Abstract
Little is known about the mechanisms of transport of neurotropic herpesviruses, such as herpes simplex virus (HSV), varicella-zoster virus, and pseudorabies virus, within neurons. For these viruses, which replicate in the nucleus, anterograde transport from the cell body of dorsal root ganglion (DRG) neurons to the axon terminus occurs over long distances. In the case of HSV, unenveloped nucleocapsids in human DRG neurons cocultured with autologous skin were observed by immunoelectron microscopy to colocalize with conventional ubiquitous kinesin, a microtubule-dependent motor protein, in the cell body and axon during anterograde axonal transport. Subsequently, four candidate kinesin-binding structural HSV proteins were identified (VP5, VP16, VP22, and US11) using oligohistidine-tagged human ubiquitous kinesin heavy chain (uKHC) as bait. Of these viral proteins, a direct interaction between uKHC and US11 was identified. In vitro studies identified residues 867 to 894 as the US11-binding site in uKHC located within the proposed heptad repeat cargo-binding domain of uKHC. In addition, the uKHC-binding site in US11 maps to the C-terminal RNA-binding domain. US11 is consistently cotransported with kinetics similar to those of the capsid protein VP5 into the axons of dissociated rat neurons, unlike the other tegument proteins VP16 and VP22. These observations suggest a major role for the uKHC-US11 interaction in anterograde transport of unenveloped HSV nucleocapsids in axons.
Herpes simplex virus (HSV) consists of four structural components: a DNA core enclosed in a capsid, a layer of proteins designated tegument, and a lipid envelope studded with virally encoded glycoproteins. In humans, HSV enters via cells lining mucous membranes and then infects the termini of dorsal root ganglion (DRG) neurons innervating the portal of entry into the body. From there, HSV is transported retrogradely in axons to the cell body, where it becomes latent. Reactivation is followed by anterograde transport into skin or mucous membranes of the same dermatome involved in the initial infection. Reactivation of HSV from latency during a patient's lifetime may be frequent, resulting in symptomatic disease or, more commonly, unrecognized lesions and asymptomatic shedding (30).
Early work showed that retrograde axonal transport of unenveloped HSV nucleocapsid in rat neurons utilizes microtubules (17). Recently, it has been reported that transport within cells in culture probably involves the microtubule-dependent motor protein dynein and possibly the HSV tegument protein UL34 (41, 46).
Previously, we have used a two-chamber system to examine the mechanism of anterograde transport of HSV from infected DRG neurons in the central chamber along axonal fascicles to autologous epidermal explants in the outer chamber. The velocity of viral transport (0.6 μm/s) indicated a mechanism based on fast anterograde transport (28). Using freeze-substitution transmission immunoelectron microscopy, we have shown that HSV is transported as unenveloped nucleocapsids coated with tegument proteins and separately from glycoproteins, which are transported in vesicles (13).
Separate axonal transport of HSV structural components was supported by experiments with nocodazole, an inhibitor of microtubule polymerization, and brefeldin A (BFA), which inhibits transport through the Golgi apparatus. The transport of all three viral components--nucleocapsid, tegument, and glycoproteins--was inhibited by nocodazole, indicating their dependence on microtubule-associated transport. However, BFA inhibited glycoprotein transport but not nucleocapsids, confirming that each is transported along a separate pathway and indicating the close association of HSV glycoproteins with Golgi membranes (23).
Fast anterograde axonal transport of another alphaherpesvirus, pseudorabies virus (PRV), has also been observed in dissociated chick DRG neurons (40). Recently, transport of glycoproteins, but not capsid proteins, into axons has been shown to be dependent on a type II integral membrane viral protein, US9, which is conserved in all alphaherpeviruses, including HSV (2, 3, 42). Deletion of this protein prevents transport of glycoproteins. These results also confirm the separate pathways of anterograde transport of nucleocapsids and glycoprotein in alphaherpesviruses.
The motor protein candidates for anterograde viral transport should therefore be those involved in microtubule-dependent fast axonal anterograde transport of organelles. These include the conventional kinesins (ubiquitous and neuronal) (12, 24, 25, 36) and the kinesin-related protein KIF3 (16, 44). Conventional kinesin is a heterotetramer consisting of two identical heavy and two identical light chains (1). The heavy chain has a three-domain structure consisting of an N-terminal motor domain, highly conserved among the kinesin superfamily; a central stalk domain containing heptad repeats; and a C-terminal tail domain (24, 25). KIF3 consists of two nonidentical heavy chains and an associated accessory protein, KAP3. Like conventional kinesin, the heavy chains have an N-terminal motor domain, a stalk, and tail domain (44).
Our hypothesis is that anterograde axonal transport of HSV nucleocapsids is mediated by a direct interaction between kinesin and viral nucleocapsid or tegument protein. This study shows that ubiquitous conventional kinesin is likely to be a major molecular motor responsible for anterograde axonal transport of HSV nucleocapsids. In addition, the HSV tegument protein US11 was shown to bind to human ubiquitous kinesin heavy chain (uKHC).
MATERIALS AND METHODS
HSV infection of neurons.
Generation of virus stocks and titrations of wild-type (wt) HSV type 1 (HSV-1) strain CW1 were performed in HEp-2 cells as previously described (23). The rat and human neuronal culture systems described above were used to examine anterograde transport of HSV in dissociated rat neonatal neurons (23) or a two-chamber system containing human fetal DRG neurons whose axons grow out through an agarose barrier to interact with epidermal explants in the outer chamber (13). Human fetal tissues were obtained after informed consent under protocols approved by the Western Sydney Area Health Services Ethics Committee. Preparation and culture of dissociated rat neonatal and human fetal DRG neurons, infection with HSV-1, and immunofluorescence, confocal, and electron microscopy were performed as previously described (13, 23). For transmission immunoelectron microscopy, cold-water fish skin gelatin (Sigma) was used to block tissue sections prior to immunolabeling. Treatment of HSV-infected dissociated rat neonatal neurons with BFA at 2 μg/ml from 3 to 24 h postinfection was performed as described previously (23).
Expression constructs.
The generation of the N-terminal oligohistidine-tagged uKHC constructs 555-772, 771-963, and 855-963 (uKHC amino acid numbering) along with untagged human kinesin light chain (KLC; residues 4 to 569) were described previously (8). Other fragments of uKHC were amplified from plasmid pET-28a/uKHC555-963 (8). PCR fragments corresponding to residues 555 to 854, 555 to 866, and 555 to 894 were ligated into the BamHI/EcoRI-digested plasmid pET-28a (Novagen). PCR fragments corresponding to residues 867 to 963 and 895 to 963 were ligated into the BamHI site of plasmid pET-28a. Oligohistidine-tagged neuronal KHC spanning amino acids 408 to 1032 was constructed by ligating an FspI/EcoRI neuronal-KHC-containing fragment from pWBC7 (25) (provided by R. Vale) into BamHI (Klenow filled in)/EcoRI-digested pET-28b (Novagen). The glutathione-S-transferase (GST)-tagged uKHC555-963 plasmid construct has been described elsewhere (8). To generate a GST-KLC plasmid construct, the NcoI (Klenow filled in)/EcoRI cDNA fragment containing human KLC4-569 from pET-28a/KLC (8) was first ligated into BamHI (Klenow filled in)/EcoRI-digested pET-28a before being ligated into the BamHI site of pGEX-2T (Amersham Pharmacia).
