Abstract
Explicit solvent molecular dynamics (MD) simulations were carried out for sarcin–ricin domain (SRD) motifs from 23S (Escherichia coli) and 28S (rat) rRNAs. The SRD motif consists of GAGA tetraloop, G-bulged cross-strand A-stack, flexible region and duplex part. Detailed analysis of the overall dynamics, base pairing, hydration, cation binding and other SRD features is presented. The SRD is surprisingly static in multiple 25 ns long simulations and lacks any non-local motions, with root mean square deviation (r.m.s.d.) values between averaged MD and high-resolution X-ray structures of 1–1.4 Å. Modest dynamics is observed in the tetraloop, namely, rotation of adenine in its apex and subtle reversible shift of the tetraloop with respect to the adjacent base pair. The deformed flexible region in low-resolution rat X-ray structure is repaired by simulations. The simulations reveal few backbone flips, which do not affect positions of bases and do not indicate a force field imbalance. Non-Watson–Crick base pairs are rigid and mediated by long-residency water molecules while there are several modest cation-binding sites around SRD. In summary, SRD is an unusually stiff rRNA building block. Its intrinsic structural and dynamical signatures seen in simulations are strikingly distinct from other rRNA motifs such as Loop E and Kink-turns.
INTRODUCTION
The sarcin–ricin domain (SRD) is a highly conserved component of 23S–28S rRNA from the large ribosomal subunit. The SRD motif forms a crucial site for the binding of elongation factors EF-Tu and EF-G (1,2). EF-Tu participates in the binding of aminoacyl-tRNA to ribosomes while EF-G catalyzes the translocation process of peptidyl-tRNA from the A site to the P site. Binding of elongation factors is inhibited by ribotoxins, such as α-sarcin (ribonuclease) and ricin (RNA N-glycosidase), which specifically modify the SRD leading to inactivation of the ribosome. Sarcin cleaves the phosphodiester backbone on the 3′-side of G2661 (for numbering see Figure 1a) (4), while ricin depurinates the 5′-adjacent A2660 (5,6). There are two SRD areas which are recognized by both toxins and elongation factors: the GAGA tetraloop, where toxin cleavage sites are positioned, and the bulged G2655 (2,7–9). The G2655 nucleotide represents the most critical site for EF-binding (8). However, the primary determinant of recognition is not the nucleotide type but the SRD conformation (8,10,11).
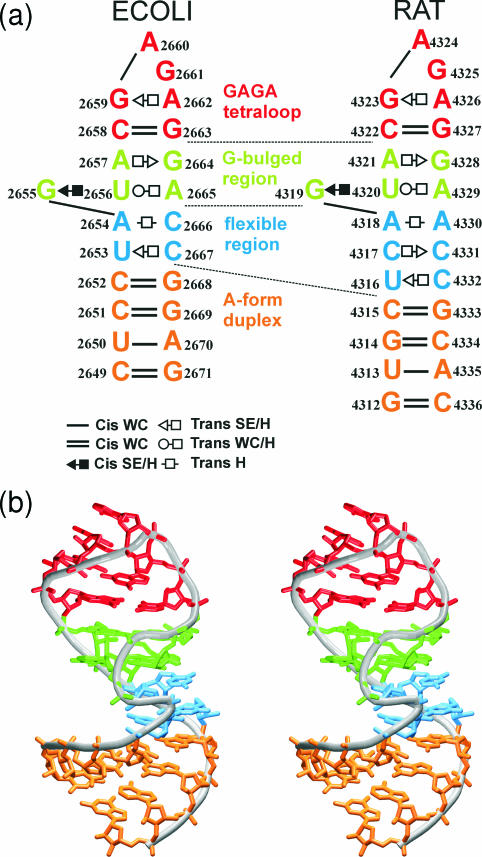
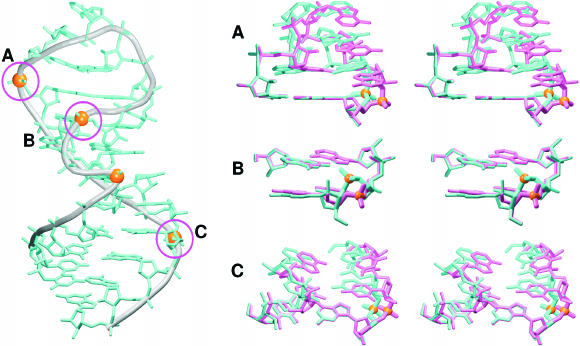
Figure 1.
(a) The studied SRD motifs. Base pairs are labeled according to Leontis et al. (3). Original numbering is used in this study. (b) Stereo view of the SRD motif (ECOLI). The S-turn is visible at the fore.
Several atomic resolution structures of the SRD motif were determined. Except for SRD motifs in the ribosomal crystal structures (12,13), there are structures of isolated SRD observed by both X-ray and NMR techniques, namely, the SRD from Escherichia coli 23S rRNA [X-ray, resolution 1.11 Å (10) and 1.04 Å (11)] and rat 28S rRNA [X-ray 2.1 Å (14) and NMR (15–17)]. There are also two mutant structures that mimic the 28S rRNA SRD motif (11). Three different analogs of the same motif were also observed in the complex with restrictocin (sarcin analog) (18). Although atomic resolution structures of SRD–EF complexes have yet to be determined, there is evidence that the SRD is in the close proximity of the EF catalytic center (19). A homology modeling approach and energy minimization method were used for preparation of an atomic model of 70S ribosomal complex containing SRD motif (20).
We report long-scale molecular dynamics (MD) simulations of two SRD structures based on crystal structures of E.coli (ECOLI) and rat (RAT) SRD motifs. Despite force field and sampling limitations (21), MD simulations represent a useful tool to study non-canonical RNA molecules (22–37). This study is a continuation of our efforts to characterize all key non-Watson–Crick ribosomal RNA motifs (24–26,28). The aim of simulations is to reveal the intrinsic properties of isolated RNA motifs and to suggest how they could affect the ribosomal structures. Despite that the surrounding components modify the dynamics of smaller rRNA fragments in ribosome, their intrinsic properties undoubtedly pre-determine their roles in the ribosome. Thus, for example, intrinsic properties of isolated K-turns and sarcin–ricin loop are strikingly different (see below). One of the advantages of simulations is that they are too short to allow any major unfolding of the isolated RNAs, so we can compare the intrinsic properties of all rRNA motifs in the ribosome-like arrangements. It is not always possible with solution experiments. Stochastic fluctuations seen in MD simulations usually represent a good indicator of the deformability of the molecules also on a longer time scale (38). In addition, simulations capture some features (such as long-residency hydration) that are not apparent in the experiments. Thus, properly executed simulations represent a legitimate tool to complement the experiments.
Description of studied SRD motifs
The SRD motifs are distorted hairpins containing a GAGA tetraloop, a G-bulged cross-strand A-stack, a flexible region and a terminal A-form duplex (Figure 1). The secondary structure of both studied SRD motifs is very similar for residues 2655–2665 (using ECOLI numbering—for the RAT numbering see Figure 1). We use the following abbreviations: WC, Watson–Crick; H, Hoogsteen; and SE, Sugar Edge.
The GAGA tetraloop exhibits common GNRA tetraloop backbone geometry. GNRA tetraloops can differ in base orientation and are characterized by the following features (14). The first and the fourth bases (G2659 and A2662) form modified trans SE/H (‘sheared’) G/A base pair (Figure 2a) which is stabilized by bifurcated H-bond interconnecting N2(G) with N7(A) and O2P(A). Another H-bond between N3(G) and N6(A) was reported in NMR studies (15,16,39). Crystallographic studies suggest that these base pairs tend to be water-mediated between N3(G) and N6(A) (40). The last three bases of the tetraloop form a triple purine stack. Its orientation to the closing WC base pair can be nearly parallel (crystal structures of SRD GAGA tetraloops) or altered (crystal structure of GUAA tetraloop) (40). Typically, a contact between O2′(G2659) and N7(G2661) is observed (14). Torsion angles in GNRA tetraloops adopt predominantly A-form values. Sugar puckering is C3′-endo in crystal structures while mixed puckering (C2′-endo and C3′-endo) was found by NMR (10,41).
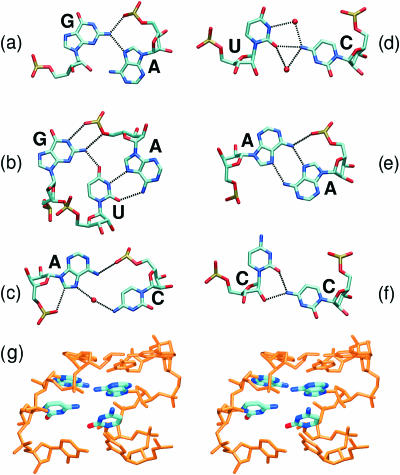
Figure 2.
