A longstanding unknown in viral RNA biology is the relationship between translation and packaging of genomic RNA. For retroviruses, an extensive body of work has characterized nuclear export of the unspliced genome-length transcript (5, 8, 15), but the cytoplasmic trafficking of the RNA has remained relatively undefined. An elegant experimental approach that was initiated over 25 years ago has been updated and extended to human retroviruses during the last year. A consensus on the relationship between translation and packaging of retroviral RNA has been reached. An unexpected finding was that retroviruses have adapted two divergent approaches to manage the cytoplasmic fate of genomic RNA. This minireview introduces the interdependent relationship between translation and packaging of retroviral RNA, postulates models of retroviral RNA trafficking in the cytoplasm, summarizes experimental results that address the models, and discusses the recent consensus.
Unspliced genome-length retroviral RNA is utilized for both translation and packaging.
Retroviruses are a unique family of RNA viruses that utilize virally encoded reverse transcriptase (RT) to replicate genomic RNA through a proviral DNA intermediate (40). The provirus is permanently integrated into the host cell chromosome and, like a cellular gene, is expressed by the host cell transcription, RNA processing, and translation machinery. The primary transcription product interacts with the cellular RNA processing machinery and, similar to a typical cellular pre-mRNA, is spliced, exported to the cytoplasm, and translated by the host protein synthesis machinery. A proportion of the pre-mRNA subverts typical RNA processing and interacts with viral and/or cellular nucleocytoplasmic shuttle proteins that activate nuclear export, despite the lack of intron removal (8). In the cytoplasm, the unspliced genome-length RNA serves two essential roles. The immediate function is to serve as an mRNA template for translation of viral proteins. Another function is to serve as a genomic RNA that is packaged into assembling virions. The Gag polyprotein facilitates the specific packaging of two copies of the unspliced genome-length RNA. The nucleocapsid domain of Gag contains redundant Cys-X2-Cys-X4-His-X4-Cys motifs that interact with the highly structured packaging signal (ψ or E), which is located in the 5′ untranslated region (UTR) (34a).
Long and structured 5′ UTRs, which are typical in retroviruses, inhibit cap-dependent ribosome scanning of cellular mRNA (33). Results of in vitro translation assays and transient transfection assays have directly validated the hypothesis that structured motifs in the human immunodeficiency virus type 1 (HIV-1) 5′ UTR inhibit protein synthesis (12, 24, 31). Distal RNA segments of HIV-1 and four other retroviruses have been shown to function as internal ribosome entry sites in bicistronic and monocistronic reporter gene assays (3, 4, 6, 10, 19, 26, 41). The data suggest that these internal ribosome entry sites function to promote synthesis of Gag and/or glyco-Gag polyprotein, although a functional role for internal ribosome entry during retroviral replication has not been demonstrated.
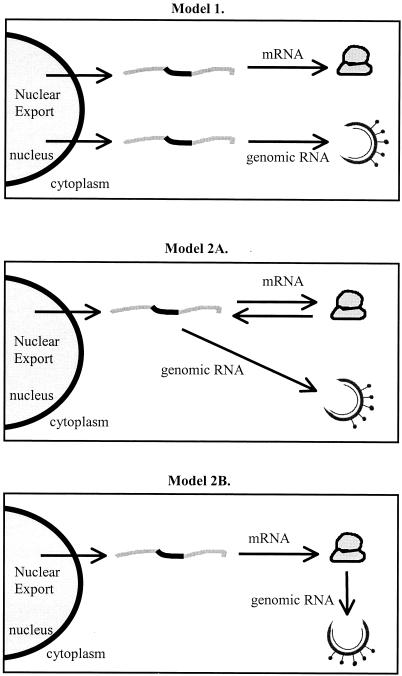
Efficient cap-dependent translation of the genome-length RNA is expected to require localized melting of RNA structure, which would distort presentation of the RNA packaging signal. Also, interaction between the nucleocapsid and the RNA packaging signal is expected to arrest ribosome scanning and inhibit efficient translation of the viral RNA (2, 30). This scenario implies that the cellular translation machinery and viral assembly complexes compete for cytoplasmic utilization of genome-length RNA. One possibility is that genome-length RNAs segregate into functionally independent RNA populations for packaging or translation (Fig. 1, model 1). Another possibility is that the genomic-length RNA functions interchangeably as an mRNA template and virion RNA (Fig. 1, model 2A). Considering that viral proteins are necessary for virion assembly, an alternative possibility is that translation is an obligate step in RNA packaging (Fig. 1, model 2B). In this model, recruitment of the mRNA template protein to the viral assembly complexes by the newly synthesized Gag protein may improve genomic RNA packaging specificity. Historically, studies using retroviral vectors have provided clues to the cytoplasmic fate of genomic RNA of the parental replication-competent retrovirus.