An untagged full-length US11 construct was generated by digestion of plasmid pRB4766, which contains HSV-1 US11 genomic DNA in pGEX-KG (4), with NcoI and its ligation into the NcoI site of pET-28a. This adds five amino acids (MGRLE) to the N terminus of US11. The untagged US11 N-terminal fragment (residues 1 to 84) and C-terminal fragment (residues 84 to 161) were amplified from plasmid pET-28a/US111-161 and ligated into NcoI/BamHI-digested or NcoI/XhoI-digested pET-28a, respectively. Oligohistidine-tagged US11 was constructed by ligating an EcoRI/FspI US11-containing fragment from pRB4766 into EcoRI/HindIII (Klenow filled in)-digested pET-28c. Between the oligohistidine tag and the US11 sequence were inserted amino acids LDSMGRLE.
Genomic DNA containing HSV-1 VP16 was provided by P. O'Hare (14, 26). The plasmid pPO54 consists of the gene for VP16 ligated into the BamHI site of pBS. An untagged VP16 construct was generated by first digesting pPO54 with BamHI and ligating it into the BamHI site of pET-28c. VP16 was subsequently released from pET-28c by digestion with BamHI/XhoI and ligation into the yeast vector pACT2 (Clontech), also cut with BamHI/XhoI. VP16 was then released with an NcoI/XhoI digest and ligated into the NcoI/XhoI site of pET-28a to allow expression of untagged VP16. The resulting fusion protein had an additional 19 amino acids (MEAPGIRDPRSSFPYQPHP) at the N terminus of VP16.
Antibodies.
Oligohistidine-tagged uKHC555-772 expressed in Escherichia coli was used to generate chicken polyclonal anti-uKHC. The cell pellet from a 1-liter bacterial culture expressing oligohistidine-tagged uKHC555-772 was resuspended in 25 ml of column running buffer (20 mM sodium diphosphate [pH 7.4], 0.5 M NaCl) and lysed by sonication. The lysate was clarified by centrifugation before being applied to a POROS 20 metal chelating column (4.6 by 10 mm; Perseptive Biosystems) which had been charged with nickel according to the manufacturer's instructions. The oligohistidine-tagged fusion protein was eluted at 5 ml/min with 15 column volumes of a gradient from 0 to 0.5 M imidazole in running buffer. Pooled fractions containing purified oligohistidine-tagged uKHC555-772 were then dialyzed against phosphate-buffered saline. Protein concentrations were determined using a Bio-Rad protein assay. A total of 2 mg of protein was then used to raise a polyclonal antibody in chickens (the service was provided by the Institute of Medical and Veterinary Science, Veterinary Services Division, Gilles Plains, Australia). Pre- and postimmune sera from a single immunized chicken were subsequently used in this study.
Other antibodies used included mouse monoclonal antibodies against US11 (32), VP16 (21) (LP1; provided by T. Minson), gC1, and KLC (L2; Chemicon International) and rabbit polyclonal antibodies against HSV-1 (Dako), VP5 (5) (provided by G. Cohen and R. Eisenberg), and VP22 (10) (AGV30; provided by P. O'Hare). A rabbit polyclonal antibody raised against the C-terminal 11 amino acids of US11 was provided by H. Marsden (20).
Generation of 35S-labeled and unlabeled infected cell lysates.
HEp-2 cells were infected with HSV-1 at a multiplicity of infection of 5 PFU/cell and incubated for 16 h (35S labeling) or 24 h (unlabeled). For labeling, the cell monolayers were washed once with phosphate-buffered saline before the addition of 100 μCi of Trans35S-label (>1,000 μCi/nmol; ICN)/ml in Dulbecco's modified Eagle's medium (minus cysteine, methionine, and glutamine; ICN) supplemented with 2 mM glutamine. The cells were labeled for 3 h at 37°C. Both labeled and unlabeled cells were harvested, washed twice with phosphate-buffered saline, and then lysed in phosphate-buffered saline (106 cells/ml) containing 1% (vol/vol) NP-40, 5 μg of leupeptin/ml, and 1 mM phenylmethylsulfonyl fluoride. The cells were vortexed for 15 s and then placed on ice for 30 s. This was repeated two more times before final incubation on ice for 30 min. The soluble fraction was harvested by centrifugation at 4°C (10,000 × g for 20 min).
Binding assays.
The oligohistidine-tagged uKHC proteins were expressed, harvested, bound to, and eluted from nickel-activated beads as previously described (8). Bacteria expressing untagged US11, VP16, or KLC were lysed in normal-salt binding buffer (150 mM NaCl, 5 mM imidazole, 20 mM Tris-HCl [pH 7.9], 0.1% [vol/vol] Triton X-100, 5 μg of leupeptin/ml, and 1 mM phenylmethylsulfonyl fluoride) for use in binding assays. The soluble fractions were harvested by centrifugation at 4°C (10,000 × g for 20 min). Bacterial lysates containing recombinant HSV-1 proteins (1 ml) or mock-infected and infected HEp-2 cell lysates (35S labeled or unlabeled; 500 μl) were added to oligohistidine-tagged uKHC fragments on nickel-activated beads. The beads were incubated overnight with rocking at 4°C and washed four times with 20 volumes of wash buffer (150 mM NaCl, 120 mM imidazole, 20 mM Tris-HCl [pH 7.9]) prior to the elution of bound protein complexes. The protein complexes were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose, and identified by immunoblotting, as previously described (8), or by autoradiography.
GST-tagged proteins were expressed in E. coli strain BL21. Bacteria were grown in Luria broth with 100 μg of ampicillin/ml to mid-log phase at 37°C prior to induction of recombinant protein expression with 0.1 mM isopropyl-β-d-thiogalactopyranoside for 3 h at 37°C. Typically, 100 ml of induced bacterial cultures was harvested by centrifugation at 4°C (2,500 × g for 10 min). The cell pellet was resuspended in 5 ml of lysis buffer (phosphate-buffered saline, 5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 2 μg of leupeptin/ml, 0.1% [vol/vol] Triton X-100) and then lysed by sonication. The soluble fraction was harvested as described above. For this binding assay, bacterial lysates containing untagged recombinant US11 fragments (expressed from the pET-28a vector as described above) were resuspended in the same lysis buffer as the GST fusion proteins. Binding and washing were carried out as described above using glutathione-Sepharose beads (Amersham Pharmacia). The beads were washed with phosphate-buffered saline lysis buffer (without protease inhibitors) prior to elution by boiling the beads in SDS-PAGE sample buffer.
RESULTS
Colocalization of kinesin and HSV.
