ectodomain was quantitatively released from the membrane by MβCD treatment (Fig. 6C versus D). Importantly, release occurred even though no TX-100 extraction was performed, and thus E1* differs from many membrane proteins in its requirement for cholesterol in the membrane to maintain membrane association. Release was dependent on cholesterol extraction, since treatment with MβCD that was precomplexed with saturating levels of cholesterol did not release any E1* (Fig. 6E). Interestingly, it appeared that at least part of the labeled pool of liposome cholesterol was able to exchange with the unlabeled cholesterol present in the MβCD complex, since a reduction in the [3H]cholesterol present in the floating liposome fraction was observed (14% versus 84%).
FIG. 6.
Extraction of membrane [3H]cholesterol, BHA, and E1* with MβCD. BHA or SFV ectodomains were mixed with complete liposomes at a final lipid concentration of 1 mM and treated at low pH for 10 min at 37°C to trigger BHA or E1* membrane binding. Samples were adjusted to neutral pH and then treated where indicated for 30 min at 37°C with either 20 mM MβCD or 20 mM MβCD precomplexed with cholesterol (MβCD/chol). Samples were then separated by sucrose gradient floatation and analyzed as described for Fig. 4. A representative example of two experiments is shown.
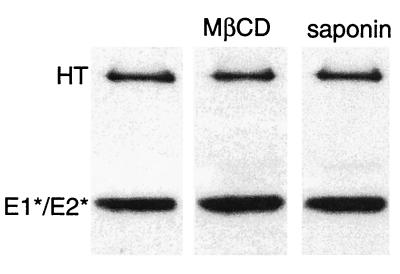
The highly efficient solubilization of E1* by either saponin or cyclodextrin treatment raised the possibility that it was removed from the membrane due to disruption of its homotrimeric structure. Direct assay of solubilized E1* demonstrated that the highly stable homotrimer conformation was preserved following either treatment (Fig. 7). Thus, the E1* homotrimer was specifically released from the target membrane by agents that solubilize or extract cholesterol from the membrane.
FIG. 7.
Extracted E1* is a homotrimer. Radiolabeled ectodomains were mixed with complete liposomes at a final lipid concentration of 1 mM, treated at pH 5.5 for 10 min at 37°C as described for Fig. 1, and then adjusted to neutral pH. The samples were extracted as indicated with 20 mM MβCD for 30 min at 37°C or with 1% saponin for 20 min on ice. The samples were then solubilized in nonreducing SDS gel buffer at 30°C and the presence of the E1 homotrimer (HT) was analyzed by SDS-PAGE as described for Fig. 1. A representative example of two experiments is shown.
Raft association is via the E1 fusion peptide.
The association of the SFV E1 ectodomain with the target bilayer has very similar properties to the fusion of the virus with the target membrane and thus presumably reflects the insertion of the SFV fusion peptide that would drive the viral fusion reaction. However, it is clear from studies of srf mutants that E1 regions outside of the fusion peptide modulate the cholesterol dependence of virus fusion and E1 conformational changes (8), suggesting that other portions of E1 may interact with the target membrane and/or cholesterol. In order to address if the E1*-raft association was mediated by the virus fusion peptide or by some other region of the low-pH form of the E1 molecule, we made use of a MAb termed MAb 1f, which maps to residues 85 to 95 of E1 by Pepscan analysis (Lena Hammar, personal communication). The putative E1 fusion peptide extends approximately from residues 80 to 96, as estimated by sequence conservation and hydrophobicity (11) and by mutagenesis studies (22, 28). We reasoned that following insertion of E1* into the membrane, the MAb 1f epitope should be inaccessible to antibody. If this region mediates raft association, it should be made accessible by saponin and MβCD treatment but not by solubilization with TX-100 in the cold. Ectodomains were treated at low pH in the presence of complete liposomes, and the membranes were floated on a sucrose step gradient. To quantify the total membrane-bound E1*, an aliquot of the floating ectodomains was solubilized with saponin and precipitated with a rabbit polyclonal antiserum. Parallel aliquots were treated under various solubilization conditions and