FIG. 1.
(Model 1) The unspliced genome-length RNA (gray lines with intronic sequences denoted in black) achieves nuclear export and segregates into functionally distinct populations of either mRNA template for translation of viral proteins on host cell ribosomes or genomic RNA that is packaged into assembling virions. (Model 2A) The genome-length RNA can function interchangeably as both mRNA and genomic RNA with no requirement for prior translation. (Model 2B) Prior translation is a requirement for generation of genomic RNA.
Coordinate viral protein synthesis is not required for packaging of vector RNA.
The ability of retroviral genomes to function as vectors was established in the early 1980s when mutated murine and avian retroviral RNAs lacking viral protein coding regions were shown to be eligible for packaging if the missing viral proteins were provided in trans (13, 36, 39). In helper cells, Ψ-negative helper RNAs function exclusively as mRNA templates for translation of viral Gag, polymerase, and envelope proteins, while Ψ-positive vector transcripts function as the genomic RNA that is packaged into the helper virus. After transduction of target cells, the helper proteins support a single cycle of vector RNA replication, and subsequently the integrated vector genome functions exclusively as mRNA for synthesis of vector-encoded protein. The ability to segregate the translation and RNA packaging functions to helper and vector RNAs gives retroviruses their utility as gene transfer vectors and indicates that in murine and avian vectors, coordinate translation of Gag structural protein is not necessary to target an RNA for packaging.
Competition assays between wild-type HIV-1 RNAs expressed from provirus and genetically marked HIV-1 vectors indicate that coordinate synthesis of HIV-1 Gag polyprotein is not necessary for lentiviral vector RNA packaging. The packaging efficiency of an HIV-1 vector RNA that contains a premature stop codon in the capsid domain of the gag open reading frame (CA, p24) was not reduced compared to that of the wild-type RNA (20). Unexpectedly, the packaging efficiency of HIV-2 vector RNA that contains a similar premature stop codon in CA was reduced to 30% of that of the wild-type HIV-2 RNA, suggesting a cotranslational preference for HIV-2 (16). A possible explanation for the difference between HIV-2 and HIV-1 is utilization of alternative RNA packaging mechanisms, which may be attributable to differences in the location of their respective RNA packaging signals. The dominant packaging signal of HIV-2 is located upstream of the major splice donor and is present in both genome-length and spliced vector RNAs, while the dominant packaging signal of HIV-1 is located downstream of the major splice donor and is present solely on the genome-length viral RNA (21). A tendency to cotranslationally package HIV-2 RNA may serve to increase the packaging specificity of genome-length RNA (16). Recent results indicated that while Ψ-positive HIV-2 vectors containing the premature CA stop codon were not efficiently packaged by wild-type HIV-2, helper RNAs containing several different deletions within the dominant HIV-2 RNA packaging signal exhibited increased packaging efficiency, apparently due to reduction of cotranslational RNA packaging (14). These results are consistent with the demonstrated utility of HIV-2 vector systems (1, 9, 34, 35) and indicate that cotranslational packaging is not a requirement for HIV-2.
Overall, the results with HIV-1 and HIV-2 vector RNAs are in agreement with the theory that coordinate Gag protein synthesis is not absolutely necessary for packaging of lentivector RNA (as would be expected by the established utility of HIV-1- and HIV-2-derived vectors). Studies using replication-competent retrovirus, summarized below, have directly established that prior translation of genome-length viral RNA is not a necessary step in the retroviral replication cycle.
Hypothetical models to describe cytoplasmic trafficking of unspliced viral RNA.
Elucidation of the interdependent relationship between translation and packaging is necessary for complete discernment of the retroviral replication cycle. Following early results with murine leukemia virus (MLV), which are discussed below, a primary goal of studies involving both simple and complex retroviruses has been to determine whether genome-length transcripts segregate in the cytoplasm as distinct populations of mRNA template for protein synthesis and genomic RNA for packaging into progeny virions (Fig. 1, model 1) or function as both mRNA and genomic RNA (Fig. 1, model 2). Because coordinate translation is a theoretical mechanism to provide additional packaging specificity, another goal has been to determine whether prior translation is a requirement for genomic RNA packaging (model 2A versus model 2B).