Non-Watson–Crick base pairs in crystal structures of studied SRD motifs. Tetraloop region, (a) trans SE/H G/A base pair; G-bulged region, (b) G/U/A triplet of cis SE/H G/U and trans WC/H U/A base pairs; flexible region, (c) trans H water-mediated A/C base pair; (d) trans SE/H water-mediated U/C base pair, (e) trans H A/A base pair and (f) trans SE/H C/C base pair. (a–d) are based on the crystal structure of ECOLI system (PDB, 1Q9A), (e and f) are based on the crystal structure of mutated RAT system (PDB, 1Q96). (g) Stereo view of the flexible region of the RAT crystal structure (PDB, 430D) with unpaired bases in A/A and C/C base pairs (highlighted).
The bulged G-motif consists of trans H/SE A2657/G2664 base pair and G2655/U2656/A2665 base triple (Figure 2b) which is formed by cis SE/H G/U and trans WC/H U/A base pairs. The motif has characteristic cross-strand stacking of adenines and the backbone kink of A2665. In contrast to Loop E A-stack, the SRD motif has an extra bulged guanine G2655 which is paired with U2656 and forms the cross-strand G-stack with G2664 from the opposite strand. The nucleotide most critical for binding of EF-G is the bulged G2655 (8).
The flexible regions contain non-Watson–Crick base pairs shown in the Figure 2c–f. The flexible regions differ in number of non-Watson–Crick base pairs (RAT, 3 bp; ECOLI, 2 bp) and their type. The ECOLI flexible region is formed by water-mediated trans H/H A2654/C2666 and trans H/SE C2667/U2653 base pairs. The RAT flexible region contains trans H/H A4318/A4330, trans H/SE C4317/C4331 and trans H/SE C4332/U4316 base pairs. Owing to lower resolution of the RAT crystal structure no interconnecting water molecules were identified in this area. A/A and mainly C/C base pairs are not coplanar in the crystal RAT structure (Figure 2g), but crystal structures of SRD mutants (1Q93 and 1Q96) (11) obtained at higher resolution revealed coplanar orientation of the interacting bases. Different flexible regions of eukaryotes and prokaryotes likely prevent the EFs of one species from functioning with the ribosomes of the other (10).
The S-turn is caused by the bulged G and consists of two turns: the first [residues U2653–A2654 (ECOLI), C4317–A4318 (RAT)] reverses the chain direction and the second [G2655–U2656 (ECOLI), G4319–U4320 (RAT)] restores the chain direction (11). The chain direction is changed by torsion angle deviations throughout the backbone of A2654 and G2655. Their C2′-endo sugar pucker places the 2′-hydroxyl groups into the major groove.
METHODS
The simulations were carried out using the AMBER6 and AMBER7 programs (42) with Cornell et al. force field (43). Systems were neutralized with sodium counterions (Na+ radius 1.868 Å and well depth 0.0028 kcal/mol) placed either by Xleap module of AMBER (simulations ECOLI1 and RAT1) or manually positioned into phosphate bisectors (simulations ECOLI2 and RAT2). Xleap places the cations iteratively into the minima of the electrostatic potential which is calculated on a grid (grid spacing 1 Å) using Coulomb's law and distance dependent dielectric constant. While the Xleap is currently the most common method to initially place cations, it possibly could lead to trapping of ions and bias the ion positions in shorter simulations. The manual placement of ions may improve sampling. However, our experience with ∼3 μs of RNA simulations suggests that trapping likely does not pose a problem and the monovalent ion distribution is not affected by the initial ion positioning in any systematic way for simulations longer than 5–10 ns.
A box of ∼6500 TIP3P water molecules (44) was added around the molecule to a depth of 10–16 Å. Simulations were carried out using the particle mesh Ewald technique (45) with 9 Å nonbonded cutoff and 2 fs time step. Equilibration started by 5000 steps of minimization followed by 250 ps of MD, with the atomic positions of the solute molecule fixed. Then, two series of minimization (1000 steps) and MD simulation (20 ps) were carried out with restraints of 50 and 25 kcal/(mol·Å2) applied to all solute atoms. This was followed by five rounds of a 1000-step minimization with solute restraints reduced by 5 kcal/(mol·Å2) in each round. Then 100 ps of unrestrained MD followed, with the system heated from 50 to 300 K. The production MD runs were carried out with constant pressure boundary conditions (relaxation time of 1.0 ps). Constant temperature of 300 K using the Berendsen weak-coupling algorithm was applied with a time constant of 1.0 ps. SHAKE (46) constraints with a tolerance of 10−8 Å were applied to all hydrogens to eliminate the fastest X-H vibrations and allow a longer simulation time step. Translational and rotational center-of-mass motion was removed every 5 ps.
Trajectories were analyzed using Carnal and Ptraj modules of AMBER as well as with in-house software. Averaged distances (over a given period) between H-bond donors and acceptors below 3.2 Å are classified as H-bonds. As any distance threshold is artificial, detail monitoring of the dynamics of possible H-bonds was carried out to assure that H-bonds with slightly longer donor–acceptor distances are not missed.
Significant motions of the molecule were analyzed using principal component analysis (PCA) (47,48) in Ptraj module. PCA is based on construction of a covariance matrix from the atomic fluctuations along the trajectory. This matrix is diagonalized, which provides a set of eigenvectors and eigenvalues. Then, filtered main motions along the most significant eigenvectors are analyzed.
Hydration and ion distributions were tracked by binding atom positions from root mean square (r.m.s.) coordinate fit frames over all solute atoms at 1 ps intervals into (0.5 Å)3 grids. This procedure gives water density around the solute and, thus, main hydration sites are visible. Then, systematic monitoring of solute–solvent and solute–ion distances is carried out. Molecular interaction potentials (MIPs) were calculated by DelPhi (49) by solving the non-linear Poisson–Boltzmann equation with implicit solvent representation and zero ionic strength. Visualization was carried out using VMD (50), Pymol (DeLano Scientific, San Carlos, CA) and InsightII (Biosym/MSI, San Diego, CA). Interaction (stacking) energies between two bases were calculated using the force field (Anal module) considering only the two bases in a given mutual geometry and neglecting the rest of the system. The atomic charges were not modified after dissection. The interaction energy is then defined as the energy difference between the given base stack and the two bases separated into infinity assuming dielectric constant of 1 (in vacuo calculation). Relaxation of the monomers is not performed. The calculations thus include only the van der Waals and electrostatic terms. The calculations provide crude insight in the stability because solvent screening is not included.
Initial structures
The RAT simulated system was based on the 29 nt RNA crystal structure (PDB code, 430D; NDB code, UR0002) determined at 2.1 Å resolution. Two terminal G/C WC base pairs were omitted. The ECOLI simulated structure was based on the 27 nt RNA crystal structure (PDB code, 483D; NDB code, UR0007) determined at 1.11 Å. Unpaired terminal nucleotides U, G and adjacent G/U base pair were removed. Additional simulations (35 ns in total) of mutated structures (containing C4322G/G4327C and C4322U/G4327A mutations) based on the rat SRD motif (11) were carried out. These control simulations exhibited very similar behavior as simulations of non-mutated structures and their results are only briefly mentioned in Supplementary Data.
RESULTS
The length of each simulation of ECOLI and RAT systems was 25 ns (total 100 ns). The simulations were very stable with instantaneous r.m.s.d. values to the crystal structures along the whole trajectories 1.5–1.7 Å (Supplementary Figure S1).
Hydrogen bonding
ECOLI simulations
All WC base pairs were stable. Non-WC base pairs are described in detail in Supplementary Table S1. Originally, G2659 and A2662 are connected by bifurcated hydrogen bond between N2(G) and N7(A)/O2P(A). During the simulation new contact between N1(G) the O2P(A) was established. In the tetraloop area, there is also H-bond contact between O2′(G2659) and N7(G2661). The G2664/A2657 base pair contains direct H-bonds between bases in the crystal: N7(A)–N2(G) and N6(A)–N3(G). Other contacts are N6(A2657)–O2′(G2664), N1(G2664)–O2P(U2656), N2(G2664)–O2P(U2656) and O6(G2664)–O2′(G2655). The last three contacts of G2664 with backbone were disrupted due to backbone switch observed in early stages of the simulation followed by modest reorientation of G2655 phosphate (see below).
The base triplet G2655/U2656/A2665 is formed by two base pairs: G/U with one base-base H-bond N2(G)–O4(U), and U/A connected by N3(U)–N7(A) and O2(U)–N6(A) H-bonds. Other H-bonds are N1(G)–O2P(A) and N2(G)–O5′(A). During the simulation a slight shear of A2665 and U2656 bases disrupted O2(U)–N6(A) H-bond (distance 4.1 Å). G and U remained coplanar while angle between A and U base planes increased from 20 to 40°.