immunoprecipitated with either MAb 1f or the previously described acid conformation-specific MAb E1a-1 (2) (Fig. 8A). MAb 1f showed inefficient precipitation in the absence of detergent (6% of the total in four experiments) and a small increase (to 36% of the total in three experiments) when the sample was solubilized with TX-100 in the cold. Precipitation was strongly increased by saponin solubilization (119% of the total in four experiments) or by treatment with MβCD (48% of the total in two experiments) (data not shown) but not by the MβCD-cholesterol complex (3% of the total in two experiments) (data not shown). In contrast, precipitation by MAb E1a-1, which was mapped to a region outside the putative fusion peptide (2), was efficient in the absence of detergent (42% of the total in three experiments) and was not strongly increased by the addition of either TX-100, saponin, or MβCD (precipitation was 42, 75, and 37% of the total, respectively, in one to three experiments) (Fig. 8A and data not shown). We then tested if these properties of E1* were dependent simply on low-pH treatment or reflected the insertion of the protein into the lipid bilayer. Ectodomains were treated at pH 5.5 in the absence of target membranes and reacted with antibodies following various solubilization conditions (Fig. 8B). MAb 1f efficiently precipitated low-pH-treated E1* (104% of total E1* in two experiments) and showed no change after saponin solubilization (102% of total in two experiments). As observed previously, the MAb E1a-1 epitope was not exposed when E1* was treated at acid pH in the absence of target membranes, in keeping with the lipid requirement for these conformational changes (Fig. 8B) (21). While we cannot exclude interactions of other regions of E1, together these results suggest that the association of E1* with cholesterol-enriched raft domains in the target bilayer is mediated by the putative fusion peptide.
DISCUSSION
The results presented here indicate that both low-pH-triggered liposome insertion and the acid-induced conformational changes in E1* required the presence of cholesterol and sphingolipid in the same target membrane bilayer. Once inserted, membrane-bound E1* was strongly associated with the DRM fraction via the putative fusion peptide. In contrast, although inserted in liposomes of identical lipid composition, neither the E1 TM domain nor the fusion peptide of influenza virus HA was associated with rafts. Analysis of liposome-bound E1* showed that it was resistant to extraction with Triton X-100 in the cold but could be efficiently solubilized using either saponin, a detergent that interacts with cholesterol, or MβCD, a reagent that extracts cholesterol from membranes. What are the general implications of these findings for the alphavirus entry pathway? One membrane domain enriched in both cholesterol and sphingolipids is the caveolar membrane (3, 16). However, cellular and liposomal raft domains can be formed independent of caveolae and caveolin 1 (1), and extensive previous data indicate that SFV enters cells via typical clathrin-coated endocytic vesicles rather than caveolae (for review, see reference 18). In addition, we found (data not shown) that SFV efficiently infected FRT cells that are deficient in caveolae and caveolin 1 protein (30), and recent studies demonstrated that SFV is internalized via a caveolin 1-negative compartment (38). Studies of endosomal lipids have demonstrated that DRMs are abundant in early and recycling endosomes (33). Together, these results fit best with a model in which SFV enters cells via receptor-mediated endocytosis, fusion is triggered at low pH, and the fusion peptide interacts with DRM domains in the endosome membrane. The involvement of both sterol and sphingolipid in the formation and interaction of the raft domain with E1 may explain previous findings showing that under some conditions sterol-depleted cells could adapt to support more efficient SFV fusion (32). It is possible that the raft properties or sphingolipid composition of these cells was altered to a more fusion-permissive state.