Mutational analyses to ascertain these issues for replication-competent retroviral genomes are problematic because proviral mRNA and genomic RNA are physically indistinguishable and contain overlapping motifs that are necessary for translation, packaging, and other steps in viral replication. (Mutational analyses of vector genomes have been highly useful for mapping RNA sequences necessary and sufficient for RNA packaging [2].) The approach used to evaluate the relationship between protein synthesis and packaging of genome-length RNA has been to employ chemical inhibitors to modulate host cell transcription or translation and evaluate relative effects on genome-length RNA translation and packaging. These experiments are advantageous for direct evaluation of genomic RNA expressed from provirus in an infected cell and have added value in illuminating additional aspects of the process of viral assembly. The following sections summarize insights gained into the cytoplasmic fate of genome-length RNA in cells infected with MLV, avian sarcoma virus (ASV), HIV-1, and HIV-2. The overall conclusion drawn from the studies is that retroviruses do not all use the same means of managing the interdependent relationship between translation and packaging. The results instead validate one model of cytoplasmic trafficking for MLV genome-length RNA and a second model for ASV, HIV-1, and HIV-2 genome-length RNAs.
Simple retroviruses: fixed fate of genomic RNA?
Beginning in 1974, Levin et al. used the transcription inhibitor actinomycin D (actD) to limit host cell RNA synthesis and study the effect on production of Rauscher MLV (17). Following a 6-h incubation with actD and [3H]uridine, synthesis of cellular [3H]RNA was reduced to 5% of that of the mock-treated control, and the actD-treated cells continued to produce morphologically normal C-type progeny virions. RT assays performed on exogenously provided template indicated that virion abundance remained relatively constant, but virion infectivity was reduced to 15% of the control level. The reduction in infectivity correlated with reductions in the abundance of virion-associated genomic RNA to 10% and virus particle-associated RT activity on an endogenous template to 20% of that of the control. These data indicated that ongoing RNA synthesis was necessary to produce MLV virions that contained MLV genome-length RNA. Examination of the radiolabeled MLV producer cells following actD treatment verified that steady-state genome-length RNA in the infected cells remained competent for continued synthesis of MLV structural proteins (18). From these data, Levin and colleagues concluded that MLV-infected cells contain two populations of unspliced viral RNAs that segregate as mRNA or as genomic RNA (Fig. 1, model 1). Subsequent analysis of RNA half-life by molecular hybridization assay after actD treatment reinforced the conclusion that two separate populations of MLV RNA exist (22). MLV virion-associated RNA exhibited a half-life of approximately 3 to 4 h, while cytoplasmic MLV genome-length RNA exhibited a half-life of at least 12 h. Paskind et al. arrived at a similar conclusion during an actD analysis of cells infected with the Moloney strain of MLV (32).
Recently, Dorman and Lever repeated Levin et al.'s studies with highly sensitive RNase protection assays (RPAs) (11). NIH 3T3 cells chronically infected with Moloney MLV were treated with actD for 8 h, and cytoplasmic and virion RNA samples were collected and subjected to RPA. Virion abundance was again estimated by the exogenous RT assay. Post-actD treatment, cytoplasmic unspliced MLV RNA was reduced to 70% of the control level. By contrast, MLV virion-associated genome-length RNA declined to 10%. The distinct actD responses observed for cytoplasmic and virion-associated genome-length RNA mirror the results of Levin and colleagues and support their conclusion of the existence of two distinct populations of genome-length MLV mRNA and genomic RNA.
Stoltzfus et al. studied another simple retrovirus, ASV, by applying separate experimental approaches to measure the half-life of ASV RNA in the cytoplasm or in virions (38). The half-life of ASV cytoplasmic unspliced RNA was evaluated in a [3H]uridine labeling experiment following actD treatment and estimated to be 6 to 7.5 h. The half-life of virion-associated ASV RNA was also estimated to be 6 to 7 h by [methyl-3H]methionine pulse-chase analysis and isotopic equilibrium assay in the absence of actD. These results in the ASV system stand in contrast to those obtained in the MLV system. The finding that genome-length ASV RNAs in the cytoplasm and in virions exhibit similar half-lives is consistent with the alternative hypothesis that ASV contains a single RNA population that functions as both mRNA and genomic RNA (Fig. 1, model 2).
