Abstract
Candida albicans is the most common etiological agent of vaginal candidiasis. Elevated host estrogen levels and the incidence of vaginal candidiasis are positively associated. Elevated estrogen levels may affect host and/or fungal cells. This study investigates the effect of 17-β-estradiol, 17-α-estradiol, ethynyl estradiol, and estriol on several C. albicans strains at concentrations ranging from 10−5 to 10−10 M. The addition of 17-β-estradiol or ethynyl estradiol to C. albicans cells caused an increase in the number of cells forming germ tubes and an increase in germ tube length in a dose- and strain-dependent manner. The addition of 17-α-estradiol or estriol did not have a significant effect on germ tube formation by the cultured cells. Exposure to exogenous estrogens did not significantly change the biomass of any C. albicans culture tested. The transcriptional profile of estrogen-treated C. albicans cells showed increased expression of CDR1 and CDR2 across several strain-estrogen concentration-time point combinations, suggesting that these genes are the most responsive to estrogen exposure. Analysis of strain DSY654, which lacks the CDR1 and CDR2 coding sequences, showed a significantly decreased number of germ tube-forming cells in the presence of 17-β-estradiol. PDR16 was the most highly up-regulated gene in strain DSY654 under these growth conditions. The cell biology and gene expression data from this study led to a model that proposes how components of the phospholipid and sterol metabolic pathways may interact to affect C. albicans germ tube formation and length.
The majority of women experience at least one episode of vulvovaginal candidasis (VVC) (44). This condition affects both normally healthy and immunocompromised individuals and has symptomatic manifestations that include a thick white curdlike discharge, intense itching, and dysuria. Candida albicans is isolated from 80% of patients with VVC. While the majority of women experience isolated episodes of VVC, others endure a recurring form of the disease. Women may also be colonized with C. albicans in an asymptomatic state.
Risk factors for VVC include douching, antibiotic therapy, and diabetes (6, 19, 37, 44). Conditions and practices that elevate estrogen levels have also been noted as risk factors for developing disease (37, 46). The elevated estrogen levels present during pregnancy result in both increased vaginal colonization with C. albicans (3, 14, 37) and increased risk of VVC (37). Exogenous estrogens such as oral contraceptives are also associated with increased C. albicans colonization (37) and VVC (46). Elevated estrogen levels are also associated with the use of hormone replacement therapy. In one study, VVC was observed in 26% of postmenopausal women using estrogen in hormone replacement therapy compared to disease in 4% of women who were not using estrogen (45).
The association between elevated estrogen and incidence of C. albicans colonization or vaginal disease may be due to the effect of estrogen on the host, on the fungal cells, or both. The effect of estrogen exposure on C. albicans has received attention in the literature over the years. Cell biological observations of the effects of estrogen on C. albicans have focused primarily on measurements of germ tube formation and length, and on culture biomass. The concentration of estrogen used in some of these studies is above physiological levels. The addition of 10−7 M 17-β-estradiol to a culture of yeast forms increased percent germination and germ tube length (30). Percent germination responded in a dose-dependent manner with concentrations ranging from 10−6 to 10−8 M (30). Another group observed increased germination upon exposure to 17-β-estradiol, although the response was greater as estrogen concentrations decreased from 10−6 to 10−8 M (49). One common feature of these studies was that they were conducted in growth medium containing serum to induce germ tube formation. The undefined and variable composition of serum complicates dissection of the estrogenic effect. A chemically defined growth medium was used to observe that the addition of 10−6 M estrogen led to an increase in biomass of a culture grown for 7 days (22).
The desire to understand the effect of estrogen exposure on C. albicans also led to a series of biochemical studies to identify a C. albicans estrogen receptor. This work resulted in the isolation and purification of Ebp1p (estrogen binding protein 1), which binds 17-β-estradiol with high affinity (38, 43). EBP1 is similar to the Saccharomyces cerevisiae gene that encodes the oxidoreductase old yellow enzyme (OYE). Subsequent experimentation showed that Ebp1p is localized to vacuoles (35, 51) and that 17-β-estradiol weakly inhibits the activity of Ebp1p (7, 35). Ebp1p binds a variety of compounds in addition to 17-β-estradiol and mediates an aromatization and reduction reaction (7).
More recently, the effects of estrogen on C. albicans have been studied at the molecular level (16, 28). Expression of CDR1 and CDR2, which encode multidrug transporters of the ABC family (31, 41), is up-regulated in the presence of estrogen at various concentrations from 10−3 to 10−9 M (16, 28, 31, 50). Cdr1p can specifically transport estradiol in an energy-dependent manner (32). An AP-1 recognition element was noted upstream of the multidrug resistance gene CDR1 in C. albicans (39). The presence of this recognition sequence raised the possibility that expression of CDR1 is influenced by host estrogen (39).