In general, raft-associated membrane proteins have TM domains or a GPI linkage that causes their lateral segregation into cholesterol- and sphingolipid-enriched domains in the Golgi compartment (4, 46). For example, the TM domain of influenza virus HA associates with DRM to mediate the membrane traffic and apical targeting of HA and the selective recruitment of raft domains into the budding virus particle (41, 42). Interestingly, although the HA TM domain clearly has an affinity for DRMs, our data indicate that the HA fusion peptide does not. The cellular membrane proteins known as SNAREs have recently been localized to cholesterol-dependent clusters that are required for efficient exocytic fusion (7, 26). In contrast to these intrinsic membrane proteins, the SFV E1 ectodomain inserts into membranes de novo upon low-pH treatment. Insertion occurs via the SFV fusion peptide, which in the crystal structure of native E1* forms a disulfide-stabilized loop at one end of the molecule (27). These characteristics of E1* show intriguing similarities to those of other soluble proteins that can interact with cholesterol-rich DRMs. Caveolin, the major structural protein of caveolae, can cycle between a soluble form and an oligomeric form associated with DRMs in the cytoplasmic face of the plasma membrane (3, 16). Reconstitution of caveolin into liposomes is cholesterol dependent (29, 34), and caveolin binds cholesterol at an approximately equimolar stoichiometry (34). The membrane interaction of caveolin is proposed to be mediated at least in part by a hairpin loop in approximately the center of the molecule (3). Perhaps an even greater similarity to the E1* membrane interaction is discernible in the cholesterol-dependent cytolysins. These soluble proteins, exemplified by θ-toxin, convert to a membrane-bound, oligomeric form in a reaction that requires cholesterol in the target membrane (49). The θ-toxin-membrane interaction is mediated by the conformational change of an α-helical region to two β-hairpin loops that insert into the membrane (43). Recent studies have shown that θ-toxin preferentially interacts with cholesterol present in membrane raft domains (52). Although no sequence similarities between E1*, caveolin, and θ-toxin are apparent, one common structural feature of these three proteins is a hairpin or loop that inserts into the membrane. Although considerable mutagenesis of the fusion peptide region of SFV has been performed (22, 28, 45), it is not yet clear which residues are important in DRM interaction or whether specific amino acids shown to be critical for fusion and homotrimer formation might also be required for raft association. More generally, might DRM association be a common feature of internal viral fusion peptides that insert into the target membrane as a loop? The flavivirus TBE virus has an internal fusion peptide loop. TBE fusion does not require sphingolipid but is strongly facilitated by cholesterol, suggesting that it may involve cholesterol but perhaps not a typical DRM (14). For viral fusion reactions that have not been reconstituted in vitro, definitive experiments are more difficult since they are based on the depletion of cellular cholesterol, a process that may generate significant nonspecific effects (19). Clearly, further studies are needed to define the roles of cholesterol, sphingolipids, DRM, and other lipid components in virus fusion and infection.
Taken in the context of our understanding of the alphavirus fusion reaction, several unexpected but important concepts are suggested by the results reported here. First, the insertion of the fusion peptide into the target membrane and its association with DRM appears to reflect specific E1*-lipid interactions rather than an explicit requirement for rafts in fusion. Thus, although replacement of sphingolipid with DPPC produced a DRM as previously reported (1), this target bilayer composition did not support E1* insertion or acid-dependent conformational changes. Conversely, although androstanol did not efficiently generate a DRM (54), it supported fairly efficient E1* insertion and was previously shown to support SFV fusion (23). These data are reminiscent of results with synaptophysin and SNARE proteins, which were directly demonstrated to interact with cholesterol in photolabeling studies but did not associate with DRM (26, 48).
Secondly, once inserted, E1* could be quantitatively removed from the liposomes by MβCD treatment in the absence of detergent. The released E1* no longer floated in sucrose gradients, and preliminary gradient centrifugation experiments suggested that much of the protein sedimented as a trimer (data not shown). These results differ from those of previous studies of the release of the immunoglobulin E receptor and the GPI-linked protein Thy-1 from cells treated with MβCD, in which only a fraction of the protein was released from cells, primarily as membrane vesicles that could be floated in sucrose gradients or sedimented by high-speed centrifugation (17, 44). It is not yet clear if the released E1* homotrimer can reinsert into bilayers, if such reinsertion would be cholesterol dependent, or if other raft-associated membrane proteins might be releasable by a similar experimental mechanism.
Lastly, data from three SFV srf mutants indicate that their decreased cholesterol requirements for fusion are conferred by single amino acid changes in E1, all of which lie outside of the fusion peptide (50) (Chatterjee and Kielian, unpublished data). srf-3, the first such mutant isolated, has a single point mutation of E1 (P226→S) and is less cholesterol dependent for fusion, E1 homotrimer formation, and E1 acid epitope exposure (8). The mutant data thus suggest that cholesterol interacts with E1 regions outside of the fusion peptide to promote fusogenic conformational changes, but unexpectedly, our present results indicate that the fusion peptide itself has a strong association with cholesterol-enriched membranes.
One possible model to encompass these findings is that regions outside of the fusion peptide regulate the E1 conformational changes to ensure that the fusion peptide will only be inserted into membranes containing the optimal lipid components. The strong correlation between the TX-100 solubility properties of E1* and those of the liposome sterol and the ability of MβCD to simultaneously extract both cholesterol and E1* suggest that sterol might be tightly associated with the membrane-interacting fusion peptide of E1. Such sterol association could be important for clustering the fusion peptide, increasing its local concentration in the membrane to promote fusion.