Complex retroviruses: two competing cytoplasmic fates of genome-length RNA.
Genetically complex retroviruses, including HIV-1, encode the Rev posttranscriptional regulatory protein, which is absolutely necessary for nuclear export of the intron-containing RNA (15). The nucleocytoplasmic shuttling ability of Rev is disrupted by actD (23). Therefore, actD plays a redundant role in curtailing cytoplasmic accumulation of genomic RNA in HIV-1-infected cells: inhibition of RNA synthesis and prevention of nuclear export.
We treated HIV-1-infected human T cells with actD and analyzed the cytoplasmic fate of HIV-1 genomic RNA by RPA (7). First, conditions were determined under which RNA synthesis was inhibited to 15% of that of the mock-treated control with no overt effect on the viability of the infected CEM(A) T cells. Second, we evaluated the effect of 4 h of actD treatment on viral particle assembly and established that virion assembly was sustained. Radioimmunoprecipitation assays indicated that HIV-1 Gag protein synthesis was sustained at 93% during the 4-h experiment. Pulse-chase experiments showed that newly synthesized Gag protein begins to be assembled into virions by 2 h postsynthesis. Assembly and release of the virions, and processing of the Gag precursor protein within the progeny virions, were also sustained despite the reduction in de novo RNA synthesis. Another critical finding was that Gag protein synthesized prior to actD treatment remains competent for assembly into HIV-1 virions for at least 4 h postsynthesis.
Quantitative RPAs on RNAs collected 4 h post-actD treatment detected an expected decrease in cytoplasmic HIV-1 genome-length RNA (to 30% of the level of the mock-treated control). Virion-associated RNA from equivalent amounts of virions that were standardized by Gag p24 enzyme-linked immunosorbent assay was also decreased (to 60%). The results show that in contrast to MLV, de novo RNA synthesis is not necessary for packaging of HIV-1 genomic RNA. The HIV-1 results are similar to the ASV findings and are consistent with the existence of a single multifunctional RNA population (Fig. 1, model 2).
Dorman and Lever analyzed RNAs from HIV-1- or HIV-2-infected Jurkat T cells after treatment with actD or leptomycin B (LMB) (11). Similar to actD, LMB inhibits Rev function and disrupts nuclear export of intron-containing HIV-1 RNA (42). Therefore, LMB provided a second approach to curtailing cytoplasmic accumulation of genome-length RNA in HIV-1-infected cells. The effect of actD or LMB on the activity of HIV-2 Rev has not been characterized.
RPAs detected coordinate decreases in HIV-1 and HIV-2 cytoplasmic and virion-associated RNA levels following actD treatment (11). For HIV-1, cytoplasmic RNA and virion RNA levels declined to 40% of those of the controls after 9 h of actD treatment. The decline in virion RNA was calculated after standardization of virion abundance by exogenous RT assay. Unexpectedly, HIV-2 cytoplasmic and virion RNA levels were sustained at 80 to 100% of control levels at 12 h post-actD treatment. The relative persistence of the HIV-2 RNA implies that cytoplasmic HIV-2 proviral transcripts are inherently more stable in the actD assay than their HIV-1 counterparts. Surprisingly, particle abundance, as measured by exogenous RT activity, declined to 65% despite the maintenance of the steady-state cytoplasmic RNA. A possible explanation is that actD treatment produces a defect in RT function or virion assembly.
LMB treatment also produced proportionate responses in HIV-1 and HIV-2 cytoplasmic and virion-associated RNA levels. HIV-1 cytoplasmic RNA was reduced to 25% and virion-associated RNA levels declined to 45% after a 12-h incubation with LMB. Again, HIV-2 genome-length cytoplasmic RNA and virion-associated RNA levels were sustained at 100 to 140% of control levels at 12 h post-LMB treatment. Similar to the actD result for HIV-2, virion abundance was reduced (to 40%) without reduction of the cytoplasmic RNA level. The possibility that HIV-2 virion assembly is disrupted by actD or LMB treatment was not addressed, but the data imply a requirement for an unknown labile cofactor that is important for virion production. The results of the actD and LMB experiments are in agreement regarding the theory that HIV-1 and HIV-2 genome-length RNAs do not segregate into functionally independent populations of mRNA and genomic RNA. The cytoplasmic fate of HIV-1 and HIV-2 genomic RNA is similar to that of ASV RNA, and the data are consistent with a single RNA population functioning as both mRNA and genomic RNA (Fig. 1, model 2).