Recently, two new drug responsive elements (DREs) were identified upstream of CDR1 and CDR2. The DRE consensus sequence is 5′ CGGA(A/T)ATCGGATATTTTTTTT 3′ (16). While this consensus sequence is found uniquely upstream of CDR1 and CDR2, a more redundant form of the sequence, 5′ WCGGWWWWCGGWWW 3′ (W is A or T), is found in the promoters of IFU5, RTA3, and HSP12 (13, 16). DRE-containing genes are expressed in response to exposure to antifungal drugs and steroids. Further investigation identified Tac1p as the transcriptional activator that binds to DREs (13). Another cis-acting regulatory element of CDR1 responsive to 17-β-estradiol and another steroid is the steroid-response element SRE2 (28). Collectively, these results suggest that estradiol might be involved in C. albicans drug resistance.
The goal of the studies presented here is to better define the effect of estrogen exposure on C. albicans. This work revisits measurements of germ tube formation and length, and culture biomass using chemically defined growth media. Various concentrations of estrogenic compounds, which include physiological levels, are added to the growth media. These studies investigate the effects of 17-β-estradiol on C. albicans and also look at estrogenic compounds that are found commonly in oral contraceptives and hormone replacement therapy. Growth conditions that demonstrate a significant effect on C. albicans compared to untreated controls are analyzed using microarrays to assess changes in gene expression that accompany the cell biological effects. These data provide a comprehensive evaluation of the phenotypic effects of estrogen exposure on cultured C. albicans cells and associate them with the corresponding changes in gene expression to provide a more integrated view of the cellular mechanisms that respond to estrogen exposure.
MATERIALS AND METHODS
C. albicans strains.
All C. albicans strains were stored at −80°C and streaked to YPD agar plates (per liter: 10 g yeast extract, 20 g peptone, 20 g glucose, 20 g Bacto agar) prior to use. Strain SC5314 (20) was used as the control in all experiments. DSY654 (Δcdr1::hisG/Δcdr1::hisG; Δcdr2::hisG/Δcdr2::hisG-URA3-hisG) (41) was a generous gift from Dominique Sanglard (Institute of Microbiology, University Hospital Lausanne). Other C. albicans strains were clinical isolates collected in a previous study from women who were either symptomatic or asymptomatic for VVC (10). Strains in this collection are given a GC designation that is followed by a number that corresponds to the patient number in the study by Cheng et al. (10). Limited clinical history information to accompany each strain can be found in that reference.
GC strains were evaluated for their ability to form germ tubes in phenol red-free RPMI 1640 medium (RPMI-free; Invitrogen catalog number 11835). Phenol red-free medium was used in all experiments described in the article, since lipophilic impurities in standard phenol red preparations have estrogenic properties (4). A single colony from a YPD agar plate of each strain was inoculated into 15 ml of synthetic complete medium (SC) (24) and grown for 16 h at 37°C with 200 rpm shaking. Strain SC5314 was included as a control. Cells were pelleted and washed three times in Dulbecco's phosphate-buffered saline without Ca2+ or Mg2+ (DPBS; Cambrex catalog number 17-513Q) and counted using a hemacytometer. Cells were inoculated at a density of 5 × 106 cells ml−1 into 15 ml RPMI-free that had been prewarmed at 37°C for 30 min. Cultures were grown for 1 h at 37°C and 200-rpm shaking and cells were then fixed in 1% (vol/vol) glutaraldehyde.
Strains were evaluated for their ability to form germ tubes relative to strain SC5314. This qualitative microscopic evaluation categorized strains as strong germ tube formers that appeared similar to SC5314, intermediate germ tube formers where germination occurred but with obviously lower frequency than for SC5314, and weak germ tube formers where the presence of germ tubes was nearly absent under the growth conditions tested. Evaluations were repeated twice to ensure reproducibility of results. From these observations, two strains were designated as strong germ tube formers, 14 as intermediate, and eight as weak. In subsequent experiments, strain SC5314 represented the strong group, strain GC15 represented the intermediate group, and strain GC29 represented the weak group.
Estrogenic compounds.
17-α-Estradiol, 17-β-estradiol, estriol, and ethynyl estradiol were purchased from Steraloids (Newport, RI). Estrogen stock solutions were dissolved in absolute ethanol and stored at 4°C. Stock solutions were made monthly. 17-β-Estradiol was selected because it is the most biologically common estrogen, 17-α-estradiol as a chemically similar control compound that lacks estrogenic activity, estriol as an estrogen that rises during pregnancy, and ethynyl estradiol because of its common use in oral contraceptives.
Evaluation of germ tube formation.
All glassware for these experiments was acid washed, rinsed thoroughly in MilliQ water, and autoclaved prior to use. A single colony of each C. albicans strain (SC5314, GC15, and GC29) was taken from a YPD agar plate and inoculated into 30 ml of SC medium. This culture was grown for 16 h at 37°C with 200-rpm shaking. Cells were collected and washed three times in DPBS and inoculated at a density of 5 × 106 cells ml−1 into the appropriate growth medium (RPMI-free or SC) that had been prewarmed for 30 min at 37°C and supplemented with estrogen. Estrogenic compounds were added to culture media at a 1:1,000 dilution to achieve final concentrations of 10−5, 10−7, and 10−10 M; an equivalent volume of absolute ethanol was added to the control (no estrogen) culture. Cultures were grown at 37°C and 200 rpm shaking.