The colocalization of kinesin and HSV-1 nucleocapsids was examined in fetal human neurons using the DRG neuron-epidermal explant two-chamber model system as previously described (13). Anterograde axonal transport of HSV-1 (CW1) (13, 23) was visualized using freeze-substitution transmission immunoelectron microscopy. Dual immunogold labeling (5- and 10-nm-diameter gold particles) of axons was performed with antibodies to uKHC and to the HSV-1 major capsid protein VP5. Anti-uKHC was shown to be specific in immunoblots for human uKHC by comparing reactivity with the immunizing antigen against bacterial lysates containing oligohistidine-tagged neuronal KHC408-1032 (data not shown).
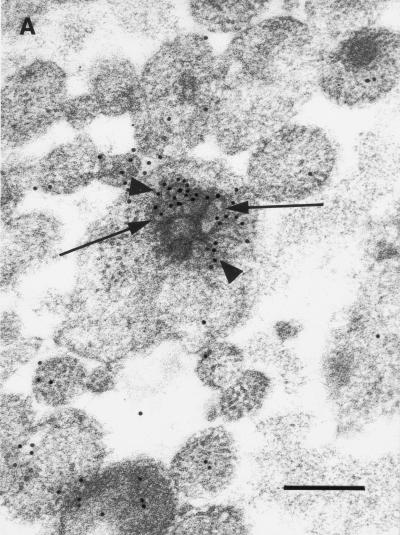
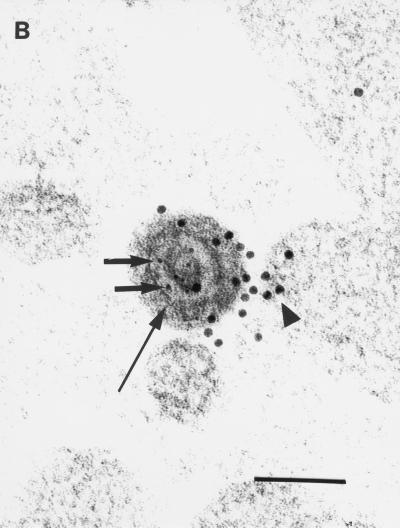
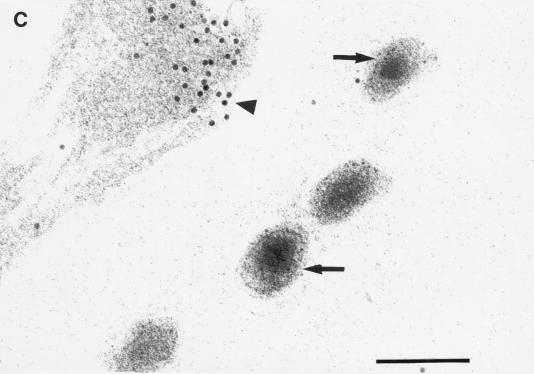
Anti-uKHC showed diffuse labeling throughout human axons, with clusters of immunolabel for uKHC observed around electron-dense axonal vesicles (Fig. 1A). Only sparse background label was found extra-axonally. With preimmune sera, only sparse background immunolabel was present on sections and there was no labeling of axonal vesicles (data not shown). Immunolabels for both uKHC and VP5 were found in and around unenveloped HSV-1 nucleocapsids within axons (Fig. 1B). Five nucleocapsids immunolabeled for VP5 were identified in over 400 sections examined. All five were colabeled for uKHC. The anti-VP5 antibody was specific, with no labeling of axons in mock-infected cultures (references 13 and 23 and data not shown). Only VP5 (not kinesin immunolabel) was present on extracellular virus (Fig. 1C). The immunolabeling for uKHC over intra-axonal virus (but not extra-axonal virus) was as intense as that around axonal vesicles, strongly suggesting that kinesin is involved in the anterograde axonal transport of HSV nucleocapsids. Therefore, tegument or outer capsid proteins are candidates for binding to conventional ubiquitous kinesin.
FIG. 1.
Transmission immunoelectron micrographs of human fetal axons labeled with 5- and 10-nm-diameter immunogold particles recognizing HSV major capsid protein VP5 and the stalk of human uKHC, respectively. Short arrows, 5-nm-diameter gold particles; arrowheads, 10-nm-diameter gold particles. Bars, 200 nm (A and C) and 100 nm (B). (A) Clustering of immunogold label for uKHC around axonal vesicles (long arrows) and diffuse labeling of other axons. Labeling with preimmune sera produced only sparse background label (data not shown). (B) Colocalization of immunogold labels against uKHC and VP5 over a dense unenveloped viral particle in an axon. The long arrow indicates the position of an axonlema. (C) Extracellullar virus labeled only with anti-VP5. In the upper left corner is an axon with diffuse 10-nm immunolabel against uKHC. There is no immunolabel for uKHC over extracellular virus, with only sparse background label outside the axons.
In vitro identification of kinesin-binding viral proteins.
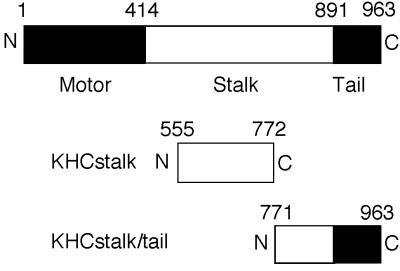
Fragments of human uKHC tagged at the N terminus with an oligohistidine sequence were used as bait to identify kinesin-binding HSV-1 proteins. In this study, they are designated KHCstalk (amino acid residues 555 to 772) and KHCstalk/tail (residues 771 to 963) (Fig. 2). KHCstalk/tail contains the putative organelle-binding site (15, 37, 38) and has recently been shown to bind the organelle receptor kinectin (27). Therefore, it was most likely that an HSV protein would bind to this region of uKHC as well, and KHCstalk should serve as a negative control for the binding experiments.
FIG. 2.
Summary of KHC structure and fusion proteins expressed in bacteria. Diagrams of the domain structure of uKHC and fragments of uKHC expressed in bacteria with oligohistidine N-terminal tags are shown. Solid and open areas of the bars delineate protein domains.