In summary, RNA analysis of genomic RNA in infected cells or progeny virus after actD treatment has been an important tool to examine two hypothetical models of genomic RNA trafficking in simple and complex retroviruses. Results with MLV genome-length RNA indicate that the unspliced transcript segregates into two functionally distinct populations in the cytoplasm (Fig. 1, model 1). In contrast, results with ASV, HIV-1, and HIV-2 genome-length RNAs support the existence of a single RNA population that functions interchangeably as both mRNA and genomic RNA (Fig. 1, model 2). These experiments did not address whether genome-length RNA is obligated to function as mRNA prior to being packaged as genomic RNA (model 2A versus model 2B). A modification of the experimental approach was implemented in the HIV-1 system to address this important open question.
Translation is not necessary to produce HIV-1 genomic RNA.
Inhibition of host cell translation was used to evaluate whether translation of the unspliced genomic-length RNA is necessary to target HIV-1 genome-length RNA for packaging into virions (7). Virion assembly in HIV-1-infected T cells is sustained during 4 h of treatment with each of three mechanistically distinct translation inhibitors (pactamycin, cycloheximide, and anisomycin). [35S]cysteine-methionine pulse-chase experiments demonstrated that HIV-1 virions are composed of a combination of steady-state and nascent Gag protein. Newly synthesized [35S]cysteine-methionine-labeled Gag commences to be assembled into virions by 2 h postsynthesis and continues to be incorporated for over 6 h. Immunoprecipitation assays verified that HIV-1 assembly and processing are sustained despite translation inhibition. These results established that de novo translation is not necessary to sustain assembly and release of the virions or processing of the Gag structural protein within the progeny virions.
Virion-associated RNA levels were evaluated by two methods. Quantitative RPAs were used to measure the abundance of packaged HIV-1 RNA transcripts. [3H]uridine pulse-labeling followed by trichloroacetic acid precipitation assay was used to determine the onset of RNA packaging following inhibitor treatment and to detect packaging of HIV-1 transcripts and any cellular RNAs. This assay revealed that the onset of RNA packaging is within 2 h of RNA synthesis and that inhibition of de novo translation does not eliminate the supply of RNA available to virions.
In response to inhibition of cellular translation to 20%, both assays detected expected coordinate decreases in steady-state cytoplasmic RNA and virion-associated RNA. At 4 h, the level of [3H]uridine-labeled RNA in virions decreased to 30 to 50% of that of the mock-treated control. RPA results, however, indicate that the packaging efficiency of HIV-1-specific RNA was sustained at 97% after treatment with anisomycin and had increased up to 165% of that of the mock-treated control following treatment with pactamycin or cycloheximide. These results are in agreement with the hypothesis that de novo translation is not necessary for packaging of HIV-1 genomic mRNA and thus nullify model 2B (Fig. 1). The data support model 2A, i.e., that HIV-1 mRNA can function interchangeably as genomic RNA without a requirement for previous translation.
Two conclusions can be made upon considering the decline in virion-associated RNA detected by the [3H]uridine labeling-trichloroacetic acid assay relative to the increase in packaged HIV RNA detected by the RPA. (i) Cellular RNAs are packaged into HIV-1 virions. Muriaux et al. recently demonstrated that MLV virions also accumulate cellular transcripts, which they attribute to incorporation of small cellular RNAs that are involved in virion assembly (25). (ii) The increased level of HIV-1-specific RNA packaging is attributable to rerouting of genome-length RNA from the translation machinery to viral assembly complexes in the face of diminished competition from the host translation machinery for genome-length RNA. Under these conditions, packaging efficiency is increased because viral assembly components compete more successfully for genomic RNA.
Perspectives.