At 85 and 300 min, C. albicans cells were removed from the culture, fixed with 1% (vol/vol) glutaraldehyde, and stored at 4°C. Percent germ tube formation was evaluated microscopically by evaluation of 100 cells per culture; three or more replicates were completed for each culture condition. Cells with germ tubes of the same or greater length as those of the mother yeast were considered positive and others were considered negative. Data were expressed as percent positive cells for each culture with three or more replicates of the experiment for each strain-estrogen combination. To assess differences in germ tube formation among the treatment groups, the mean of the observations was calculated and an analysis of variance was conducted using the MIXED procedure in SAS (SAS Institute). Germ tube length was evaluated microscopically using a micrometer. Fifty cells were measured for each culture; three replicates were completed for each culture condition. Mean germ tube length was calculated for each culture condition and analyzed with the MIXED procedure in SAS.
Evaluation of culture biomass.
Cultures were inoculated as described above and grown for 120 and 480 min at 37°C and 200 rpm shaking. Filter membranes (0.2 μm pore size; Fisher) were baked at 80°C for 2 h and immediately weighed on an analytical balance. At the specified time interval, cells from 20 ml of culture volume were collected by filtration on the weighed membranes. Membranes were dried by baking overnight at 80°C and immediately reweighed. Biomass was calculated as the difference between the filter weights. The mean biomass of four replicates was used in calculating the differences between the various treatment groups by using a mixed-model analysis.
Microarray analysis.
Preparation of starter and experimental cultures of C. albicans strains followed the method described above. To evaluate the effect of 17-β-estradiol on C. albicans gene expression, cultures of strain SC5314 were grown for either 10 or 85 min in either SC or RPMI-free with an estrogen concentration of either 10−5 or 10−10 M. Control flasks contained an equivalent volume of absolute ethanol. Cells were collected by filtration, and filters were flash-frozen in a dry ice/ethanol bath and stored at −80°C until RNA was extracted. RNA extraction used a hot phenol method (11). Construction of the PCR product-based microarray and methods for target labeling and data analysis were described previously (52).
Microarray hybridizations were carried out using a standard reference design with a dye swap for each comparison. The “control” cells served as the “reference” for the appropriate comparisons with either SC- or RPMI-free-grown cells. Two biological replicates were completed. Microarray fluorescence data were inspected for normalization using MA plots. Data were normalized with a log2 transformation. Specific estrogen treatments were compared using the MIXED procedure in SAS. The estimate value for each gene is a means separation between the control and estrogen-treated groups. The estimate value and significance for each gene in a comparison were calculated using the least squares estimation using the LSMEANS option in SAS. Fold change was calculated as twice the estimate value.
The PCR product microarray was based on the assembly 6 annotation of the C. albicans sequencing project. orf6 identities were matched to their closest orf19 counterparts using the conversion tool at http://candida.bri.nrc.ca/candida/. orf19 identities were verified at http://agabian.ucsf.edu/canoDB/. When discrepancies occurred, orf19 names were assigned based on a BLAST search at http://www.ncbi.nlm.nih.gov/. Gene names and descriptions were based on descriptions from the Agabian lab (http://agabian.ucsf.edu/canoDB/) and CGD (http://www.candidagenome.org/) websites.
Real-time RT-PCR analysis.
Differential gene expression results from microarray analysis were validated using real-time reverse transcription (RT)-PCR. Specific methods for this analysis were described previously (52). Primers for real-time RT-PCR analysis were designed using PrimerExpress (Applied Biosystems). Each primer had a Tm of between 59°C and 60°C and an amplicon size of between 50 and 100 bp. Primer sequences are shown in Table 1. Primer specificity was verified with a dissociation profile of each amplicon. A standard curve for each primer set was derived using 1:10, 1:25, 1:50, 1:100, 1:250, and 1:500 dilutions of the cDNA. The slopes of the standard curves were within 10% of 100% efficiency. CT values were determined using the Autoanalyze feature of the ABI PRISM 7000 sequence detection system software.
TABLE 1.