Acknowledgments
We thank Deborah Brown for many helpful discussions, references, and experimental suggestions about detergent-resistant membranes and for her insightful comments on the manuscript. We thank Lena Hammar and Holland Cheng for generously providing MAb 1f and for helpful discussions. We thank Jennifer Gruenke and Judy White for the generous gift of BHA and C-HA1 antibody and for helpful advice on their use. We thank all the members of our lab for their comments and input, Duncan Wilson and the members of our lab for critical reading of the manuscript, and Christina Eng for assistance with tissue culture.
This work was supported by grants to M.K. from the Public Health Service (R01 GM52929, R01 GM57454), by the Jack K. and Helen B. Lazar Fellowship in Cell Biology, and by Cancer Center Core support grant NIH/NCI P30-CA13330. D.L.G. was supported through the Medical Scientist Training Program (NIH T32 GM07288).
REFERENCES
- 1.Ahmed, S. N., D. A. Brown, and E. London. 1997. On the origin of sphingolipid/cholesterol-rich detergent-insoluble cell membranes: physiological concentrations of cholesterol and sphingolipid induce formation of a detergent-insoluble, liquid-ordered lipid phase in model membranes. Biochemistry 36:10944-10953. [DOI] [PubMed] [Google Scholar]
- 2.Ahn, A., M. R. Klimjack, P. K. Chatterjee, and M. Kielian. 1999. An epitope of the Semliki Forest virus fusion protein exposed during virus-membrane fusion. J. Virol. 73:10029-10039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, R. G. W. 1998. The caveolae membrane system. Annu. Rev. Biochem. 67:199-225. [DOI] [PubMed] [Google Scholar]
- 4.Brown, D. A., and E. London. 1998. Functions of lipid rafts in biological membranes. Annu. Rev. Cell Dev. Biol. 14:111-136. [DOI] [PubMed] [Google Scholar]
- 5.Brown, D. A., and E. London. 2000. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J. Biol. Chem. 275:17221-17224. [DOI] [PubMed] [Google Scholar]
- 6.Brown, D. A., and J. K. Rose. 1992. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 68:533-544. [DOI] [PubMed] [Google Scholar]
- 7.Chamberlain, L. H., R. D. Burgoyne, and G. W. Gould. 2001. SNARE proteins are highly enriched in lipid rafts in PC12 cells: implications for the spatial control of exocytosis. Proc. Natl. Acad. Sci. USA 98:5619-5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatterjee, P. K., M. Vashishtha, and M. Kielian. 2000. Biochemical consequences of a mutation that controls the cholesterol dependence of Semliki Forest virus fusion. J. Virol. 74:1623-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christian, A. E., M. P. Haynes, M. C. Phillips, and G. H. Rothblat. 1997. Use of cyclodextrins for manipulating cellular cholesterol content. J. Lipid Res. 38:2264-2272. [PubMed] [Google Scholar]
- 10.Doms, R. W., A. Helenius, and J. White. 1985. Membrane fusion activity of the influenza virus hemagglutinin. J. Biol. Chem. 260:2973-2981. [PubMed] [Google Scholar]
- 11.Garoff, H., A.-M. Frischauf, K. Simons, H. Lehrach, and H. Delius. 1980. Nucleotide sequence of cDNA coding for Semliki Forest virus membrane glycoproteins. Nature 288:236-241. [DOI] [PubMed] [Google Scholar]
- 12.Gibbons, D. L., A. Ahn, P. K. Chatterjee, and M. Kielian. 2000. Formation and characterization of the trimeric form of the fusion protein of Semliki Forest virus. J. Virol. 74:7772-7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibbons, D. L., and M. Kielian. 2002. Molecular dissection of the Semliki Forest virus homotrimer reveals two functionally distinct regions of the fusion protein. J. Virol. 76:1194-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heinz, F. X., and S. L. Allison. 2000. Structures and mechanisms in flavivirus fusion. Adv. Virus Res. 55:231-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helenius, A., E. Fries, and J. Kartenbeck. 1977. Reconstitution of Semliki Forest virus membrane. J. Cell Biol. 75:866-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikonen, E., and R. G. Parton. 2000. Caveolins and cellular cholesterol balance. Traffic 1:212-217. [DOI] [PubMed] [Google Scholar]