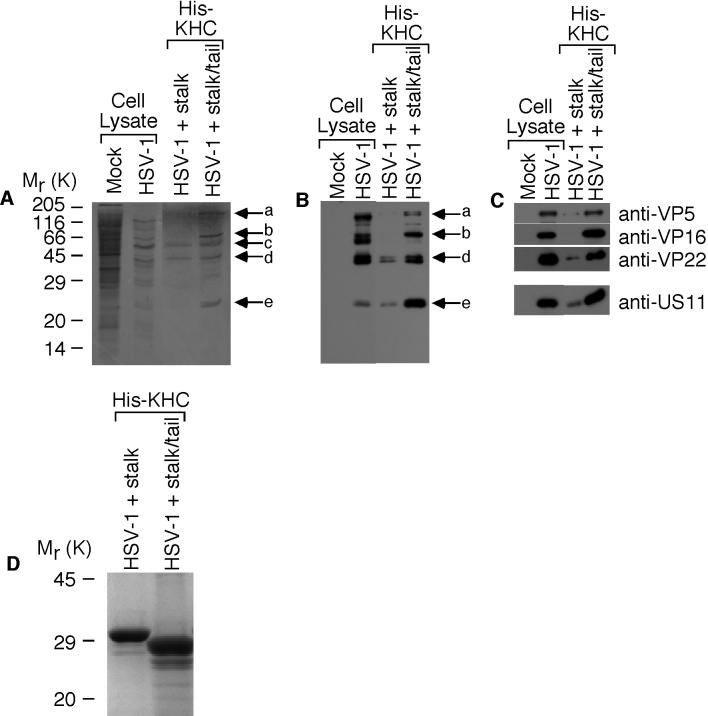
His-KHC fragments attached to nickel-activated beads were incubated with a lysate of 35S-labeled mock-infected or HSV-1-infected HEp-2 cells. The eluted complexes were separated by SDS-PAGE before transfer to nitrocellulose, followed by autoradiography (Fig. 3A). This process revealed five labeled proteins from HSV-1-infected cells coeluting with KHCstalk/tail (Fig. 3A). Of the five bands designated a through e, bands c (Fig. 3A), d (Fig. 3A and B), and e (Fig. 3B) appeared to also coelute with KHCstalk (Fig. 3). All except band c were subsequently confirmed to be HSV-1 proteins by immunoblotting the same samples with anti-HSV-1 (Fig. 3B). Band c may be a cellular protein or a viral protein that is not detected or is poorly detected by anti-HSV-1. Using electrophoretic mobility as a starting point, appropriate antibodies were chosen to identify the four viral proteins (a, b, d, and e) which preferentially bind to KHCstalk/tail. Immunoblotting identified band a as the major capsid protein VP5 (150 kDa) and bands b, d, and e as the tegument proteins VP16 (65 kDa), VP22 (38 kDa), and US11 (21 kDa), respectively (Fig. 3C). Similar amounts of His-KHCstalk and His-KHCstalk/tail were present in these experiments (Fig. 3D).
FIG. 3.
Identification of kinesin-binding HSV-1 proteins. 35S-labeled HSV-1-infected HEp-2 cell lysates were incubated with oligohistidine-tagged uKHC fragments bound to nickel-activated beads. Eluted kinesin-viral-protein complexes were separated by SDS-14% PAGE, transferred to nitrocellulose, and analyzed by autoradiography and immunoblotting. (A) Detection by autoradiography of five 35S-labeled proteins, designated a to e, bound to KHCstalk/tail. (B) Detection with polyclonal antibody to HSV-1 identified bands a, b, d, and e as viral proteins preferentially bound to KHCstalk/tail. (C) The viral protein bands were identified with specific antibodies as the structural proteins VP5 (a), VP16 (b), VP22 (d), and US11 (e). In each immunoblot, mock-infected and HSV-1-infected HEp-2 cell lysates are shown to illustrate antibody specificity. (D) Detection of oligohistidine-tagged KHCstalk and KHCstalk/tail fragments in eluted kinesin-viral-protein complexes. Samples were separated by SDS-12% PAGE and stained with Coomassie blue.
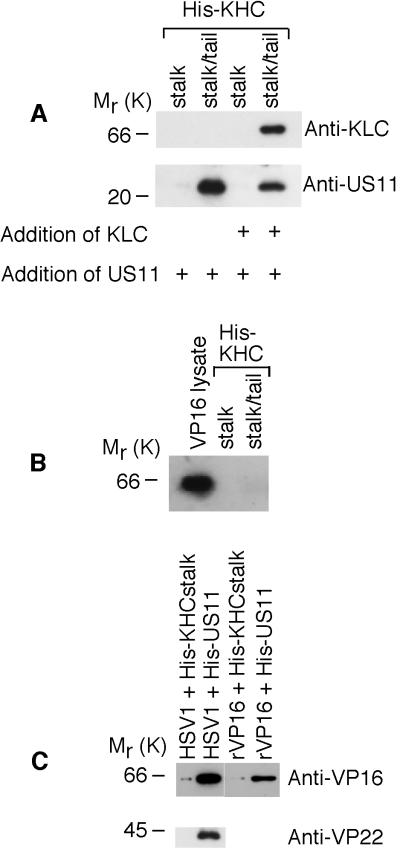
Previous findings have strongly suggested that anterograde axonal transport of HSV-1 nucleocapsids is dependent on tegument proteins which surround the nucleocapsid during transport (23). Therefore, access of kinesin to nucleocapsid VP5 in axons is unlikely. The tegument proteins VP16 and VP22 are also unlikely candidates, as transport of VP16 and VP22, but not VP5 or unenveloped nucleocapsids, into most axons is inhibited by BFA (23) (see data below). On this basis, US11 was considered the most likely viral protein to be directly interacting with uKHC. To establish whether US11 bound directly to uKHC, untagged US11 was expressed in E. coli. The addition of untagged US11 to His-KHC showed that US11 binds specifically to KHCstalk/tail (Fig. 4A, lower blot). This also shows that the binding of US11 to uKHC can occur in the absence of other viral proteins, although it may well be modulated in vivo by viral or cellular factors. A complex of uKHC and KLC was generated by preincubating KHCstalk/tail with untagged KLC (Fig. 4A, upper blot) as described previously (8). This kinesin heavy-light chain complex also binds US11, though at a decreased level compared with heavy chain alone (Fig. 4A, lower blot). The reduction in US11 binding is probably due to steric effects and not competitive inhibition, since the binding sites in KHCstalk/tail for KLC (residues 771 to 813) (8) and US11 (Fig. 5A) do not overlap.
FIG. 4.
Interaction of US11 with uKHC and with VP5, VP16, and VP22. Shown are immunoblots of protein complexes eluted from nickel-activated beads and run on SDS-14% PAGE. (A) Lysates of bacteria expressing untagged KLC or US11 were incubated, as indicated (+), with KHCstalk and KHCstalk/tail. US11 bound only to KHCstalk/tail in the absence of other HSV-1 proteins (lower blot). Overnight preincubation of the uKHC fragments with KLC, which binds only to KHCstalk/tail (upper blot), prior to addition of US11 also showed binding of US11 (lower blot). (B) In a similar experiment, recombinant untagged VP16 did not bind to either His-KHC fragment. Expression of VP16 in bacterial lysates was confirmed. (C) Unlabeled HSV-1-infected HEp-2 cell lysate was incubated with KHCstalk and oligohistidine-tagged US11. The viral proteins VP16 and VP22 coeluted with US11 only. Recombinant untagged VP16 was incubated with His-US11 or His-KHCstalk. Recombinant VP16 coeluted with His-US11 only.