Real-time RT-PCR primers used in this studya
| orf6 | orf19 | Gene | Primer name | Sequence (5′-3′) |
|---|---|---|---|---|
| orf6.9037 | orf19.6000 | CDR1 | qCDR1-2F | 5′ AGG CAA TTA GTC AAG ACT CTT CTT CA 3′ |
| qCDR1-3R | 5′ AAG TAT TTC AAT AAA CCT GCT GAC GAG 3′ | |||
| CDR2b | qCDR2-4F | 5′ TGC AAA AGT CAC TTG TGC ACC TA 3′ | ||
| qCDR2-4R | 5′ TGA ACA AGT ACC ATC ACT ATT TGT GGA 3′ | |||
| TEF1c | qRTTEF1F | 5′ CCA CTG AAG TCA AGT CCG TTG A 3′ | ||
| qRTTEF1R | 5′ CAC CTT CAG CCA ATT GTT CGT 3′ | |||
| ALS4c | qRTALS4-F | 5′ TCT GCA ACA CGA GTC AGC TCA 3′ | ||
| qRTALS4-R | 5′ CCG CAC CAA CAC AAG CAT ATA T 3′ | |||
| EBP1d | qEBP1-F | 5′ ATC CTA ATG CTG CTA AAC ATG CAC 3′ | ||
| qEBP1-R | 5′ GCC AAT CTA TTA GCA CCA ACA ACT T 3′ | |||
| orf6.6398 | orf19.1429 | SOH1 | q1429-F | 5′ TCT TTG ATG AAT GAC ATG GTT AAG C 3′ |
| q1429-R | 5′ CTT GGT CAT TCG CAT TCA GTT G 3′ | |||
| orf6.3437 | orf19.1172 | PHO84 | q1172-F | 5′ AAG CTG CCA TTG ATG TCC ATG 3′ |
| q1172-R | 5′ AAG AAG CCT TTG GTG GAG CA 3′ | |||
| orf6.6993 | orf19.6844 | ICL1 | q6844-F | 5′ TGC TTC CAC TTC TAA CGA ACC AT 3′ |
| q6844-R | 5′ TCC ATA GGA TAA TCA GCC AAA TCT G 3′ | |||
| orf6.3288 | orf19.3618 | YWP1 | orf-3618F | 5′ ATT CCA GCT TCA TTA GCT GCT TTC T 3′ |
| orf-3618R | 5′ ATA TTC TTC AAG GGA AAC AAA TGT GA 3′ | |||
| orf6.6934 | orf19.3111 | PRA1 | q3111-F | 5′ GCG ACA GTG GCT CTG ATT CA 3′ |
| q3111-R | 5′ GAT GAG AAC TTG AGG CTG TGC TAC T 3′ | |||
| orf6.648 | orf19.1027 | PDR16 | q1027-F | 5′ GAG CTA CTA AAT GGC ATG AAT CTG AG 3′ |
| q1027-R | 5′ GGT TCG GAT ATA CCA AAT TCA CGT 3′ | |||
| orf6.3947 | orf19.909 | STP4 | q909-F | 5′ CCT GTG TCA AGA CCA AGA AGT ACG T 3′ |
| q909-R | 5′ ATT AGA CAG TAT AGC TGG ATG CAG TTG 3′ | |||
| orf6.5917 | orf19.23 | RTA3 | qRT-1923F | 5′ CAA TGA AGA ATC GAC GTT GCA 3′ |
| qRT-1923R | 5′ TCA TTC CTT ATT CTC TTG AAT AGG ATC AC 3′ | |||
| orf6.854 | orf19.3188 | TAC1 | qRT-3188F | 5′ GAA AGA TCG TGT TAA AGA TGC CAA C 3′ |
| qRT-3188R | 5′ GAG AGA AAA TGC AAG TAA TTG AAT GAG TAC 3′ |
Gene sequences were obtained from the C. albicans genome annotation data at http://agabian.ucsf.edu/canoDB/.
The CDR1 probe on the microarray hybridizes to both CDR1 and CDR2, so gene-specific real-time RT-PCR primers were designed to differentiate between the signals.
Primers were defined by Green et al. (21). The ALS4 probe on the array hybridizes to both ALS2 and ALS4 so signals must be differentiated from each other using gene-specific real-time RT-PCR primers.
EBP1 is not represented on the microarray. EBP1 gene expression data were derived using real-time RT-PCR.
Relative gene expression was quantified using the ΔΔCT method (34), where ΔΔCT = (CT,Estrogen − CT,TEF1) − (CT,Control − CT,TEF1). The change (n-fold) in the target gene, normalized to TEF1, was calculated for each sample by using the equation 2−ΔΔCT. Independent replicate RNA preparations were tested for each target gene. Each RNA sample was reverse transcribed twice. Triplicate real-time PCRs were run on each of the four cDNA preparations. The mean of the CT values for the triplicate reactions was calculated and used to derive a 2−ΔΔCT value. The mean and standard error of the four 2−ΔΔCT values are reported.
RESULTS
Effect of estrogen exposure on germ tube formation and length.
The effect of 17-β-estradiol on germ tube formation was evaluated using three different C. albicans strains (SC5314, GC15, and GC29) and two different growth media (RPMI-free and SC). The three strains were selected because of their different tendencies to form germ tubes in RPMI-free (see Materials and Methods). Different growth media were used to avoid identification of effects that are specific for a single growth condition. SC and RPMI-free were selected because they are defined and, therefore, do not contain complex or variable components such as serum that may confound experimental results. Cultures were supplemented with one of three different 17-β-estradiol concentrations: 10−5, 10−7, or 10−10 M, with the lowest concentration approximating physiological estrogen levels in nonpregnant women.