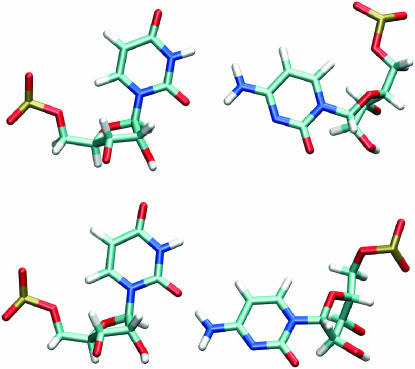
In the flexible region, (see also Hydration) the A2654/C2666 base pair has only N6(A)–O2P(C) and C8(A)–O1P(A) contacts. During the simulation the second contact disappeared. The X-ray base pair U2653/C2667 is connected by one H-bond between O2(U) and N4(C). In simulations, C2667 fluctuates leading to alternating participation of both hydrogens of cytosine aminogroup in the H-bond (Figure 3). The H-bond is almost equally shared by both hydrogens (55% population for H41 binding). Similar U/C geometries were observed by others (36).
Figure 3.
Different geometries of U/C base pair observed in MD simulations.
RAT simulations
Compared with ECOLI structure, the crystal geometry of RAT G4323/A4326 base pair is a little bit different. Again, there are two H-bonds N2(G)–N7(A) and N2(G)–O2P(A), but there is a relatively short distance of 3.5 Å between N3(G) and N6(A) not observed in the ECOLI structure. During the simulation this base pair relaxed to a similar geometry as described above for ECOLI simulations. The A4321/G4328 base pair is connected by two H-bonds, N2(G)–N7(A) and N3(G)–N6(A). Similarly to ECOLI simulation, the close contacts between G4328 and sugar-phosphate backbone N1(G4328)–O2P(U4320), N2(G4328)–O2P(U4320), O6(G4328)–O2′(G4319) and N1(G4328)–O2′(G4319) are not observed during the simulation. Another N6(A4321)–O2′(G4328) close contact was formed. The crystal geometry of G4319/U4320/A4329 triplet and its simulation behavior are very similar to the ECOLI triplet, with the angle between U4320 and A4329 base planes ∼33° in simulations.
The flexible region in the X-ray structure is markedly perturbed but is swiftly repaired in the simulation. The U4316/C4332 base pair exhibits similar behavior as the U2653/C2667 of ECOLI (see above). Base pairs A4318/A4330 and C4317/C4331 have bases vertically separated by ∼1 and 3 Å, respectively (Figure 2g). The A4318/A4330 base pair relaxes into nice in-plane geometry during equilibration, with two symmetrical N7(A)–N6(A) H-bonds and another N6(A4318)–O2P(A4330) contact. Bases of C4317/C4331 have no H-bond contact in the crystal and do not interact with the sugar-phosphate backbone. The O2(C4317)–N4(C4331) distance is 3.2 Å but the atoms are above each other. Again, this base pair is repaired in early stages of the simulations with N4(C4331)–O2(C4317) and N4(C4331)–O2′(C4317) H-bonds. Another arrangement with two symmetrical N4(C)–N3(C) H-bonds forms in the interval 5–9 ns in simulation RAT2. Geometries of A/A and C/C base pairs found in simulations are consistent with geometries observed in more accurate crystal structures of mutated RAT SRD, e.g. 1Q96 (Supplementary Table S1).
Sugar puckering, glycosidic torsions and backbone angles
The backbone dynamics was very similar in ECOLI and RAT simulations.
In agreement with the initial crystal structures, sugar puckering is preferentially C3′-endo (0–25°), except of S-turn residues A2654 and G2655, characterized by C2′-endo sugar puckering (165–170°). Glycosidic torsion angles for majority of residues are in the anti range of 185–230°. However, the bulged G2655 (and G4319 in RAT) was switched from high anti conformation (X-ray χ angle ∼260°) to syn conformation (χ angle of 320°) during the first 500 ps of all simulations. While the value of 320° is formally within the syn region it is still rather close to the average value typical for bulged G-motif and rather far from the common syn value (∼45°). The observed shift by ∼60° is related to a backbone switch of P(U2656) described below. This event leads to geometrical changes in the sugar-phosphate backbone of G2655 while the position of the base is not affected.
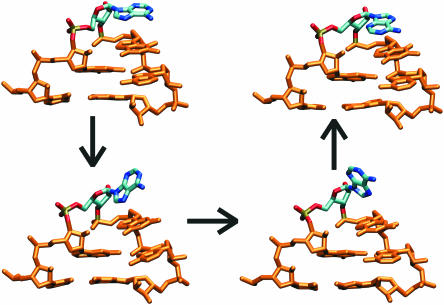
Changes of sugar pucker and χ angle were observed in the tetraloop region. First, rotation of the A2660 (and A4324 in RAT) base localized in the tetraloop apex around the glycosidic bond occurred in almost all simulations (Figure 4) due to change of χ(A2660) from anti to syn (Supplementary Figure S2). This occurred at 19.5, 19 and 5.5 ns in ECOLI1, RAT1 and ECOLI2 simulations, respectively, with no switch in RAT2. After the switch, only very short attempts to regain the anti position were seen. Both anti to syn orientations further exhibit two χ substates with oscillations on a scale of several hundreds picoseconds (Supplementary Table S2). Anti conformation is characterized by values of 180 and 200°. Syn conformation is characterized by substates around 340 and 50°. The value of 340° is typical for the parallel orientation of the triple purine stack A2660-G2661-A2662 (and the equivalent RAT bases) with respect to the adjacent WC base pair (standard X-ray orientation). χ angles of G2661 and A2662 are in this case around 200°. χ angle of 50° for A2660, and 250° for G2661 and A2662 lead to altered orientation known from the crystal structure of GUAA tetraloop (40) (Supplementary Figure S3). The triple purine stack was found in the altered orientation (population of 13% of the simulation time) predominantly when A2660 adopted syn conformation. Changes in sugar puckering were observed entirely for A2660 and only in simulations RAT1 and ECOLI2. The switch from C3′-endo to C2′-endo is connected with A2660 reorientation. In case of RAT1 simulation sugar pucker was changed at 19 ns and remained C2′-endo till the end of the simulation, while sugar pucker oscillated several times in ECOLI2 simulation (45% population for C2′-endo since 5.5 ns—Supplementary Figure S2).
Figure 4.
Reorientation of A2660 base (highlighted). Original anti conformation is in the upper left corner, the alternative syn conformation in the upper right corner, intermediates in the base-flipping process are below.
Virtual torsion angles (51) and are typically in the intervals of 160–170° and 220–240° in the SRD crystal structures, except of the S-turn, the opposite kink and the tetraloop apex. There are few and only modest backbone changes observed in almost all ECOLI and RAT simulations. At the beginning of all simulations switch of phosphate U2656 (and RAT U4320) has been observed (Figure 5B), associated with disruption of O2P(U2656)–N2(G2664), and O2′(G2655)–O6(G2664) contacts and change of glycosidic torsion angle of bulged G2655 (see above). In the crystal structure, the phosphate geometry is characterized by α2656, β2656 and γ2656 (290°, 150° and 40°, respectively). During the early stages of all simulations the phosphate group is reoriented to form a more compact phosphate cluster in the S-turn motif. Then, two compensatory substates with identical phosphate positions are seen, with α2656, γ2656 (75°, 180°) or (200°, 60°) and β2656 = 180°. Note again that only the local phosphate position is affected.
Figure 5.
Backbone switches observed in MD simulations. Switching phosphates are presented as orange balls. Labeling of phosphates in magenta circles is consistent with presented stereo views: G2663 switch (A); U2656 switch (B); and G2668 switch (C). Crystal structure is in cyan, the averaged MD structure in magenta, the backbone around the switched phosphate in bold.
Another switch in the tetraloop region concerns phosphate group of G2663 (and RAT G4327). The crystal G2663 is characterized by torsion angles ζ2662, α2663, β2663 and γ2663 (300°, 145°, 220° and 180°, respectively). These angles change to (330°, 295°, 160° and 70°) during the equilibration (canonical RNA) with no effect on the base and phosphate group positions. During the simulation, switch to (70°, 230°, 80° and 180°) is observed (Figure 5A) when the shifted stacking geometry occurs (see below).