- 17.Ilangumaran, S., and D. C. Hoessli. 1998. Effects of cholesterol depletion by cyclodextrin on the sphingolipid microdomains of the plasma membrane. Biochem. J. 335:433-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kielian, M. 1995. Membrane fusion and the alphavirus life cycle. Adv. Virus Res. 45:113-151. [DOI] [PubMed] [Google Scholar]
- 19.Kielian, M., P. K. Chatterjee, D. L. Gibbons, and Y. E. Lu. 2000. Specific roles for lipids in virus fusion and exit: examples from the alphaviruses, p. 409-455. In H. Hilderson and S. Fuller (ed.), Subcellular biochemistry, vol. 34. Fusion of biological membranes and related problems. Plenum Publishers, New York, N.Y. [DOI] [PubMed] [Google Scholar]
- 20.Kielian, M., and A. Helenius. 1985. pH-induced alterations in the fusogenic spike protein of Semliki Forest virus. J. Cell Biol. 101:2284-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kielian, M., S. Jungerwirth, K. U. Sayad, and S. DeCandido. 1990. Biosynthesis, maturation, and acid activation of the Semliki Forest virus fusion protein. J. Virol. 64:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kielian, M., M. R. Klimjack, S. Ghosh, and W. A. Duffus. 1996. Mechanisms of mutations inhibiting fusion and infection by Semliki Forest virus. J. Cell Biol. 134:863-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kielian, M. C., and A. Helenius. 1984. The role of cholesterol in the fusion of Semliki Forest virus with membranes. J. Virol. 52:281-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kilsdonk, E. P., P. G. Yancey, G. W. Stoudt, F. W. Bangerter, W. J. Johnson, M. C. Phillips, and G. H. Rothblat. 1995. Cellular cholesterol efflux mediated by cyclodextrins. J. Biol. Chem. 270:17250-17256. [DOI] [PubMed] [Google Scholar]
- 25.Klimjack, M. R., S. Jeffrey, and M. Kielian. 1994. Membrane and protein interactions of a soluble form of the Semliki Forest virus fusion protein. J. Virol. 68:6940-6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lang, T., D. Bruns, D. Wenzel, D. Riedel, P. Holroyd, C. Thiele, and R. Jahn. 2001. SNAREs are concentrated in cholesterol-dependent clusters that define docking and fusion sites for exocytosis. EMBO J. 20:2202-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lescar, J., A. Roussel, M. W. Wien, J. Navaza, S. D. Fuller, G. Wengler, and F. A. Rey. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137-148. [DOI] [PubMed] [Google Scholar]
- 28.Levy-Mintz, P., and M. Kielian. 1991. Mutagenesis of the putative fusion domain of the Semliki Forest virus spike protein. J. Virol. 65:4292-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, S., K. S. Song, and M. P. Lisanti. 1996. Expression and characterization of recombinant caveolin. Purification by polyhistidine tagging and cholesterol-dependent incorporation into defined lipid membranes. J. Biol. Chem. 271:568-573. [PubMed] [Google Scholar]
- 30.Lipardi, C., R. Mora, V. Colomer, S. Paladino, L. Nitsch, E. Rodriguez-Boulan, and C. Zurzolo. 1998. Caveolin transfection results in caveolae formation but not apical sorting of glycosylphosphatidylinositol (GPI)-anchored proteins in epithelial cells. J. Cell Biol. 140:617-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu, Y. E., and M. Kielian. 2000. Semliki Forest virus budding: assay, mechanisms, and cholesterol requirement. J. Virol. 74:7708-7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marquardt, M. T., and M. Kielian. 1996. Cholesterol-depleted cells that are relatively permissive for Semliki Forest virus infection. Virology 224:198-205. [DOI] [PubMed] [Google Scholar]
- 33.Mukherjee, S., and F. R. Maxfield. 2000. Role of membrane organization and membrane domains in endocytic lipid trafficking. Traffic 1:203-211. [DOI] [PubMed] [Google Scholar]