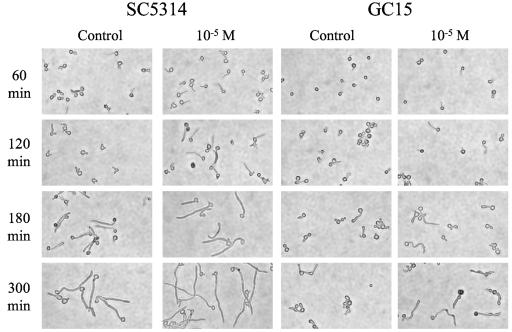
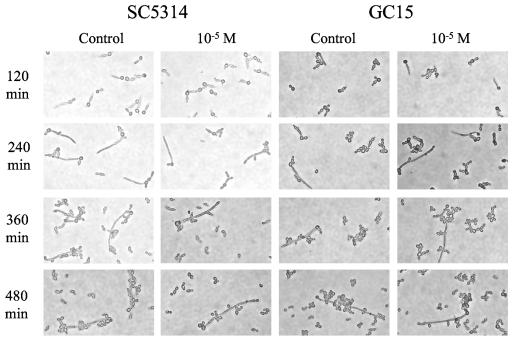
Figure 1 shows the morphology of strains SC5314 and GC15 following culture in RPMI-free; similar images are shown in Fig. 2 for cells grown in SC. Growth in RPMI-free resulted in germ tube formation for strains SC5314 and GC15; cells of strain GC29 (not shown) remained as yeast forms even after several hours of incubation. In SC, strains SC5314 and GC15 formed pseudohyphae (Fig. 2), while GC29 grew as yeast forms (not shown). As incubation progressed in SC, the elongated pseudohyphae gave rise to yeast forms (Fig. 2). This pattern of growth did not lend itself well to counting or measuring the length of elongated forms, so these efforts were pursued only with RPMI-free.
FIG. 1.
Light micrographs of C. albicans cells from strains SC5314 or GC15 grown in RPMI-free for the indicated times in the presence of 10−5 M 17-β-estradiol or an equivalent volume of absolute ethanol (control).
FIG. 2.
Light micrographs of C. albicans cells from strains SC5314 or GC15 grown in SC for the indicated times in the presence of 10−5 M 17-β-estradiol or an equivalent volume of absolute ethanol (control).
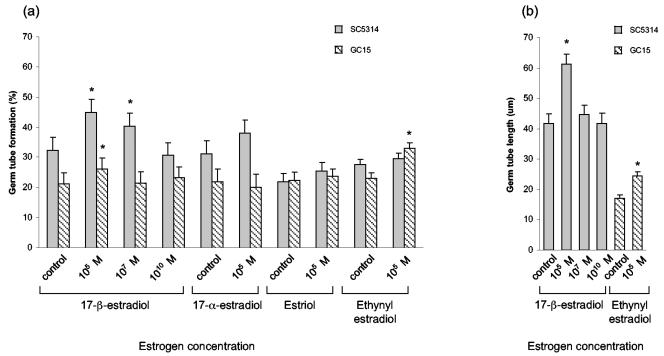
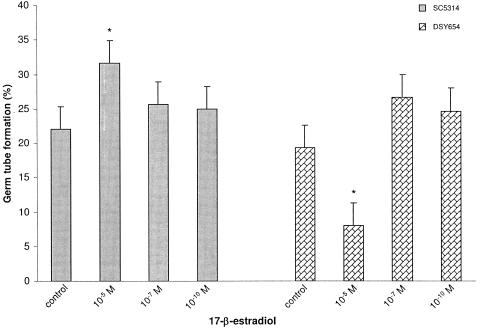
Measurement of percent germ tube formation in RPMI-free showed that strain SC5314 formed more germ tubes at the time points evaluated than strain GC15, although the overall difference between the strains was not statistically significant (P = 0.18) (Fig. 3a). The addition of 17-β-estradiol to the culture medium of SC5314 significantly increased percent germ tube formation at the 10−5 M (P = 0.0001) (Fig. 3a) and 10−7 M concentrations (P = 0.005), but not at the physiological estrogen concentration (P = 0.49). Increased germ tube formation was also observed for strain GC15 at the 10−5 M concentration (P = 0.03), but not at lower concentrations (Fig. 3a). Strain GC29 showed a slight increase (<0.5%) in germ tube formation for strain GC29 at 10−5 M 17-β-estradiol, but this value was not significantly different from that of the control (P = 0.89). The addition of 17-α-estradiol to the culture medium at the highest concentration (10−5 M) did not increase germ tube formation of any strain, suggesting that the observed effects for 17-β-estradiol are specific for that compound (Fig. 3a). The addition of estriol at 10−5 M did not affect germ tube formation (Fig. 3a), however, addition of ethynyl estradiol at 10−5 M increased germ tube formation for strain GC15 (P = 0.03), but not for SC5314 (P = 1.0; Fig. 3a).
FIG. 3.
(a) Histogram summarizing germ tube formation data at 85 min for experiments using different C. albicans strains and various concentrations of estrogenic compounds. Strain GC29 was tested in all growth conditions and gave values of <0.5%. (b) Histogram summarizing germ tube length data at 300 min for strains SC5314 and GC15. Strain GC29 was not tested.