There is also an oscillation of S-turn phosphate A2654 (A4318) (torsion angles ɛ2653, ζ2653 and α2654). The phosphate group fluctuates between two positions spending with 1–2 ns period (Supplementary Figure S4a). Strikingly, this dynamics agrees well with bistability of phosphate groups positioned in the S-turn motif (A2654 and G2655) visible in the high-resolution crystal structure (Supplementary Figure S4b).
In the duplex region, the following irreversible phosphate switches were observed: G2668 switch at 16 ns (ECOLI1) (Figure 5C), A2670 switch at 19 ns (ECOLI2) and C4332 switch at 14 ns (RAT1). All the flipped geometries are characterized by α = 140° and γ = 185°, while the standard geometry is described by α = 290° and γ = 60° (β is in both cases ∼180°). This flipped angle combination is typical for the AII-RNA backbone family, which is an established (X-ray) A-RNA duplex substate (52). Such topology is seen in both crystal structures of ECOLI for phosphate U2650. The changes of selected torsion angles connected with U2656, G2663 and G2668 switches in ECOLI1 simulation are shown in Supplementary Figure S5.
Stacking energies and shifted tetraloop substate
Stacking energies were calculated along the trajectories (for definition see Methods). Total interaction energies and van der Waals (VDW) energy terms were analyzed. The VDW term corresponds to what is in experimental studies discussed as base stacking overlap. Force field calculation is actually more exact than any visual overlap inspection commonly reported in X-ray studies. Most interesting changes were observed in the tetraloop (only ECOLI numbering is used in the text). Analysis of stacking identified two distinct geometries (observed in both ECOLI and RAT systems): (i) standard (X-ray) geometry characterized by stabilizing vertical contacts of G2659-G2663, C2658-G2659 and A2662-G2663, and (ii) shifted geometry, where all previously mentioned contacts are destabilized. The shifted geometry brings an increased overlap of guanines G2659 and G2663, which leads to improved VDW term. Note that the VDW term is considered to be decisive for stabilization in a polar environment. The contact between A2662 and G2663 is almost lost (Supplementary Figure S6). The shifted substate was observed in the following intervals: 15–25 ns in ECOLI1, 7–12 ns and 17–22 ns in ECOLI2, 4–11 ns and 14–25 ns in RAT1 and 19–23 ns in RAT2. Note that the G2659-G2663 stack participates in the four-guanine stack. When the G2659-G2663 stacking interaction is weaker, the vertical contact between G2663 and G2664 is improved and vice versa. Stacking interaction G2655-G2664 is not affected. The shifted geometry is correlated with G2663 phosphate switch (see above). Described interaction energies are summarized in Supplementary Table S3 and the full account of base stacking is given in the Supplementary Data.
Flexibility analysis
Dynamical behavior of the ECOLI and RAT structures was analyzed in terms of r.m.s.d., atomic and nucleotide positional fluctuations and PCA. These analyses were compared with data for other ribosomal motifs: Loop E (26,28) and K-turn (25). Simulated SRD structures are characterized by low instantaneous r.m.s.d. values (∼1.5 Å), comparable with r.m.s.d. values observed for Loop E. R.m.s.d. values of flexible K-turns, which behave as oscillating molecular elbows, are much higher (Supplementary Data). The PCA reveals a lack of any non-local movements in the simulated SRD motifs. This contrasts SRD even with the Loop E that shows clearly visible major (deep) groove breathing nanosecond-scale oscillations (several angstroms) as the leading dynamical movement. This means that Loop E is easily deformable in the direction of major groove width compression or extension. The main SRD dynamics principal component (first mode) is just the local tetraloop shift reported above, and the second mode represents mainly moderate fast thermal fluctuations of the terminal duplex base pairs. The G-bulged motif and the flexible region are motionless. It means that these regions have no easily accessible alternative substates or a broader range of low-energy conformations available around the X-ray structure, otherwise they would be spontaneously sampled in the multiple 25 ns simulations. Also atomic positional fluctuations and r.m.s.d. values per nucleotide confirm the local geometrical changes observed in the tetraloop region: movements of A2660 and the modest tetraloop re-stacking. All these analyses illustrate an amazing lack of non-local motions in the simulated SRD fragments, to our opinion not seen in any longer RNA simulations so far. This feature of SRD is unexpected, as other RNA motifs behave differently in analogous simulations and the SRD base pairing pattern in the flexible region looks visually weak. The simulated SRD systems stay firmly locked in their starting geometries. Full extensive account of the analyses is given in the Supplementary Data, including comparison with Loop E and Kink-turns.
Molecular interaction potential
Both crystal structures exhibit very similar minimal MIP values of −16.5 kT (ECOLI) and −17.6 kT (RAT), with similar distribution of electronegative sites. The most electronegative sites are positioned between phosphate groups (O2P atoms) of U2656 (U4320) and A2657 (A4321) (Supplementary Figure S15a). Other electronegative sites appeared at a contour level around −13 kT and are mainly localized in the major groove of the flexible regions. MIP maps were calculated for series of averaged MD structures (Supplementary Figure S15b and c for two representative MIP maps). The minimal MD MIP values are around −14.5 kT and the complete results are given in Table 1.
Table 1.
Important electronegative sites observed in majority of averaged ECOLI and RAT structures (averages over each 1 ns) and characterized by contour levels of −12 and −14 kT (boldface)
| Name of MIP site | Region | Atoms (ECOLI) | Atoms (RAT) |
|---|---|---|---|
| I | Tetraloop | N3(G2659), N7(A2662) | N3(G4323), N7(A4326) |
| IIa | Tetraloop | Phosphates G2659, A2660, G2661 | Phosphates G4323, A4324, G4325 |
| IIb | Tetraloop | Phosphates C2658, G2659 | Phosphates C4322, G4323 |
| IIc | Tetraloop | Phosphates A2657, C2658 | Phosphates A4321, C4322 |
| IId | Tetraloop/G-bulged A-stack | Phosphates U2656, A2657 | Phosphates U4320, A4321 |
| IIIa | Tetraloop/four-G stack | Phosphates A2662, G2663 | Phosphates A4326, G4327 |
| IIIb | Tetraloop/four-G stack | Phosphates G2663, G2664 | Phosphates G4327, G4328 |
| IIIc | four-G stack | N7,O6(G2663), N7,O6(G2664) | N7,O6(G4327), N7,O6(G4328) |
| IIId | G-bulged A-stack | Phosphates G4328, A4329, O6(G4319) | |
| IV | Flexible region | Phosphates C2652, U2653, O2′(A2654) | Phosphates U4316, C4317, O2′(A4318) |
| IVuc, IVcc | Flexible region | Phosphates C2666, C2667, N3(U2653), O4(U2653) | Phosphates C4331, C4332, N3(U4316), O4(U4316), N3(C4317) |
| IVaa | Flexible region | N1(A4318), phosphate A4330 | |
| V | G-bulged A-stack/flexible region | N3(A2654), N3(G2655) | N3(A4318), N3(G4319) |
| VIa | Flexible region | Phosphates A4330, C4331 | |
| VIb | Flexible region/regular duplex | Phosphates C2667, G2668 |
Interaction with ions and long-residency hydration
Crystallographic studies of SRD structures did not provide much information about ion positions, except the highly resolved crystal structure of ECOLI revealing ion binding (thallium) near A2665 and C2666 (10). This ion forms a lattice contact with a symmetry-related molecule and MD simulations did not show any ion-binding site in this region.
In simulations, several ion-binding sites with inner-shell occupancy higher than 10% (for both ECOLI and RAT data see Table 2) were identified. The binding sites are localized in the tetraloop and flexible regions, mainly at N7 atoms of guanine. They clearly coincide with MIP electronegative sites.
Table 2.
Ion-binding sites with inner-shell occupancies above 15% in at least one simulation
| Ion-binding site | Occupancy (%) | Maximum binding time (ns) | ||||||
|---|---|---|---|---|---|---|---|---|
| ECOLI1 | ECOLI2 | RAT1 | RAT2 | ECOLI1 | ECOLI2 | RAT1 | RAT2 | |
| Tetraloop and G-bulged A-stack region | ||||||||
| N7(G2659,G4323) | 84 | 47 | 52 | 83 | 2.6 (12) | 3.0 (9) | 2.0 (9) | 3.4 (11) |
| N7(G2663,G4327), N7(G2664,G4328) | 31 | 6 | 9 | 43 | 1.8 | 0.5 | 0.4 | 0.7 |
| O6(G2655,G4319), O2P(G2664,G4328) | 9 | 9 | 21 | 7 | 0.9 | 0.3 | 0.8 | 0.7 |
| Flexible region | ||||||||
| O4(U2653) | 19 | 9 | x | x | 2.2 | 0.6 | x | x |
| O4(U4316) | x | x | 20 | 21 | x | x | 0.9 | 0.8 |
| O2,N3(C4317) | x | x | 24 | 6 | x | x | 4.4 | 0.8 |
| Duplex region | ||||||||
| N7(G2668) | 52 | 55 | x | x | 1.7 | 1.5 | x | x |
| N7(A2670) | 25 | 23 | x | x | 1.2 | 0.8 | x | x |
| N7(G4333) | x | x | 16 | 12 | x | x | 0.9 | 0.5 |
Occupancies exceeding 20% are in boldface. Maximum binding times represent a period of uninterrupted inner-shell binding while numbers in parentheses indicate a period when an ion was found in the described ion-binding site and the surrounding area (i.e. including fluctuations not exceeding 300 ps). Sites marked by ‘x’ are not observable either in ECOLI or in RAT systems.