Since 1974, chemical inhibitors have been applied to study the interdependent relationship between the processes of translation and packaging of genome-length RNA. Recent experiments using highly quantitative RPAs have validated the early results and extended the findings to human retroviruses. Taken together, analyses of genetically simple MLV and ASV or complex HIV-1 and HIV-2 indicate that retroviruses have adapted at least two approaches to manage competition for genome-length RNA from the translational machinery and viral assembly components. Cytoplasmic trafficking of MLV genome-length RNA is distinct from that of ASV, HIV-1, and HIV-2. The half-life of MLV genomic RNA is shorter than that of MLV cytoplasmic genome-length RNA, a finding that supports model 1 (Fig. 1). Model 1 posits that MLV RNA segregates into two distinct populations that function independently as genomic RNA for packaging into progeny virions or mRNA template for protein synthesis (17, 18). There remains the possibility that packaged MLV RNA is inherently less stable than cytoplasmic transcripts. An approach to either corroborate or disprove this hypothesis would be to apply the translation inhibitor approach to the MLV system.
In contrast to that of MLV, genome-length RNAs of ASV, HIV-1, and HIV-2 exhibit dual functionality within the cytoplasm of an infected cell (Fig. 1, model 2A). Experiments with HIV-1-infected T cells and translation inhibitors indicate that prior mRNA translation is not a requirement for generation of an HIV-1 genomic RNA (7), thus eliminating model 2B. The observation that HIV-1 packaging efficiency not only is sustained but can be increased upon translation inhibition indicates that cytoplasmic HIV-1 mRNA transcripts can be rerouted from the translation machinery to assembly complexes. Although analogous experiments have not been performed with ASV or HIV-2 proviruses, studies using ASV or HIV-2 vector RNAs are in agreement with data from studies of replication-competent HIV-1 indicating that coordinate translation is not absolutely required for vector RNA packaging and are consistent with model 2A with regard to their replication-competent parental viruses (16, 37). We also noted, however, that studies with HIV-2 vectors indicate that HIV-2 RNA tends to undergo cotranslational RNA packaging (16).
Why is the pattern of cytoplasmic trafficking of genome-length RNA different for MLV? Complex retroviruses like HIV-1 utilize the well-characterized Rev-responsive element (RRE) in viral RNA and the cognate viral Rev protein to recruit the host's CRM1 nuclear export protein to activate nuclear export of intron-containing RNA (15). An RRE-like RNA element that is believed to be important for recruitment of cellular Rev-like proteins has been observed in the 3′ UTR of ASV (29). MLV has not been shown to contain an RRE-like element in the 3′ UTR, and recent studies have focused on the possibility that nuclear export of MLV genome-length RNA is mediated by sequences in proximity of a gag splice site (27, 28). We hypothesize that MLV genome-length RNA accesses a specialized nucleocytoplasmic trafficking pathway that dictates cytoplasmic utilization of MLV genome-length RNA as either mRNA or genomic RNA.
Analysis of the cytoplasmic fate of retroviral genomic RNAs has spanned 3 decades and has yielded important insights into the interdependent relationship between translation and packaging during retroviral replication. These insights have application for improvement of retroviral vector systems for gene transfer applications. Specific inhibition of HIV-1 vector RNA translation is predicted to increase the packaging efficiency of the vector genome and potentially augment the titer of vector virus produced from the helper cells.
Acknowledgments
We thank Karen Beemon, Patrick Green, C. Martin Stoltzfus, and Stacey Hull for insightful comments on the manuscript.
This work was supported by National Institutes of Health grants R29AI40851 (to K.B.-L.) and P30CA16058 (to The Ohio State University Comprehensive Cancer Center).