Germ tube lengths were measured on cells from the cultures described above. Measurements were taken on cells for which an increased germ tube formation percentage was documented in the presence of exogenous estrogen. The addition of 10−5 M 17-β-estradiol resulted in increased germ tube length for SC5314 cells (Fig. 3b). The difference in germ tube length between treated and untreated (control) cells was large enough to be visible in the micrographs shown in Fig. 1. Lower concentrations of 17-β-estradiol did not produce significantly longer germ tubes than the control cells (Fig. 3b). A significant effect on germ tube length (P = 0.011) was noted for the addition of 10−5 M ethynyl estradiol to cells of strain GC15 (Fig. 3b).
Effect of estrogen exposure on culture biomass.
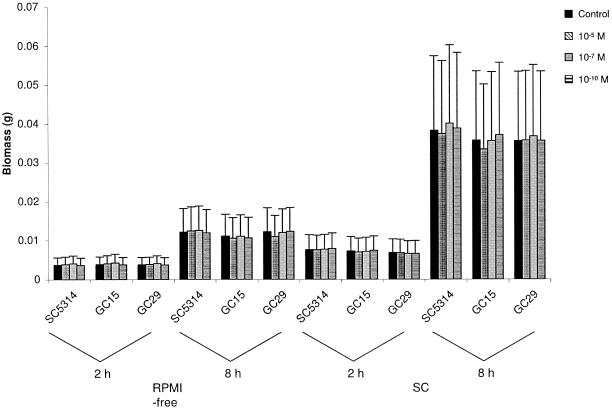
The effect of 17-β-estradiol on culture biomass was examined because of a previous report that suggested an increase in culture biomass in the presence of estrogenic compounds (22). In our studies, strains SC5314, GC15, and GC29 were grown in either RPMI-free or SC. The biomass of filtered, dried cells was measured at two different time points (see Materials and Methods). For cells grown in RPMI-free, the first measurement was taken at 2 h, when nearly all the germ tubes were longer than the mother yeast cell, and the second was taken at 8 h. For SC cultures, biomass measurements were taken at 2 h and 8 h, which corresponded to early and mid-logarithmic growth, respectively. The addition of 17-β-estradiol did not significantly change biomass compared to the control (estrogen-negative) cultures in either growth medium (Fig. 4).
FIG. 4.
Histogram showing biomass data for analysis of C. albicans strains SC5314, GC15, and GC29 at 2 h and 8 h in RPMI-free and SC. All experiments compared different concentrations of 17-β-estradiol to a negative control sample.
Microarray analysis of differential gene expression in estrogen-treated C. albicans cells.
Microarray analysis was used to detect differential gene expression in estrogen-treated C. albicans cells. A summary of the experimental comparisons that were made is shown in Table 2. These experiments were designed to detect gene expression changes that occurred almost immediately after estrogen exposure (10 min) and those that occurred at later time points, when differences in germ tube length were visible microscopically (85 min). Most comparisons were made using cells grown in RPMI-free, but cells grown in SC were also tested. The effects of different concentrations of estrogen were evaluated by the addition of physiological (10−10 M) or supraphysiological (10−5 M) estrogen concentrations. Gene expression changes in response to the addition of ethynyl estradiol were studied because this compound significantly increased germ tube formation for strain GC15. Finally, after analysis of initial results with wild-type strains (see below), microarray analysis was completed on strain DSY654, which lacks Cdr1p and Cdr2p function (41).
TABLE 2.
Summary of microarray comparisons to evaluate effects of estrogen exposure on C. albicans gene expression
| Expt no. | Growth medium | Estrogenic compound | Time exposed (min) | Condition 1a | Condition 2b | Comparison code |
|---|---|---|---|---|---|---|
| I | RPMI-free | 17-β-Estradiol | 10 | SC5314 control | SC5314, 10−5 M | A |
| RPMI-free | 17-β-Estradiol | 10 | SC5314 control | SC5314, 10−10 M | B | |
| SC | 17-β-Estradiol | 10 | SC5314 control | SC5314, 10−5 M | C | |
| SC | 17-β-Estradiol | 10 | SC5314 control | SC5314, 10−10 M | D | |
| II | RPMI-free | 17-β-Estradiol | 85 | SC5314 control | SC5314, 10−5 M | E |
| RPMI-free | 17-β-Estradiol | 85 | SC5314 control | SC5314, 10−10 M | F | |
| III | RPMI-free | Ethynyl estradiol | 85 | GC15 control | GC15, 10−5 M | G |
| IV | RPMI-free | Ethynyl estradiol | 10 | GC15 control | GC15, 10−5 M | H |
| V | RPMI-free | 17-β-Estradiol | 10 | SC5314 control | SC5314, 10−5 M | I |
| RPMI-free | 17-β-Estradiol | 85 | SC5314 control | SC5314, 10−5 M | J | |
| RPMI-free | 17-β-Estradiol | 10 | DSY654 control | DSY654, 10−5 M | K | |
| RPMI-free | 17-β-Estradiol | 85 | DSY654 control | DSY654, 10−5 M | L |
Untreated strain controls.