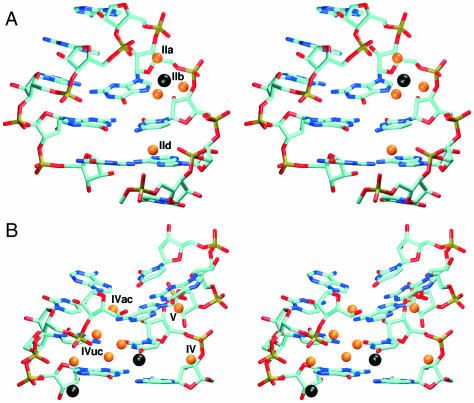
The most occupied ion-binding site of ECOLI structure was found in the tetraloop region near N7(G2659) (Figure 6A). Occupancy of this site by ions (inner-shell binding) is 47–84% and several ions were exchanged in this site during each simulation. One ion was localized in this area for 12 ns. However, its direct unperturbed contacts with N7 did not exceed 3 ns, and in-between the ion was visiting phosphate groups 4–7 Å away. Other important ion-binding site was found in the four-guanine stack, interconnecting N7 and O6 atoms of G2663 and G2664. The occupancy was 50% in the interval of 0–15 ns of ECOLI1 simulation while no ions were seen there in period 15–25 ns. We expect that this ion-binding site disappeared due to changes in the tetraloop geometry at 15 ns. In ECOLI2 simulation, this site was not substantially occupied. Other three ion-binding sites were found in the flexible region and the duplex part. One with 20% occupancy is positioned near O4(U2653) (Figure 6B). The second one is positioned near N7(G2668) with occupancy around 55% (Figure 6B) and the last one is near N7(A2670) with occupancy of 25%.
Figure 6.
Hydration and ion-binding sites observed in MD simulations. Ions and water molecules (only oxygens are shown) are black and orange balls, respectively. (A) Tetraloop area—ion positioned near N7(G2659), waters at IIa, IIb and IId positions, (B) ECOLI flexible region—ions near O4(U2653) and N7(G2668); waters at IV, IVac, IVuc and V sites.
Hydration sites correspond to positions of the most electronegative MIP sites. Maximal water binding times did not exceed 4–5 ns and were typically around 1–2 ns for majority of long-residing water molecules. Positions of many MD hydration sites are in a very good agreement with X-ray water molecules. The important hydration sites are described in the Table 3, stereo figures presenting global hydration patterns around the ECOLI and RAT molecules are available in the Supplementary Figure S16. Hydration of RAT is very similar to the ECOLI (Supplementary Table S6).
Table 3.
Hydration sites visible at the contour level of 30 (7× higher bulk density) in ECOLI and RAT simulations
| Name of common hydration site | Atoms (ECOLI) | Atoms (RAT) | Maximum residency time (ns) | Average residency time (ns) |
|---|---|---|---|---|
| I | N3(G2659), N6(A2662), N7(A2662) | N3(G4323), N6(A4326), N7(A4326) | 2.5 | 0.5–1.0 |
| IIa–IIc | 2.0 | 0.5–1.0 | ||
| IId | N2(G2664), O2P(U2656), O2P(A2657) | N2(G4328), O2P(U4320), O2P(A4321) | 1.0 | 0.5 |
| N3(A2657), O2′(A2657) | N3(A4321), O2′(A4321) | 0.7 | 0.1–0.5 | |
| IV | O2P(C2652), O2P(U2653), O2′(A2654) | O2P(U4316), O2P(C4317), O2′(A4318) | 0.7 | 0.1–0.5 |
| V | N3(A2654), N3(G2655) | N3(A4318), N3(G4319) | 2.0a/4.0b | 1.0a/1.0–2.0b |
| S | O2P(U2653), O2P(G2655) | O2P(C4317), O2P(G4319) | 0.5 | 0.1–0.3 |
| S | O1P(U2653), O1P(G2655) | O1P(C4317), O1P(G4319) | 0.5 | 0.1–0.3 |
| Name of specific ECOLI hydration site | Atoms | Maximum binding time (ns) | Average binding time (ns) | |
| IVuc | N3(U2653), N4(C2667), O2P(C2666) | 4.8 | 1.0-2.0 | |
| IVac | N6(A2654), N7(A2654), N4(C2666) | 1.0 | 0.5 | |
| IIIc | O6(G2659), O6(G2663), O2P(A2662), ion | 1.1 | 0.5 | |
| VI | N7(G2668), O2P(C2667), ion | 1.2 | 0.5 | |
Hydration sites with longest residency times are in boldface.
aECOLI simulations.
bRAT simulations.
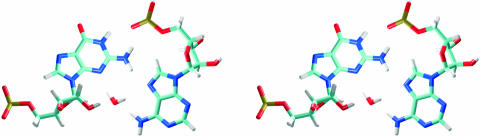
In ECOLI simulation, there is water-mediated base pair G2659/A2662 (hydration site I). Water molecule interconnects atoms N3(G) and N6,N7(A) (Figure 7) with binding times 0.5–2 ns. In the major groove of the tetraloop region and the two neighboring base pairs, there is a set of water molecules interconnecting adjacent phosphates (sites IIa–IIc). Several of them are also in the close contact with the ion positioned near N7(G2659) (Figure 6A). Maximal binding time does not exceed 2 ns. Longer hydration times are observed for waters contacting an ion. Another interesting hydration site in this area is between N2(G2664) and O2P(A2657) atoms (site IId) (Figure 6A). In the crystal structure, these atoms are in close contact, but during the equilibration period the distance between N2(G) and O2P(A) increases and hydration site is formed.
Figure 7.
Water-mediated tetraloop G/A base pair in MD simulations. Water molecule interconnects N3(G) and N6,N7(A) atoms.
Hydration site was found also in the four-guanine stack region, but only if this site is occupied by ion (site IIIc). In this case water molecule interconnects the ion with O6 atoms of G2659 and G2663, also with participation of O2P(A2662). In the minor groove side of A2657/G2664 base pair, there is a hydration site formed by N3(A) and O2′(A) atoms. There are two water-mediated base pairs in the flexible region (Figure 6B). The contact between U2653/C2667 is mediated by one direct hydrogen bond [N4(C)–O2(U)] and one water bridge between N4(C) and N3(U) (site IVuc) with binding times 1–5 ns. Water positioned in this site is also in close contact with O2P(C2666). The second water-mediated base pair is A2654/C2666 without any direct hydrogen bond connection [except fluctuations when N4(C) and N7(A) were connected by H-bond]. This base pair is stabilized by water molecule between N4(C) and N6,N7(A) in the minor groove (site IVac).
One of the most important hydration sites is positioned between G2655/U2656/A2665 triplet and the adjacent A2654/C2666 base pair (site V) (Figure 6B). This hydration site is formed by N3 atoms of A2654 and G2655. Note that only phosphate position was affected by the modest χ angle change at G2655 (see above) and we suggest that this minor local structural change is not influencing hydration and ion-binding sites involving G2655. The S-turn creates several hydration sites stabilizing clustered phosphate groups. Hydration site IV is close to the previous site (both sites are bridged by other water molecules), and interconnects O2′(A2654) with O2P(U2653) and O2P(C2652) (Figure 6B).
DISCUSSION AND CONCLUSIONS
We have carried out MD simulations of two SRD motifs (ECOLI and RAT) to provide a basic characterization of structural and dynamical properties of this ribosomal motif. Behavior of ECOLI and RAT simulations was strikingly similar. Although there are some minor differences between the crystal structures used as the starting models, (the different H-bonding in the tetraloop G/A base pair and distorted RAT flexible region in lower-resolution RAT crystal structure) all simulations resulted into identical SRD behavior.