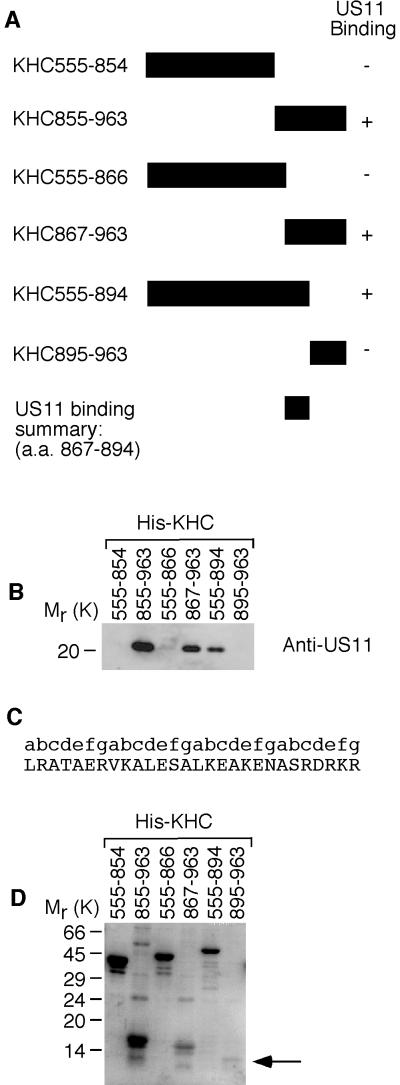
FIG.5.
Identification of the US11-binding site in uKHC. (A) Summary of US11 binding to fragments of uKHC expressed in bacteria with an oligohistidine N-terminal tag. a.a., amino acids. (B) Immunoblot of protein complexes eluted from nickel-activated beads and run on SDS-14% PAGE. Detection was with mouse monoclonal anti-US11. (C) Amino acid sequence of US11-binding site in human uKHC (residues 867 to 894) with repeating heptad motif labeled a to g.The presence of heptad repeats was determined using the COILS server (http://www.ch.embnet.org/software/COILS_form.html), which employs the algorithm of Lupas et al. (19). (D) Eluates from the blot in panel B were run on SDS-14% PAGE and stained with Coomassie blue to detect His-KHC fragments. The position of KHC895-963 is indicated by an arrow.
In contrast, untagged VP16 expressed in E. coli does not bind directly to either KHCstalk or KHCstalk/tail (Fig. 4B). The presence of VP16 and VP22 coeluting with KHCstalk/tail can be explained by the observation that these proteins also coelute in pull-down experiments when His-US11 is used as bait (Fig. 4C). VP5 could not be detected in the same experiment. Recombinant VP16 (in the absence of other viral proteins) also coelutes with KHCstalk/tail, which suggests that VP16 binds directly to US11 (Fig. 4C). Therefore, in the pull-down experiments with KHCstalk/tail, the primary interaction occurs between uKHC and US11, while secondary interactions occur among US11, VP16, and VP22. A direct interaction between uKHC and either VP5 or VP22 was not tested and cannot yet be excluded.
Locations of regions within US11 and uKHC which interact.
A series of uKHC fragments tagged at the N termini with oligohistidine were constructed to further delineate the US11-binding site in uKHC (Fig. 5A). Untagged recombinant US11 was added to these uKHC fragments immobilized on nickel-activated beads. Subsequent elution and visualization by SDS-PAGE and immunoblotting revealed that US11 binds to fragments 855 to 963, 867 to 963, and 555 to 894 (amino acid numbering) of uKHC (Fig. 5B). No US11 binding was detected with uKHC fragments 555 to 854, 555 to 866, and 895 to 963 (Fig. 5B). These results indicate that the US11-binding site in uKHC maps to residues 867 to 894 (Fig. 5A). This region, which contains four contiguous heptad repeats (Fig. 5C), is located within the predicted cargo-binding domain of KHC (37). This putative cargo-binding domain (residues 833 to 900) has previously been shown to interact with the endoplasmic reticulum membrane protein kinectin (27). The presence of uKHC fusion proteins in eluates from nickel-activated beads was confirmed by SDS-PAGE and Coomassie blue staining (Fig. 5D). Even though fragment 895 to 963 was barely detectable, making direct binding of US11 difficult to assess, it was considered not essential for US11 binding, since fragment 555 to 894 (region 895 to 963 truncated) readily binds US11 (Fig. 5B).
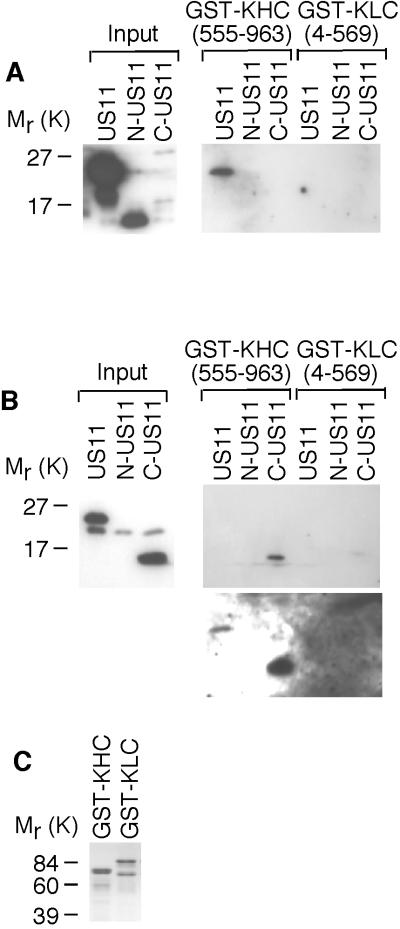
Binding of uKHC to US11 fragments was ascertained using full-length US11 (residues 1 to 161), an N-terminal fragment of US11 (N-US11; residues 1 to 84), and a C-terminal fragment of US11 (C-US11; residues 84 to 161). Untagged recombinant US11 was added to GST-KHC (fragment 555 to 963, which contains the US11-binding site) or GST-KLC immobilized on glutathione-Sepharose beads. Complexes formed on beads were eluted by boiling and analyzed by SDS-PAGE and immunoblotting. Two antibodies were employed to detect US11 fragments. Immunoblotting with a mouse monoclonal anti-US11 antibody, which recognizes an epitope in the N terminus of US11 (32), showed that full-length US11, but not N-US11, binds to uKHC (Fig. 6A). As expected, C-US11 was not detected in lysates with this antibody (Fig. 6A). Immunoblotting with a rabbit polyclonal antibody, which recognizes the C-terminal 11 amino acids of US11 (20), showed that both full-length US11 and C-US11 bind to uKHC (Fig. 6B). This antibody detects only full-length US11 and C-US11 in bacterial lysates (Fig. 6B).
FIG. 6.
Identification of the uKHC-binding site in US11. Protein complexes were eluted from glutathione-Sepharose beads and run on SDS-14% PAGE. (A) Immunoblotting with mouse monoclonal anti-US11 detected full-length US11 bound only to GST-KHC. (B) Immunoblotting with rabbit polyclonal anti-US11 detected only C-US11 bound to GST-KHC (upper blot). Overexposure of the blot also detects full-length US11 bound to GST-KHC (lower blot). (C) Eluates from glutathione-Sepharose beads were run on SDS-14% PAGE and stained with Coomassie blue to detect GST fusion proteins.
KLC appears not to be directly involved in binding US11, as no binding of US11 to GST-KLC was detected (Fig. 6A and B). KHC binding to other cargo, such as vesicles (38) and the receptor kinectin (27), also occurs in the absence of KLC. The presence of GST-KHC and GST-KLC in binding experiments was confirmed by SDS-PAGE and Coomassie blue staining of boiled glutathione-Sepharose beads (Fig. 6C).