REFERENCES
- 1.Arya, S. K., M. Zamani, and P. Kundra. 1998. Human immunodeficiency virus type 2 lentivirus vectors for gene transfer: expression and potential for helper virus-free packaging. Hum. Gene Ther. 9:1371-1380. [DOI] [PubMed] [Google Scholar]
- 2.Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214:177-218. [DOI] [PubMed] [Google Scholar]
- 3.Berlioz, C., and J.-L. Darlix. 1995. An internal ribosomal entry mechanism promotes translation of murine leukemia virus gag polyprotein precursors. J. Virol. 69:2214-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berlioz, C., C. Torrent, and J.-L. Darlix. 1995. An internal ribosomal entry signal in the rat VL30 region of the Harvey murine sarcoma virus leader and its use in dicistronic retroviral vectors. J. Virol. 69:6400-6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boris-Lawrie, K., T. M. Roberts, and S. Hull. 2001. Retroviral RNA elements integrate components of post-transcriptional gene expression. Life Sci. 69:2697-2709. [DOI] [PubMed] [Google Scholar]
- 6.Buck, C. B., X. Shen, M. A. Egan, T. C. Pierson, C. M. Walker, and R. F. Siliciano. 2001. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J. Virol. 75:181-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butsch, M., and K. Boris-Lawrie. 2000. Translation is not required to generate virion precursor RNA in human immunodeficiency virus type 1-infected T cells. J. Virol. 74:11531-11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cullen, B. R. 2000. Nuclear RNA export pathways. Mol. Cell. Biol. 20:4181-4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Costa, J., H. Brown, P. Kundra, A. Davis-Warren, and S. Arya. 2001. Human immunodeficiency virus type 2 lentiviral vectors: packaging signal and splice donor in expression and encapsidation. J. Gen. Virol. 82:425-434. [DOI] [PubMed] [Google Scholar]
- 10.Deffaud, C., and J.-L. Darlix. 2000. Rous sarcoma virus translation revisited: characterization of an internal ribosome entry segment in the 5′ leader of the genomic RNA. J. Virol. 74:11581-11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorman, N., and A. Lever. 2000. Comparison of viral genomic RNA sorting mechanisms in human immunodeficiency virus type 1 (HIV-1), HIV-2, and Moloney murine leukemia virus. J. Virol. 74:11413-11417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geballe, A. P., and M. K. Gray. 1992. Variable inhibition of cell-free translation by HIV-1 transcript leader sequences. Nucleic Acids Res. 20:4291-4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilboa, E., M. Kolbe, K. Noonan, and R. Kucherlapati. 1982. Construction of a mammalian transducing vector from the genome of Moloney murine leukemia virus. J. Virol. 44:845-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffin, S. D., J. F. Allen, and A. M. Lever. 2001. The major human immunodeficiency virus type 2 (HIV-2) packaging signal is present on all HIV-2 RNA species: cotranslational RNA encapsidation and limitation of Gag protein confer specificity. J. Virol. 75:12058-12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hope, T. J. 1999. The ins and outs of HIV Rev. Arch. Biochem. Biophys. 365:186-191. [DOI] [PubMed] [Google Scholar]
- 16.Kaye, J. F., and A. M. Lever. 1999. Human immunodeficiency virus types 1 and 2 differ in the predominant mechanism used for selection of genomic RNA for encapsidation. J. Virol. 73:3023-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levin, J. G., P. M. Grimley, J. M. Ramseur, and I. K. Berezesky. 1974. Deficiency of 60 to 70S RNA in murine leukemia virus particles assembled in cells treated with actinomycin D. J. Virol. 14:152-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levin, J. G., and M. J. Rosenak. 1976. Synthesis of murine leukemia virus proteins associated with virions assembled in actinomycin D-treated cells: evidence for persistence of viral messenger RNA. Proc. Natl. Acad. Sci. USA 73:1154-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez-Lastra, M., C. Gabus, and J.-L. Darlix. 1997. Characterization of an internal ribosomal entry segment within the 5′ leader of avian reticuloendotheliosis virus type A RNA and development of novel MLV-REV-based retroviral vectors. Hum. Gene Ther. 8:1855-1865. [DOI] [PubMed] [Google Scholar]
- 20.McBride, M. S., M. D. Schwartz, and A. T. Panganiban. 1997. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J. Virol. 71:4544-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCann, E. M., and A. M. Lever. 1997. Location of cis-acting signals important for RNA encapsidation in the leader sequence of human immunodeficiency virus type 2. J. Virol. 71:4133-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Messer, L. I., J. G. Levin, and S. K. Chattopadhyay. 1981. Metabolism of viral RNA in murine leukemia virus-infected cells: evidence for differential stability of viral message and virion precursor RNA. J. Virol. 40:683-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer, B. E., and M. H. Malim. 1994. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev. 8:1538-1547. [DOI] [PubMed] [Google Scholar]