The GAGA tetraloop is the most dynamical part of the SRD motif. The first and fourth bases of the tetraloop form trans SE/H G/A base pair, water-mediated in all simulations. It contains one bifurcated H-bond connecting N2(G) with N7(A) and O2P(A). N3(G) and N6(A) atoms, which typically form an H-bond in trans SE/H G/A base pairs, create a long-residency hydration site together with N7(A). Highly resolved crystal structures of E.coli SRD (483D and 1Q9A) show an increased distance between N3(G) and N6(A) atoms but no crystal water molecules are indicated in the position observed in simulations. X-ray water molecules are positioned close to N6(A) but are not deep enough to form contacts with N7(A) and N3(G). NMR structure (15,16) and low-resolution crystal structure of SRD (14) do not suggest this hydration site as the distance between N3(G) and N6(A) is 3.4 Å. Several theoretical studies of GNRA tetraloops suggest the presence of water molecule in G/A base pair [GCAA (53–55) and GUGA (56)]. The tetraloop region also contains the most occupied ion-binding site, the N7(G2659, G4323 RAT) atom, with 50–80% inner-shell occupancies.
The simulations show changes in the sugar puckering and glycosidic torsion angle for A2660 (A4324 RAT) at the tetraloop apex. In crystal structures, sugar pucker for tetraloop residues is C3′-endo, while NMR experiments suggest mixed population of C3′-endo/C2′-endo (17,41). In simulations, A2660 ribose repuckered to C2′-endo and this was connected with A2660 anti to syn flipping. While C3′-endo pucker was partly populated for A2660 in syn conformation, C2′-endo never occurred when A2660 adopted the original anti conformation. Flip of the apex base was also observed in simulations of GCAA tetraloop using implicit solvent model (57) but the presence of syn bases in unbound GNRA tetraloops is not supported by any current atomic resolution experiments. However, experiments suggest that GNRA tetraloops adopt an unfolded geometry upon binding of elongation factors and/or toxins, as in the SRD–restrictocin complex (18). The unfolded geometry is characterized by distinct geometrical changes of almost all tetraloop bases. The χ angle of A2660 is shifted to ∼260° (high anti) while adjacent bases (G2661 and A2662) are flipped away with the χ angle between 30 and 70° (syn). We cannot rule out that the A2660 χ dynamics may be partly affected by force field imperfectness, but it has no non-local effect on the simulated structures.
Mutual shifts were observed between G2659/A2662 and closing C2658/G2663 base pairs. The shifted substate is reversible with population ∼40% and is characterized by an increased overlap of guanine aromatic rings while the vertical interaction between A2662 and G2663 is lost. Similar geometry was found in the crystal structures of GYRA tetraloops and 2 out of 13 X-ray structures of GRRA tetraloops with resolution at least 2.4 Å (40). Thus, we suggest that it is a viable substate. Simulations of GCAA tetraloops carried out by Sorin et al. (57) showed extended geometry of G/A base pair characterized by flipped out adenine. These simulations, however, neglected explicit solvent, which is an important contact mediator in the tetraloop G/A base pair. The shifted geometry could represent an intermediate structure in the unfolding process; such unfolded tetraloop geometry was observed in the SRD–restrictocin complex (18).
The simulations suggest presence of a number of long-residency hydration sites in SRD structures. Common feature of the flexible region is formation of water bridges instead of direct H-bonds. Despite the differences in the RAT and ECOLI flexible regions, both systems exhibit many similar features. Both areas are characterized by electronegative potential site with minimum around −15 kT. This minimum, however, is much weaker than in some other RNA systems, like 5S rRNA Loop E (28) (∼−21 kT), Hepatitis delta virus ribozyme (23) or kissing-loop motif (27) (both ∼−30 kT). Electronegative sites in the three later systems are occupied permanently by (often multiple) cations in simulations (22,23,26,27,29). In case of SRD motif, it is supposed that cations are not necessary for the stability of this motif (10) and this is supported by the simulations.
Except for the changes in the tetraloop, positions of bases remained well conserved during the simulations. The sugar-phosphate backbone exhibits few localized flips. Some flips basically agree with SRD or other RNA crystal structures (the fluctuations of phosphate A2654 in the S-turn motif, and the common phosphate AII-RNA switches observed in the duplex part). Other switches are not observed in the SRD crystals. The switch of G2663 is connected with the shifted tetraloop geometry while the switch of U2656 leads to formation of more compact phosphate cluster in the S-turn and presence of several hydration sites in this area. While the G2663 switch is reversible with ∼50% population, the U2656 switch occurs in early stages of all simulations and is irreversible. This switch has no effect on position of bases and likely reduces repulsion in the most electronegative site positioned between U2656 and A2657 phosphate groups (Supplementary Figure S15a). Importantly, we evidenced no cumulative backbone flips similar to those recently reported for more than 10 ns B-DNA simulations (58,59). (Behavior of RAT simulations was again close to identical.)
The simulation outcome is affected by force field approximations and limited nanosecond-scale sampling. Regarding the sampling, the simulations show unusually good agreement with the starting structures and lack of any structural changes or oscillations. While it does not rule out presence of additional substates, these would have to be separated from the X-ray like geometry by >5–6 kcal/mol free energy barrier, otherwise they would be sampled (25). Multiple 25 ns scale simulations are entirely capable to reveal all significant monovalent ion-binding sites (27,28,58) and long-residency hydration sites, thus the ion binding and hydration picture is qualitatively converged. The agreement with X-ray structures indicates a good performance of the force field. Cornell et al. force field provides balanced description of stacking and H-bonding of bases, including interactions utilizing the 2′ hydroxyl group (60,61). This stabilizes simulations of RNA molecules with an extended network of base–base interactions (such as SRD). The instantaneous r.m.s.d. of simulated structures with respect to the X-ray structures oscillates around 1.5–1.7 Å. The r.m.s.d. between the averaged simulated ECOLI structure and the high-resolution X-ray structure is 1.0 and 1.4 Å before and after the tetraloop rearrangements, respectively. For comparison, r.m.s.d. between the high-resolution and low-resolution (the 70S ribosome) ECOLI SRD X-ray structures is ∼1.7 Å. Notable is the swift reparation of the perturbed RAT flexible region when starting the simulation from the lower-resolution RAT X-ray structure. The force field description is assumed to be less accurate for the backbone; however, the present study indicates (except of few local changes) good performance of the force field over a wide range of RNA backbone topologies. It still does not rule out occurrence of force field artifacts on a longer time scale. The most likely region to experience them would be the apical tetraloop and the A2660 dynamics could indicate their onset. Nevertheless, this and other recent studies indicate that conventional explicit solvent AMBER RNA simulations can be rather safely used to study even complicated RNA molecules on a time scale around 25 ns, which is sufficient to discern many qualitative structural and dynamical features of these molecules (22–29,62). No other force field has so far been tested for non-canonical RNA molecules. Accuracy of simulations on a longer time scale remains to be established.
Theoretical SRD structures are in a relatively good agreement with SRD geometry observed in the Haloarcula marismortui 50S ribosomal subunit (12), considering that these systems differ in flexible region sequences. We also compared the theoretical ECOLI structure with the geometry observed in the E.coli 70S ribosome structure (13). The flexible region and the tetraloop area (when compared separately) are in a good agreement. However, the global shape of both SRD structures is slightly different due to geometrical difference in the G/U/A base triplet. This triplet is coplanar in the 70S ribosome, but in both theoretical and highly resolved crystal structures an inclination between U and A base planes is observed (∼35° in MD simulations and ∼20° in the crystal) while G and U bases remain coplanar. This difference could originate in the lower resolution of the 70S ribosome crystal. Comparison of cryo-EM maps (63) and available X-ray structures indicates rotation of the SRD around its helical axis (by ∼17°) with a rotational center in the upper half of the SRD helical axis. The origin of the motion was not determined and the authors supposed that the SRD was moving as a rigid unit. The motion could be induced by twisting movement of two distinct lobes of the stalk base region (domains II and VI). Thus, considering the available ribosomal structures, the SRD domain also appears to be stiff.
In summary, the simulations reveal that the SRD is a unique RNA shape. Our data support the statement that the SRD is structurally conserved (20). The central part of the SRD molecule is unusually stiff, which is quite surprising in the flexible region containing consecutive at first sight poorly bound base pairs. These pairs are stabilized by water bridges. Note that long-residency waters (binding times above 1 ns) are not seen around standard duplex regions but are associated with some non-Watson–Crick RNA motifs and compactly folded RNA molecules (23,24,26,28,62). The SRD is a salient example of such molecules. The SRD is perhaps the most rigid (on nanosecond-scale) rRNA motif simulated so far, being amazingly different from Kink-turns that act as large-scale flexible molecular elbows (24,25). Once formed, SRD is firmly locked in its geometry and has no easily accessible alternative substates or wider free energy basin. SRD is even stiffer than the rather rigid 5S rRNA loop E which shows substantial major groove width flexibility. In a sharp contrast to Loop E (and some other RNAs such as kissing-loop complex and hepatitis delta virus ribozyme), SRD is not associated with any deep electrostatic potential pockets and is not a major cation-binding motif.