Distribution and kinetics of US11 and other HSV proteins in dissociated rat neurons.
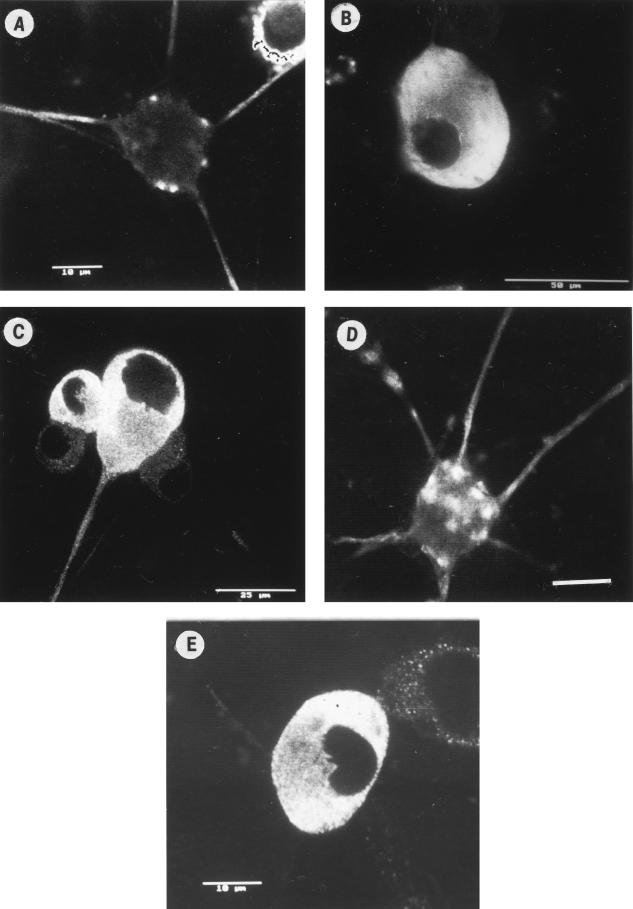
The appearance and distribution of structural HSV protein antigens from the tegument (US11) and nucleocapsid (VP5) and their anterograde transport into axons of dissociated rat neonatal DRG was followed by fixation at serial times of 6, 10, 12, 17, and 24 h. The distribution of HSV antigens was visualized by immunofluorescence and confocal microscopy as previously described (23).
First, US11 antigen from HSV-1 (CW1 clinical strain) (13, 23) was transported anterogradely into the distal axon (a criterion for its proposed role in anterograde axonal transport) (Fig. 7A) from the cytoplasm of the cell body (Fig. 7B). In neurons infected with wt HSV-1, VP5 antigen was diffusely distributed in the cytoplasm of the cell body at 10 to 12 h and then transported throughout the axon from 17 to 24 h (Fig. 7C), as previously described (23). The kinetics of transport of US11 and VP5 from wt HSV-1 into axons were similar, with both antigens being present in axons from 17 to 24 h, consistent with colocalization of US11 with nucleocapsids (Fig. 7A and C). The distribution and kinetics of gC in the cell body and axons were similar to those previously reported (23), with gC present in axons from 13 to 24 h postinfection (data not shown). Incubation of HSV-infected neurons with BFA from 3 to 24 h postinfection differentially inhibited anterograde transport of tegument proteins into axons. As previously described, BFA completely inhibited transport of gC, but not VP5, into axons. Transport of VP16 and VP22 into axons was only observed (at reduced intensity) in 20 to 30% of infected neurons, i.e., in 70 to 80% of infected neurons, no VP16 or VP22 was observed in the axons (23). However, BFA had no effect on US11 compared to reduced axonal transport of VP16 (Fig. 7D and E). Therefore, in the presence of BFA, VP5 and US11, but not VP16 or VP22, were cotransported into the majority of axons.
FIG. 7.
Transport of structural HSV antigens into axons of infected dissociated rat neonatal neurons in vitro. Dissociated DRG neurons were infected with wt HSV-1 (CW1) at 10 PFU/neuron in the absence (A to C) or presence (D and E) of BFA. The appearance and transport into the principal axon of HSV-1 tegument (US11) and nucleocapsid (VP5) antigens was followed by serial fixation over 6 to 24 h postinfection, and then immunofluorescence and confocal microscopy were performed as previously described (23). US11 was present in axons at 24 h (A) but not at 10 h (B). VP5 was also present in axons at 24 h (C) but not at 10 h (reference23 and data not shown). After treatment of neurons with BFA, US11 (D), but not VP16 (E), was present at 24 h in the majority of axons. Bars, 10 (A and E), 50 (B), and 25 (C and D) μm.
DISCUSSION
In this study, we have shown the molecular motor conventional ubiquitous kinesin is cotransported with HSV nucleocapsids in the axons of human fetal neurons. Furthermore, a direct interaction between uKHC and the HSV-1 tegument protein US11 was identified in vitro.
US11 appears to be a multifunctional protein. It is a structural protein proposed to be part of the tegument of HSV (32). In addition, US11 has RNA-binding activity, stably associates with 60S ribosomal subunits, has a role in posttranscriptional regulation of gene expression, and localizes to the nucleoli (6, 7, 20, 33, 34). The amino half of US11 is required for transactivation of gene expression (35). The RNA-binding properties of US11 are dependent on the 20 to 24 RXP repeats in the carboxy half of the protein (31). This domain, as shown in this study, also contains the uKHC-binding site. This polyproline domain may acquire a type II helix configuration, with arginine residues on one face to interact with negatively charged phosphates of RNA (31, 35). The other faces of the helix are either hydrophobic (proline repeats) or a mixture of hydrophobic, uncharged, and acidic side chains and presumably provide specificity to the binding of RNA (31) and the heptad repeat US11-binding site in uKHC.
The US11 protein is well conserved between HSV-1 and -2 (63% homology), with the greater variation at the N terminus. There are no US11 homologues in other neurotropic herpesviruses, such as varicella-zoster virus or PRV, suggesting they may use different proteins for anterograde axonal transport. However, there are precedents within the herpesvirus family for similar functions on different proteins (e.g., entry into cells via gD in HSV (29) and via gE-gI and gB in varicella-zoster virus (11).