Treated strains and concentrations of estrogenic compound.
Each experimental comparison was assigned a letter in Table 2 (called comparison codes) so the experiments can be distinguished from each other in the subsequent data tables and text. These codes were used since some microarray comparisons are essentially the same (A and I and E and J, for example) (Table 2). Real-time RT-PCR was used to validate microarray results. Genes with a ≥2.0-fold change in gene expression on the microarray also showed the same direction of expression change (up- or down-regulation, for example) and a similar relative magnitude of change in the real-time analysis. This comparison provided our biological cutoff of 2.0-fold change for the microarray data. All genes listed in data tables have a statistical significance cutoff at P ≤ 0.05. Table 3 lists genes that were differentially expressed in more than one experimental condition, while Table A1 in the supplemental material lists genes that were differentially expressed in only one experimental condition. Real-time RT-PCR data are included in Table A2 in the supplemental material.
TABLE 3.
List of genes differentially expressed in more than one experimental conditiona
| Gene group | orf6 | orf19 | Gene | Description | Change in gene expression (n-fold) under indicated conditions
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Growth medium | Phenol red-free RPMI 1640
|
SC
|
|||||||||||||||
| Estrogen | 17-β-Estradiol
|
Ethynyl estradiol
|
17-β-Estradiol
|
||||||||||||||
| C. albicans strain | SC5314
|
GC15
|
DSY654
|
SC5314
|
|||||||||||||
| Time (min) | 10
|
85
|
10
|
85
|
10
|
85
|
10
|
||||||||||
| Concn (M) | 10−5 | 10−5 | 10−10 | 10−5 | 10−5 | 10−10 | 10−5 | 10−5 | 10−5 | 10−5 | 10−5 | 10−10 | |||||
| Comparison code (defined in Table 2) | A | I | B | E | J | F | H | G | K | L | C | D | |||||
| Up-regulated | orf6.9037 | orf19.6000 | CDR1 | Multidrug resistance protein Cdr1p | 4.5 | 3.7 | 2.8 | 2.0 | 7.2 | 2.9 | 3.2 | ||||||
| orf6.648 | orf19.1027 | PDR16 | Potential protein response to drug and sterol biosynthesis | 2.1 | 3.9 | 3.9 | 7.3 | ||||||||||
| orf6.7574 | orf19.6586 | Hypothetical protein (313 amino acids) | 2.2 | 3.0 | |||||||||||||
| orf6.7217 | orf19.3322 | DUT1 | dUTP pyrophosphatase | 2.5 | 3.1 | ||||||||||||
| orf6.854 | orf19.3188 | TAC1 | Transcriptional activator of drug-responsive genes (HAL92) | 2.6 | 2.5 | 2.1 | |||||||||||
| orf6.3947 | orf19.909 | STP4 | Potential zinc finger protein | 2.2 | 3.1 | 4.7 | |||||||||||
| Down- regulated | orf6.6093 | orf19.2170 | PHM7 | Potential transmembrane protein similar to S. cerevisiae PHM7 | −2.3 | −2.0 | |||||||||||
| orf6.6000 | orf19.6459 | Potential diacylglycerol pyrophosphate phosphatase | −2.0 | −3.0 | |||||||||||||
| orf6.3031 | orf19.3646 | CTR1 | High-affinity copper transporter | −2.7 | −2.1 | ||||||||||||
| orf6.4984 | orf19.10979 | Hypothetical protein (190 amino acids) | −2.5 | −2.2 | |||||||||||||
| orf6.5674 | orf19.6140 | Potential ferric reductase fragment | −2.5 | −2.0 | |||||||||||||
| orf6.5675 | orf19.6139 | Potential ferric reductase fragment | −2.7 | −2.3 | |||||||||||||
| orf6.6934 | orf19.3111 | PRA1 | pH-regulated cell surface antigen (FBP1) | −2.7 | −2.7 | ||||||||||||
For each gene, the orf6 and orf19 identification numbers are shown along with the gene name derived from database searches as described in Materials and Methods. Experimental comparisons are indicated using the headings at the right of the table. The comparison code (defined in Table 2) is also shown to aid identification of each set of experimental conditions. For each gene, the change (n-fold) in gene expression is shown. Genes that were up-regulated in estrogen-treated C. albicans cells and genes that were down-regulated in estrogen-treated cells (up-regulated in the control culture) are shown as separate groups. Negative n-fold change values indicate down-regulation of these genes in the estrogen-treated cells.
The initial microarray experiment (experiment I, comparisons A and B) (Table 2) used C. albicans strain SC5314 in RPMI-free supplemented with either supraphysiological or physiological concentrations of 17-β-estradiol. These growth conditions were examined first because they showed the most marked differences in germ tube formation and length (Fig. 3). The first set of experiments focused on 10 min of estrogen exposure to detect early changes in gene expression. Incubation in SC (experiment I, comparisons C and D) (Table 2) was also included in order to identify genes that were differentially expressed in response to estrogen exposure, independent of growth medium effects. Differential expression of CDR1, which encodes a drug efflux pump, was common between two of the four experimental comparisons (Table 3). These results were consistent with previous reports of the up-regulation of CDR1 following 17-β-estradiol exposure (16, 28, 31, 50).