Supplementary Material
Acknowledgments
This work was supported by Wellcome Trust International Senior Research Fellowship in Biomedical Science in Central Europe GR067507, grants GA203/05/0388 and GA203/05/0009 by Grant Agency of the Czech Republic, and research projects and grants LC512, AVOZ50040507 and MSM0021622413 by Ministry of Education of the Czech Republic. Funding to pay the Open Access publication charges for this article was provided by Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hausner T.P., Atmadja J., Nierhaus K.H. Evidence that the G2661 region of 23S ribosomal-RNA is located at the ribosomal-binding sites of both elongation-factors. Biochimie. 1987;69:911–923. doi: 10.1016/0300-9084(87)90225-2. [DOI] [PubMed] [Google Scholar]
- 2.Moazed D., Robertson J.M., Noller H.F. Interaction of elongation-factors Ef-G and Ef-Tu with a conserved loop in 23S RNA. Nature. 1988;334:362–364. doi: 10.1038/334362a0. [DOI] [PubMed] [Google Scholar]
- 3.Leontis N.B., Stombaugh J., Westhof E. The non-Watson–Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Endo Y., Wool I.G. The site of action of alpha-sarcin on eukaryotic ribosomes—the sequence at the alpha-sarcin cleavage site in 28 S-ribosomal ribonucleic-acid. J. Biol. Chem. 1982;257:9054–9060. [PubMed] [Google Scholar]
- 5.Endo Y., Mitsui K., Motizuki M., Tsurugi K. The mechanism of action of ricin and related toxic lectins on eukaryotic ribosomes—the site and the characteristics of the modification in 28-S ribosomal-RNA caused by the toxins. J. Biol. Chem. 1987;262:5908–5912. [PubMed] [Google Scholar]
- 6.Endo Y., Tsurugi K. RNA N-glycosidase activity of ricin a-chain—mechanism of action of the toxic lectin ricin on eukaryotic ribosomes. J. Biol. Chem. 1987;262:8128–8130. [PubMed] [Google Scholar]
- 7.Gluck A., Wool I.G. Determination of the 28 S ribosomal RNA identity element (G4319) for alpha-sarcin and the relationship of recognition to the selection of the catalytic site. J. Mol. Biol. 1996;256:838–848. doi: 10.1006/jmbi.1996.0130. [DOI] [PubMed] [Google Scholar]
- 8.Munishkin A., Wool I.G. The ribosome-in-pieces: binding of elongation factor EF-G to oligoribonucleotides that mimic the sarcin/ricin and thiostrepton domains of 23S ribosomal RNA. Proc. Natl Acad. Sci. USA. 1997;94:12280–12284. doi: 10.1073/pnas.94.23.12280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perez-Canadillas J.M., Santoro J., Campos-Olivas R., Lacadena J., del Pozo A.M., Gavilanes J.G., Rico M., Bruix M. The highly refined solution structure of the cytotoxic ribonuclease alpha-sarcin reveals the structural requirements for substrate recognition and ribonucleolytic activity. J. Mol. Biol. 2000;299:1061–1073. doi: 10.1006/jmbi.2000.3813. [DOI] [PubMed] [Google Scholar]
- 10.Correll C.C., Wool I.G., Munishkin A. The two faces of the Escherichia coli 23 S rRNA sarcin/ricin domain: The structure at 1.11 angstrom resolution. J. Mol. Biol. 1999;292:275–287. doi: 10.1006/jmbi.1999.3072. [DOI] [PubMed] [Google Scholar]
- 11.Correll C.C., Beneken J., Plantinga M.J., Lubbers M., Chan Y.L. The common and the distinctive features of the bulged-G motif based on a 1.04 angstrom resolution RNA structure. Nucleic Acids Res. 2003;31:6806–6818. doi: 10.1093/nar/gkg908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ban N., Nissen P., Hansen J., Moore P.B., Steitz T.A. The complete atomic structure of the large ribosomal subunit at 2.4 angstrom resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 13.Schuwirth B.S., Borovinskaya M.A., Hau C.W., Zhang W., Vila-Sanjurjo A., Holton J.M., Cate J.H.D. Structures of the bacterial ribosome at 3.5 angstrom resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 14.Correll C.C., Munishkin A., Chan Y.L., Ren Z., Wool I.G., Steitz T.A. Crystal structure of the ribosomal RNA domain essential for binding elongation factors. Proc. Natl Acad. Sci. USA. 1998;95:13436–13441. doi: 10.1073/pnas.95.23.13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szewczak A.A., Moore P.B. The sarcin ricin loop, a modular RNA. J. Mol. Biol. 1995;247:81–98. doi: 10.1006/jmbi.1994.0124. [DOI] [PubMed] [Google Scholar]
- 16.Szewczak A.A., Moore P.B., Chan Y.L., Wool I.G. The conformation of the sarcin ricin loop from 28S ribosomal-RNA. Proc. Natl Acad. Sci. USA. 1993;90:9581–9585. doi: 10.1073/pnas.90.20.9581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warren J.J., Moore P.B. Application of dipolar coupling data to the refinement of the solution structure of the sarcin–ricin loop RNA. J. Biomol. NMR. 2001;20:311–323. doi: 10.1023/a:1011214214552. [DOI] [PubMed] [Google Scholar]
- 18.Yang X.J., Gerczei T., Glover L., Correll C.C. Crystal structures of restrictocin-inhibitor complexes with implications for RNA recognition and base flipping. Nature Struct. Biol. 2001;8:968–973. doi: 10.1038/nsb1101-968. [DOI] [PubMed] [Google Scholar]
- 19.Chan Y.L., Correll C.C., Wool I.G. The location and the significance of a cross-link between the sarcin/ricin domain of ribosomal RNA and the elongation factor-G. J. Mol. Biol. 2004;337:263–272. doi: 10.1016/j.jmb.2004.01.020. [DOI] [PubMed] [Google Scholar]
- 20.Tung C.S., Sanbonmatsu K.Y. Atomic model of the Thermus thermophilus 70S ribosome developed in silico. Biophys. J. 2004;87:2714–2722. doi: 10.1529/biophysj.104.040162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheatham T.E., Young M.A. Molecular dynamics simulation of nucleic acids: Successes, limitations, and promise. Biopolymers. 2000;56:232–256. doi: 10.1002/1097-0282(2000)56:4<232::AID-BIP10037>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 22.Auffinger P., Bielecki L., Westhof E. The Mg2+ binding sites of the 5S rRNA loop E motif as investigated by molecular dynamics simulations. Chem. Biol. 2003;10:551–561. doi: 10.1016/s1074-5521(03)00121-2. [DOI] [PubMed] [Google Scholar]
- 23.Krasovska M.V., Sefcikova J., Spackova N., Sponer J., Walter N.G. Structural dynamics of precursor and product of the RNA enzyme from the hepatitis delta virus as revealed by molecular dynamics simulations. J. Mol. Biol. 2005;351:731–748. doi: 10.1016/j.jmb.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 24.Razga F., Spackova N., Reblova K., Koca J., Leontis N.B., Sponer J. Ribosomal RNA kink-turn motif—a flexible molecular hinge. J. Biomol. Struct. Dyn. 2004;22:183–193. doi: 10.1080/07391102.2004.10506994. [DOI] [PubMed] [Google Scholar]
- 25.Razga F., Koca J., Sponer J., Leontis N.B. Hinge-like motions in RNA kink-turns: the role of the second A-minor motif and nominally unpaired bases. Biophys. J. 2005;88:3466–3485. doi: 10.1529/biophysj.104.054916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reblova K., Spackova N., Koca J., Leontis N.B., Sponer J. Long-residency hydration, cation binding, and dynamics of loop E/helix IV rRNA-L25 protein complex. Biophys. J. 2004;87:3397–3412. doi: 10.1529/biophysj.104.047126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reblova K., Spackova N., Sponer J.E., Koca J., Sponer J. Molecular dynamics simulations of RNA kissing-loop motifs reveal structural dynamics and formation of cation-binding pockets. Nucleic Acids Res. 2003;31:6942–6952. doi: 10.1093/nar/gkg880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reblova K., Spackova N., Stefl R., Csaszar K., Koca J., Leontis N.B., Sponer J. Non-Watson–Crick basepairing and hydration in RNA motifs: Molecular dynamics of 5S rRNA loop E. Biophys. J. 2003;84:3564–3582. doi: 10.1016/S0006-3495(03)75089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Auffinger P., Bielecki L., Westhof E. Symmetric K+ and Mg2+ ion-binding sites in the 5S rRNA loop E inferred from molecular dynamics simulations. J. Mol. Biol. 2004;335:555–571. doi: 10.1016/j.jmb.2003.10.057. [DOI] [PubMed] [Google Scholar]