Initial findings from a pull-down experiment with US11 suggest that it also interacts with one or both of the structural HSV proteins VP16 and VP22. Furthermore, a direct interaction between US11 and VP16 in the absence of other viral proteins was demonstrated. This US11-VP16 interaction may represent an important structural interaction for US11 in the tegument of the virus and may play a regulatory role in infected cells. VP16 has also been shown to bind to other tegument proteins, including vhs (39) and VP22 (9), and it has been shown to be a component of viral tegument protein-glycoprotein complexes (48). The presence of VP22 in pull-down experiments with US11 would likely be a result of its interaction with VP16 rather than a direct interaction with US11. This is supported by work in progress using a US11 deletion mutant in the pull-down experiments, where both the 20- (US11) and 40-kDa (VP22) bands are lost in comparison to the parental strain (R. J. Diefenbach, M. Miranda-Saksena, E. Diefenbach, and A. L. Cunningham, unpublished results). The fact that VP5 appears to coelute with uKHC and not US11 suggests that, like US11, it might bind directly to uKHC. Whether VP5 has a role in axonal transport requires further investigation, but as anterograde axonal transport of nucleocapsids occurs in the presence of a tegument protein coat, this seems likely to prevent access of VP5 to uKHC (13, 23, 47). Related studies of the interaction of dynein with HSV-1 UL34 also identified VP5 coeluting with dynein. The presence of VP5 was deemed to be either nonspecific or the result of a secondary interaction with either UL34 or UL31 (45, 46).
The presence of US11 in the tegument (32) and the similar kinetics of anterograde transport of US11 and nucleocapsid VP5 are consistent with the hypothesis that the observed US11-uKHC interaction has a major role in mediating anterograde transport of nucleocapsids into and along axons. US11 is dispensable for growth in a cell in culture (18), and US11 null mutants spread and are virulent on inoculation into the central nervous system and establish latency in mice (22). Final confirmation of this hypothesis will require completion of studies with a series of US11 deletion mutants and rescuants in a DRG culture system and ultimately studies in an animal model system.
Recent reports of anterograde transport of PRV show bursts of very rapid transport (>5 μm/s), variation in velocity with a Gaussian distribution, and frequent reversal of direction (40). These findings suggest a complex interaction among microtubules, molecular motors, and neurotropic herpesviruses during anterograde transport. Therefore, at least one anterograde (kinesin) and one retrograde (dynein) motor are probably operative (although two anterograde motors with similar transport velocities could be involved). Our data suggest that the uKHC-US11 interaction is likely to have a major role in anterograde transport and represent the first report of a direct molecular interaction between a herpesvirus protein and a neuronal molecular motor. However, there may be other interactions between HSV proteins and other kinesins which could provide synergy or redundancy.
This study also raises many questions about the mechanisms of tegument formation, the location of US11 in the tegument, and homologous interactions mediating the transport of other neurotropic viruses. Inhibition of the interactions between kinesins and HSV by peptides and/or peptidomimetic analogues could provide a new strategy for antiviral treatment for this and other neurotropic viruses (43). Incorporation of specific deletions of the kinesin-interacting regions of HSV proteins into attenuated live HSV vaccine candidates could also be useful to prevent clinical recrudescence.
Acknowledgments
Monica Miranda-Saksena, Eve Diefenbach, and David J. Holland contributed equally to this work.
This work was supported by an Australian National Health and Medical Research Council grant (no. 107374).
We thank Carol Robinson for all the photographic assistance and Claire Wolczak for help in preparation of the manuscript.
REFERENCES
- 1.Bloom, G. S., M. C. Wagner, K. K. Pfister, and S. T. Brady. 1988. Native structure and physical properties of bovine brain kinesin and identification of the ATP-binding subunit polypeptide. Biochemistry 27:3409-3416. [DOI] [PubMed] [Google Scholar]
- 2.Brideau, A. D., J. P. Card, and L. W. Enquist. 2000. Role of pseudorabies virus Us9, a type II membrane protein, in infection of tissue culture cells and the rat nervous system. J. Virol. 74:834-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brideau, A. D., M. G. Eldridge, and L. W. Enquist. 2000. Directional trans-neuronal infection by pseudorabies virus is dependent on an acidic internalization motif in the Us9 cytoplasmic tail. J. Virol. 74:4549-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cassady, K. A., M. Gross, and B. Roizman. 1998. The herpes simplex virus US11 protein effectively compensates for the γ134.5 gene if present before activation of protein kinase R by precluding its phosphorylation and that of the α subunit of eukaryotic translation initiation factor 2. J. Virol. 72:8620-8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen, G. H., M. Ponce de Leon, H. Diggelmann, W. C. Lawrence, S. K. Vernon, and R. J. Eisenberg. 1980. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J. Virol. 34:521-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diaz, J. J., M. D. Dodon, N. Schaerer-Uthurralt, D. Simonin, K. Kindbeiter, L. Gazzolo, and J. J. Madjar. 1996. Post-transcriptional transactivation of human retroviral envelope glycoprotein expression by herpes simplex virus Us11 protein. Nature 379:273-277. [DOI] [PubMed] [Google Scholar]
- 7.Diaz, J. J., D. Simonin, T. Masse, P. Deviller, K. Kindbeiter, L. Denoroy, and J. J. Madjar. 1993. The herpes simplex virus type 1 US11 gene product is a phosphorylated protein found to be non-specifically associated with both ribosomal subunits. J. Gen. Virol. 74:397-406. [DOI] [PubMed] [Google Scholar]
- 8.Diefenbach, R. J., J. P. Mackay, P. J. Armati, and A. L. Cunningham. 1998. The C-terminal region of the stalk domain of ubiquitous human kinesin heavy chain contains the binding site for kinesin light chain. Biochemistry 37:16663-16670. [DOI] [PubMed] [Google Scholar]
- 9.Elliott, G., G. Mouzakitis, and P. O'Hare. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69:7932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elliott, G., and P. O'Hare. 1997. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell 88:223-233. [DOI] [PubMed] [Google Scholar]
- 11.Grose, C. 1991. Glycoproteins of varicella-zoster virus and their herpes simplex virus homologs. Rev. Infect. Dis. 13(Suppl. 11):S960-S963. [DOI] [PubMed] [Google Scholar]
- 12.Hirokawa, N., R. Sato-Yoshitake, N. Kobayashi, K. K. Pfister, G. S. Bloom, and S. T. Brady. 1991. Kinesin associates with anterogradely transported membranous organelles in vivo. J. Cell Biol. 114:295-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holland, D. J., M. Miranda-Saksena, R. A. Boadle, P. J. Armati, and A. L. Cunningham. 1999. Anterograde transport of herpes simplex virus nucleocapsid, tegument, and glycoproteins in axons of human fetal neurons. J. Virol. 73:8476-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes, T. A., S. La Boissiere, and P. O'Hare. 1999. Analysis of functional domains of the host cell factor involved in VP16 complex formation. J. Biol. Chem. 274:16437-16443. [DOI] [PubMed] [Google Scholar]