- 24.Miele, G., A. Mouland, G. P. Harrison, E. Cohen, and A. M. Lever. 1996. The human immunodeficiency virus type 1 5′ packaging signal structure affects translation but does not function as an internal ribosome entry site structure. J. Virol. 70:944-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 98:5246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohlmann, T., M. Lopez-Lastra, and J.-L. Darlix. 2000. 2000. An internal ribosome entry segment promotes translation of the simian immunodeficiency virus genomic RNA. J. Biol. Chem. 275:11899-11901. [DOI] [PubMed] [Google Scholar]
- 27.Oshima, M., T. Odawara, K. Hanaki, H. Igarashi, and H. Yoshikura. 1998. cis elements required for high-level expression of unspliced Gag-containing message in Moloney murine leukemia virus. J. Virol. 72:6414-6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oshima, M., T. Odawara, T. Matano, H. Sakahira, Y. Kuchino, A. Iwamoto, and H. Yoshikura. 1996. Possible role of splice acceptor site in expression of unspliced gag-containing message of Moloney murine leukemia virus. J. Virol. 70:2286-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paca, R. E., R. A. Ogert, C. S. Hibbert, E. Izaurralde, and K. L. Beemon. 2000. Rous sarcoma virus DR posttranscriptional elements use a novel RNA export pathway. J. Virol. 74:9507-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paillart, J. C., L. Berthoux, M. Ottmann, J.-L. Darlix, R. Marquet, B. Ehresmann, and C. Ehresmann. 1996. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DNA synthesis. J. Virol. 70:8348-8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parkin, N. T., E. A. Cohen, A. Darveau, C. Rosen, W. Haseltine, and N. Sonenberg. 1988. Mutational analysis of the 5′ non-coding region of human immunodeficiency virus type 1: effects of secondary structure on translation. EMBO J. 7:2831-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paskind, M. P., R. A. Weinberg, and D. Baltimore. 1975. Dependence of Moloney murine leukemia virus production on cell growth. Virology 67:242-248. [DOI] [PubMed] [Google Scholar]
- 33.Pelletier, J., and N. Sonenberg. 1985. Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell 40:515-526. [DOI] [PubMed] [Google Scholar]
- 34.Poeschla, E., J. Gilbert, X. Li, S. Huang, A. Ho, and F. Wong-Staal. 1998. Identification of a human immunodeficiency virus type 2 (HIV-2) encapsidation determinant and transduction of nondividing human cells by HIV-2-based lentivirus vectors. J. Virol. 72:6527-6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a.Rein, A., L. E. Henderson, and J. G. Levin. 1998. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 23:297-301. [DOI] [PubMed] [Google Scholar]
- 35.Sadaie, M. R., M. Zamani, S. Whang, N. Sistron, and S. K. Arya. 1998. Towards developing HIV-2 lentivirus-based retroviral vectors for gene therapy: dual gene expression in the context of HIV-2 LTR and Tat. J. Med. Virol. 54:118-128. [DOI] [PubMed] [Google Scholar]
- 36.Shimotohno, K., and H. M. Temin. 1981. Formation of infectious progeny virus after insertion of herpes simplex thymidine kinase gene into DNA of an avian retrovirus. Cell 26:67-77. [DOI] [PubMed] [Google Scholar]
- 37.Sonstegard, T. S., and P. B. Hackett. 1996. Autogenous regulation of RNA translation and packaging by Rous sarcoma virus Pr76gag. J. Virol. 70:6642-6652. [PMC free article] [PubMed] [Google Scholar]
- 38.Stoltzfus, C. M., K. Dimock, S. Horikami, and T. A. Ficht. 1983. Stabilities of avian sarcoma virus RNAs: comparison of subgenomic and genomic species with cellular mRNAs. J. Gen. Virol. 64:2191-2202. [DOI] [PubMed] [Google Scholar]
- 39.Tabin, C. J., J. W. Hoffmann, S. P. Goff, and R. A. Weinberg. 1982. Adaptation of a retrovirus as a eucaryotic vector transmitting the herpes simplex virus thymidine kinase gene. Mol. Cell. Biol. 2:426-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Temin, H. M. 1992. Origin and general nature of retroviruses, p. 1-15. In J. A. Levy (ed.), The Retroviridae. Plenum Press, New York, N.Y.
- 41.Vagner, S., A. Waysbort, M. Marenda, M. C. Gensac, F. Amalric, and A. C. Prats. 1995. Alternative translation initiation of the Moloney murine leukemia virus mRNA controlled by internal ribosome entry involving the p57/PTB splicing factor. J. Biol. Chem. 270:20376-20383. [DOI] [PubMed] [Google Scholar]
- 42.Wolff, B., J. J. Sanglier, and Y. Wang. 1997. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol. 4:139-147. [DOI] [PubMed] [Google Scholar]