- 34.Murata, M., J. Peränen, R. Schreiner, F. Wieland, T. V. Kurzchalia, and K. Simons. 1995. VIP21/caveolin is a cholesterol-binding protein. Proc. Natl. Acad. Sci. USA 92:10339-10343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen, D. H., and J. E. K. Hildreth. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 74:3264-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nieva, J. L., R. Bron, J. Corver, and J. Wilschut. 1994. Membrane fusion of Semliki Forest virus requires sphingolipids in the target membrane. EMBO J. 13:2797-2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishikawa, M., S. Nojima, T. Akiyama, U. Sankawa, and K. Inoue. 1984. Interaction of digitonin and its analogs with membrane cholesterol. J. Biochem. 96:1231-1239. [DOI] [PubMed] [Google Scholar]
- 38.Pelkmans, L., J. Kartenbeck, and A. Helenius. 2001. Caveolar endocytosis of simian virus 40 reveals a new two-step vesicular-transport pathway to the ER. Nat. Cell Biol. 3:473-483. [DOI] [PubMed] [Google Scholar]
- 39.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2Å resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 40.Rousso, I., M. B. Mixon, B. K. Chen, and P. S. Kim. 2000. Palmitoylation of the HIV-1 envelope glycoprotein is critical for viral infectivity. Proc. Natl. Acad. Sci. USA 97:13523-13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheiffele, P., A. Rietveld, T. Wilk, and K. Simons. 1999. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J. Biol. Chem. 274:2038-2044. [DOI] [PubMed] [Google Scholar]
- 42.Scheiffele, P., M. G. Roth, and K. Simons. 1997. Interaction of influenza virus haemagglutinin with sphingolipid-cholesterol membrane domains via its transmembrane domain. EMBO J. 16:5501-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shatursky, O., A. P. Heuck, L. A. Shepard, J. Rossjohn, M. W. Parker, A. E. Johnson, and R. K. Tweten. 1999. The mechanism of membrane insertion for a cholesterol-dependent cytolysin: a novel paradigm for pore-forming toxins. Cell 99:293-299. [DOI] [PubMed] [Google Scholar]
- 44.Sheets, E. D., D. Holowka, and B. Baird. 1999. Critical role for cholesterol in Lyn-mediated tyrosine phosphorylation of FɛRI and their association with detergent-resistant membranes. J. Cell Biol. 145:877-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shome, S. G., and M. Kielian. 2001. Differential roles of two conserved glycine residues in the fusion peptide of Semliki Forest virus. Virology 279:146-160. [DOI] [PubMed] [Google Scholar]
- 46.Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature 387:569-572. [DOI] [PubMed] [Google Scholar]
- 47.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thiele, C., M. J. Hannah, F. Fahrenholz, and W. B. Huttner. 2000. Cholesterol binds to synaptophysin and is required for biogenesis of synaptic vesicles. Nat. Cell Biol. 2:42-49. [DOI] [PubMed] [Google Scholar]
- 49.Tweten, R. K. 1995. Pore-forming toxins of the gram-positive bacteria, p. 207-230. In J. A. Roth, C. A. Bolin, K. A. Brogden, C. Minion, and M. J. Wannemuehler (ed.), Virulence mechanisms of bacterial pathogens. American Society for Microbiology, Washington, D.C.
- 50.Vashishtha, M., T. Phalen, M. T. Marquardt, J. S. Ryu, A. C. Ng, and M. Kielian. 1998. A single point mutation controls the cholesterol dependence of Semliki Forest virus entry and exit. J. Cell Biol. 140:91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vincent, S., D. Gerlier, and S. N. Manie. 2000. Measles virus assembly within membrane rafts. J. Virol. 74:9911-9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waheed, A. A., Y. Shimada, H. F. Heijnen, M. Nakamura, M. Inomata, M. Hayashi, S. Iwashita, J. W. Slot, and Y. Ohno-Iwashita. 2001. Selective binding of perfringolysin O derivative to cholesterol-rich membrane microdomains (rafts). Proc. Natl. Acad. Sci. USA 98:4926-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilschut, J., J. Corver, J. L. Nieva, R. Bron, L. Moesby, K. C. Reddy, and R. Bittman. 1995. Fusion of Semliki Forest virus with cholesterol-containing liposomes at low pH: a specific requirement for sphingolipids. Mol. Membr. Biol. 12:143-149. [DOI] [PubMed] [Google Scholar]
- 54.Xu, X., and E. London. 2000. The effect of sterol structure on membrane lipid domains reveals how cholesterol can induce lipid domain formation. Biochemistry 39:843-849. [DOI] [PubMed] [Google Scholar]