- 30.Cojocaru V., Nottrott S., Klement R., Jovin T.M. The snRNP 15.5K protein folds its cognate K-turn RNA: a combined theoretical and biochemical study. RNA. 2005;11:197–209. doi: 10.1261/rna.7149605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Golebiowski J., Antonczak S., Fernandez-Carmona J., Condom R., Cabrol-Bass D. Closing loop base pairs in RNA loop–loop complexes: structural behavior, interaction energy and solvation analysis through molecular dynamics simulations. J. Mol. Model. (Online) 2004;10:408–417. doi: 10.1007/s00894-004-0216-7. [DOI] [PubMed] [Google Scholar]
- 32.Cojocaru V., Klement R., Jovin T.M. Loss of G-A base pairs is insufficient for achieving a large opening of U4 snRNA K-turn motif. Nucleic Acids Res. 2005;33:3435–3446. doi: 10.1093/nar/gki664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Auffinger P., Westhof E. RNA solvation: a molecular dynamics simulation perspective. Biopolymers. 2000;56:266–274. doi: 10.1002/1097-0282(2000)56:4<266::AID-BIP10027>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 34.Pan Y.P., Priyakumar U.D., MacKerell A.D. Conformational determinants of tandem GU mismatches in RNA: Insights from molecular dynamics simulations and quantum mechanical calculations. Biochemistry. 2005;44:1433–1443. doi: 10.1021/bi047932q. [DOI] [PubMed] [Google Scholar]
- 35.Li W., Ma B.Y., Shapiro B.A. Molecular dynamics simulations of the denaturation and refolding of an RNA tetraloop. J. Biomol. Struct. Dyn. 2001;19:381–396. doi: 10.1080/07391102.2001.10506748. [DOI] [PubMed] [Google Scholar]
- 36.Sanbonmatsu K.Y., Joseph S. Understanding discrimination by the ribosome: Stability testing and groove measurement of codon–anticodon pairs. J. Mol. Biol. 2003;328:33–47. doi: 10.1016/s0022-2836(03)00236-5. [DOI] [PubMed] [Google Scholar]
- 37.Sanbonmatsu K.Y., Joseph S., Tung C.S. Simulating movement of tRNA into the ribosome during decoding. Proc. Natl Acad. Sci. USA. 2005;102:15854–15859. doi: 10.1073/pnas.0503456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lankas F., Sponer J., Langowski J., Cheatham T.E. DNA basepair step deformability inferred from molecular dynamics simulations. Biophys. J. 2003;85:2872–2883. doi: 10.1016/S0006-3495(03)74710-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jucker F.M., Heus H.A., Yip P.F., Moors E.H.M., Pardi A. A network of heterogeneous hydrogen bonds in GNRA tetraloops. J. Mol. Biol. 1996;264:968–980. doi: 10.1006/jmbi.1996.0690. [DOI] [PubMed] [Google Scholar]
- 40.Correll C.C., Swinger K. Common and distinctive features of GNRA tetraloops based on a GUAA tetraloop structure at 1.4 angstrom resolution. RNA. 2003;9:355–363. doi: 10.1261/rna.2147803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rife J.P., Stallings S.C., Correll C.C., Dallas A., Steitz T.A., Moore P.B. Comparison of the crystal and solution structures of two RNA oligonucleotides. Biophys. J. 1999;76:65–75. doi: 10.1016/S0006-3495(99)77178-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pearlman D.A., Case D.A., Caldwell J.W., Ross W.S., Cheatham T.E., Debolt S., Ferguson D., Seibel G., Kollman P. Amber, a package of computer-programs for applying molecular mechanics, normal-mode analysis, molecular-dynamics and free-energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun. 1995;91:1–41. [Google Scholar]
- 43.Cornell W.D., Cieplak P., Bayly C.I., Gould I.R., Merz K.M., Ferguson D.M., Spellmeyer D.C., Fox T., Caldwell J.W., Kollman P.A. A 2nd generation force-field for the simulation of proteins, nucleic-acids, and organic-molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 44.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 45.Darden T., York D., Pedersen L. Particle Mesh Ewald—an N.Log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 46.Ryckaert J.P., Ciccotti G., Berendsen H.J.C. Numerical integration of the cartesian equations of motion of a system with constrains: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 47.Orozco M., Perez A., Noy A., Luque F.J. Theoretical methods for the simulation of nucleic acids. Chem. Soc. Rev. 2003;32:350–364. doi: 10.1039/b207226m. [DOI] [PubMed] [Google Scholar]
- 48.Wlodek S.T., Clark T.W., Scott L.R., McCammon J.A. Molecular dynamics of acetylcholinesterase dimer complexed with tacrine. J. Am. Chem. Soc. 1997;119:9513–9522. [Google Scholar]
- 49.Gilson M.K., Sharp K.A., Honig B.H. Calculating the electrostatic potential of molecules in solution—method and error assessment. J. Comput. Chem. 1988;9:327–335. [Google Scholar]
- 50.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 51.Duarte C.M., Wadley L.M., Pyle A.M. RNA structure comparison, motif search and discovery using a reduced representation of RNA conformational space. Nucleic Acids Res. 2003;31:4755–4761. doi: 10.1093/nar/gkg682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneider B., Moravek Z., Berman H.M. RNA conformational classes. Nucleic Acids Res. 2004;32:1666–1677. doi: 10.1093/nar/gkh333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sarzynska J., Kulinski T. Dynamics and stability of GCAA tetraloop with 2-aminopurine and purine substitutions. J. Biomol. Struct. Dyn. 2005;22:425–439. doi: 10.1080/07391102.2005.10507014. [DOI] [PubMed] [Google Scholar]
- 54.Sarzynska J., Nilsson L., Kulinski T. Effects of base substitutions in an RNA hairpin from molecular dynamics and free energy simulations. Biophys. J. 2003;85:3445–3459. doi: 10.1016/S0006-3495(03)74766-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zichi D.A. Molecular-dynamics of RNA with the OPLS force-field—aqueous simulation of a hairpin containing a tetranucleotide loop. J. Am. Chem. Soc. 1995;117:2957–2969. [Google Scholar]
- 56.Sarzynska J., Kulinski T., Nilsson L. Conformational dynamics of a 5S rRNA hairpin domain containing loop D and a single nucleotide bulge. Biophys. J. 2000;79:1213–1227. doi: 10.1016/S0006-3495(00)76376-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sorin E.J., Engelhardt M.A., Herschlag D., Pande V.S. RNA simulations: probing hairpin unfolding and the dynamics of a GNRA tetraloop. J. Mol. Biol. 2002;317:493–506. doi: 10.1006/jmbi.2002.5447. [DOI] [PubMed] [Google Scholar]
- 58.Varnai P., Zakrzewska K. DNA and its counterions: a molecular dynamics study. Nucleic Acids Res. 2004;32:4269–4280. doi: 10.1093/nar/gkh765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beveridge D.L., Barreiro G., Byun K.S., Case D.A., Cheatham T.E., Dixit S.B., Giudice E., Lankas F., Lavery R., Maddocks J.H., et al. Molecular dynamics simulations of the 136 unique tetranucleotide sequences of DNA oligonucleotides. I. Research design and results on d(C(p)G) steps. Biophys. J. 2004;87:3799–3813. doi: 10.1529/biophysj.104.045252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sponer J., Jurecka P., Hobza P. Accurate interaction energies of hydrogen-bonded nucleic acid base pairs. J. Am. Chem. Soc. 2004;126:10142–10151. doi: 10.1021/ja048436s. [DOI] [PubMed] [Google Scholar]
- 61.Sponer J.E., Spackova N., Leszczynski J., Sponer J. Principles of RNA base pairing: structures and energies of the trans Watson–Crick/sugar edge base pairs. J. Phys. Chem. B. 2005;109:11399–11410. doi: 10.1021/jp051126r. [DOI] [PubMed] [Google Scholar]
- 62.Schneider C., Brandl M., Suhnel J. Molecular dynamics simulation reveals conformational switching of water-mediated uracil-cytosine base-pairs in an RNA duplex. J. Mol. Biol. 2001;305:659–667. doi: 10.1006/jmbi.2000.4338. [DOI] [PubMed] [Google Scholar]
- 63.Gabashvili I.S., Agrawal R.K., Spahn C.M.T., Grassucci R.A., Svergun D.I., Frank J., Penczek P. Solution structure of the E.coli 70S ribosome at 11.5 A resolution. Cell. 2000;100:537–549. doi: 10.1016/s0092-8674(00)80690-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.