- 15.Kirchner, J., S. Seiler, S. Fuchs, and M. Schliwa. 1999. Functional anatomy of the kinesin molecule in vivo. EMBO J. 18:4404-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondo, S., R. Sato-Yoshitake, Y. Noda, H. Aizawa, T. Nakata, Y. Matsuura, and N. Hirokawa. 1994. KIF3A is a new microtubule-based anterograde motor in the nerve axon. J. Cell Biol. 125:1095-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kristensson, K., E. Lycke, and J. Sjostrand. 1971. Spread of herpes simplex virus in peripheral nerves. Acta Neuropathol. (Berlin) 17:44-53. [DOI] [PubMed] [Google Scholar]
- 18.Longnecker, R., and B. Roizman. 1987. Clustering of genes dispensable for growth in culture in the S component of the HSV-1 genome. Science 236:573-576. [DOI] [PubMed] [Google Scholar]
- 19.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 20.MacLean, C. A., F. J. Rixon, and H. S. Marsden. 1987. The products of gene US11 of herpes simplex virus type 1 are DNA-binding and localize to the nucleoli of infected cells. J. Gen. Virol. 68:1921-1937. [DOI] [PubMed] [Google Scholar]
- 21.McLean, C., A. Buckmaster, D. Hancock, A. Buchan, A. Fuller, and A. Minson. 1982. Monoclonal antibodies to three non-glycosylated antigens of herpes simplex virus type 2. J. Gen. Virol. 63:297-305. [DOI] [PubMed] [Google Scholar]
- 22.Meignier, B., R. Longnecker, P. Mavromara-Nazos, A. E. Sears, and B. Roizman. 1988. Virulence of and establishment of latency by genetically engineered deletion mutants of herpes simplex virus 1. Virology 162:251-254. [DOI] [PubMed] [Google Scholar]
- 23.Miranda-Saksena, M., P. Armati, R. A. Boadle, D. J. Holland, and A. L. Cunningham. 2000. Anterograde transport of herpes simplex virus type 1 in cultured, dissociated human and rat dorsal root ganglion neurons. J. Virol. 74:1827-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Navone, F., J. Niclas, N. Hom-Booher, L. Sparks, H. D. Bernstein, G. McCaffrey, and R. D. Vale. 1992. Cloning and expression of a human kinesin heavy chain gene: interaction of the COOH-terminal domain with cytoplasmic microtubules in transfected CV-1 cells. J. Cell Biol. 117:1263-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niclas, J., F. Navone, N. Hom-Booher, and R. D. Vale. 1994. Cloning and localization of a conventional kinesin motor expressed exclusively in neurons. Neuron 12:1059-1072. [DOI] [PubMed] [Google Scholar]
- 26.O'Reilly, D., O. Hanscombe, and P. O'Hare. 1997. A single serine residue at position 375 of VP16 is critical for complex assembly with Oct-1 and HCF and is a target of phosphorylation by casein kinase II. EMBO J. 16:2420-2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ong, L. L., A. P. Lim, C. P. Er, S. Kuznetsov, and H. Yu. 2000. Kinectin-kinesin binding domains and their effects on organelle motility. J. Biol. Chem. 275:32854-32860. [DOI] [PubMed] [Google Scholar]
- 28.Penfold, M. E., P. Armati, and A. L. Cunningham. 1994. Axonal transport of herpes simplex virions to epidermal cells: evidence for a specialized mode of virus transport and assembly. Proc. Natl. Acad. Sci. USA 91:6529-6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajcani, J., and A. Vojvodova. 1998. The role of herpes simplex virus glycoproteins in the virus replication cycle. Acta Virol. 42:103-118. [PubMed] [Google Scholar]
- 30.Roizman, B., and A. E. Sears. 1996. Herpes simplex viruses and their replication, p. 2231-2295. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 31.Roller, R. J., L. L. Monk, D. Stuart, and B. Roizman. 1996. Structure and function in the herpes simplex virus 1 RNA-binding protein US11: mapping of the domain required for ribosomal and nucleolar association and RNA binding in vitro. J. Virol. 70:2842-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roller, R. J., and B. Roizman. 1992. The herpes simplex virus 1 RNA binding protein US11 is a virion component and associates with ribosomal 60S subunits. J. Virol. 66:3624-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roller, R. J., and B. Roizman. 1991. Herpes simplex virus 1 RNA-binding protein US11 negatively regulates the accumulation of a truncated viral mRNA. J. Virol. 65:5873-5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roller, R. J., and B. Roizman. 1990. The herpes simplex virus Us11 open reading frame encodes a sequence-specific RNA-binding protein. J. Virol. 64:3463-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaerer-Uthurralt, N., M. Erard, K. Kindbeiter, J. J. Madjar, and J. J. Diaz. 1998. Distinct domains in herpes simplex virus type 1 US11 protein mediate post-transcriptional transactivation of human T-lymphotropic virus type I envelope glycoprotein gene expression and specific binding to the Rex responsive element. J. Gen. Virol. 79:1593-1602. [DOI] [PubMed] [Google Scholar]
- 36.Schnapp, B. J., T. S. Reese, and R. Bechtold. 1992. Kinesin is bound with high affinity to squid axon organelles that move to the plus-end of microtubules. J. Cell Biol. 119:389-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seiler, S., J. Kirchner, C. Horn, A. Kallipolitou, G. Woehlke, and M. Schliwa. 2000. Cargo binding and regulatory sites in the tail of fungal conventional kinesin. Nat. Cell Biol. 2:333-338. [DOI] [PubMed] [Google Scholar]
- 38.Skoufias, D. A., D. G. Cole, K. P. Wedaman, and J. M. Scholey. 1994. The carboxyl-terminal domain of kinesin heavy chain is important for membrane binding. J. Biol. Chem. 269:1477-1485. [PubMed] [Google Scholar]
- 39.Smibert, C. A., B. Popova, P. Xiao, J. P. Capone, and J. R. Smiley. 1994. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J. Virol. 68:2339-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith, G. A., S. P. Gross, and L. W. Enquist. 2001. Herpesviruses use bidirectional fast-axonal transport to spread in sensory neurons. Proc. Natl. Acad. Sci. USA 98:3466-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sodeik, B., M. W. Ebersold, and A. Helenius. 1997. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 136:1007-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomishima, M. J., and L. W. Enquist. 2001. A conserved alpha-herpesvirus protein necessary for axonal localization of viral membrane proteins. J. Cell Biol. 154:741-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsiang, H., E. Lycke, P. E. Ceccaldi, A. Ermine, and X. Hirardot. 1989. The anterograde transport of rabies virus in rat sensory dorsal root ganglia neurons. J. Gen. Virol. 70:2075-2085. [DOI] [PubMed] [Google Scholar]
- 44.Yamazaki, H., T. Nakata, Y. Okada, and N. Hirokawa. 1995. KIF3A/B: a heterodimeric kinesin superfamily protein that works as a microtubule plus end-directed motor for membrane organelle transport. J. Cell Biol. 130:1387-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye, G. J., and B. Roizman. 2000. The essential protein encoded by the UL31 gene of herpes simplex virus 1 depends for its stability on the presence of UL34 protein. Proc. Natl. Acad. Sci. USA 97:11002-11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye, G. J., K. T. Vaughan, R. B. Vallee, and B. Roizman. 2000. The herpes simplex virus 1 UL34 protein interacts with a cytoplasmic dynein intermediate chain and targets nuclear membrane. J. Virol. 74:1355-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou, Z. H., D. H. Chen, J. Jakana, F. J. Rixon, and W. Chiu. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 73:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu, Q., and R. J. Courtney. 1994. Chemical cross-linking of virion envelope and tegument proteins of herpes simplex virus type 1. Virology 204:590-599. [DOI] [PubMed] [Google Scholar]