The second set of microarray experiments focused on an 85-min time point to detect gene expression changes that occurred later in germ tube formation. Physiological and supraphysiological concentrations of 17-β-estradiol were used with strain SC5314 in RPMI-free (experiment II, comparisons E and F) (Table 2). Up-regulation of CDR1 was observed in the supraphysiological estrogen condition (E) (Table 3).
The focus of the subsequent microarray experiments was strain GC15 and supraphysiological concentrations of ethynyl estradiol (experiments III and IV, comparisons G and H) (Table 2) because these growth conditions produced increased germ tube formation and length (Fig. 3). Similar to previous microarray experiments, the 10-min and 85-min time points were studied; CDR1 was up-regulated at both time points (Table 3). A greater number of differentially expressed genes were detected at the 10-min time point than later in germ tube growth. In contrast to previous microarray comparisons, more differentially expressed genes were observed in this experiment and many of these were down-regulated in the estrogen-treated culture (Table 3).
The up-regulation of CDR1 at supraphysiological estrogen concentrations was consistent across the various microarray analyses that were conducted (Table 3). One possible interpretation of the microarray data is that exposure to estrogen increases expression of CDR1, leading to production of Cdr1p, which pumps estrogen out of the C. albicans cell and minimizes its effect on expression of other genes. CDR1 shares 84% sequence identity with CDR2, which also encodes a drug efflux pump (41). On the microarray used in this analysis, cDNA derived from CDR1 or CDR2 transcripts could hybridize with the PCR product-derived array probe.
In order to determine whether the apparent increase in CDR1 expression was due to a single gene or both genes, real-time RT-PCR primers that distinguish between the two genes were designed (Table 1). Real-time RT-PCR analysis showed that expression of both CDR1 and CDR2 increased in C. albicans cells exposed to estrogen (see Table A2 in the supplemental material). In order to remove the effect of these drug efflux pumps from our analysis, the experiments were conducted with strain DSY654, in which both genes were deleted (41).
Prior to microarray analysis, strain DSY654 was grown in RPMI-free to determine the effect of 17-β-estradiol addition on germ tube formation after 85 min of exposure (Fig. 5). Strain SC5314 was included in the analysis as a control. The addition of 17-β-estradiol to SC5314 cells showed increased germ tube formation at the highest estrogen concentration, with the effect diminishing as the estrogen concentration decreased. These results were similar to those obtained in previous experiments (Fig. 3). In contrast to the case with SC5314, the addition of 17-β-estradiol to strain DSY654 decreased germ tube formation significantly (P = 0.03) (Fig. 5).
FIG. 5.
Histogram summarizing germ tube formation data at 85 min for strains SC5314 and DSY654 treated with 17-β-estradiol.
These growth conditions were used for growing cells for microarray analysis (experiment V, comparisons I, J, K, and L) (Table 2). At the 10-min time point, three genes were differentially expressed in DSY654 in the presence of 17-β-estradiol (Table 3). This number increased dramatically to 41 differentially expressed genes at the 85-min time point (Table 3; see Table A1 in the supplemental material). The identity of the genes suggested that this effect could be explained, in part, by cellular morphology. Inhibition of germ tube/hypha formation by 17-β-estradiol resulted in the differential expression of many genes known to be associated with hyphal growth such as SAP4, SAP5, SAP6, ECE1, HYR1, ALS3, and HWP1 (1, 5, 18, 25, 47). In the absence of CDR1 and CDR2, the PDR16 gene was the most highly differentially expressed gene in the analysis (Table 3). This gene encodes a 369-amino-acid protein with similarity to Pdr16p in S. cerevisiae (15). In S. cerevisiae, Pdr16p is a phosphatidylinositol transfer protein that, when deleted, results in alterations in lipid biosynthesis and increased sensitivity to various antifungal drugs (33, 48).
Microarray analysis of strain DSY654 also showed up-regulation of TAC1, which encodes a transcriptional regulator of CDR1 and CDR2 (13). RTA3, which is regulated by TAC1, was also up-regulated in DSY654 (13, 16) (see Table A1 in the supplemental material). IFU5, which is also regulated by TAC1, was up-regulated in DSY654, but just missed the 2.0-fold biological cutoff for inclusion in Table A1 in the supplemental material. Several hypothetical proteins, for which no function has been deduced, were also up-regulated in strain DSY654 following exposure to 17-β-estradiol (Table 3) (see Table A1 in the supplemental material).
Real-time RT-PCR analysis of EBP1 expression.
Initial searches for a C. albicans estrogen-binding protein led to biochemical purification of the protein encoded by the gene EBP1 (estrogen binding protein 1) (38, 43). Completion of the C. albicans genome sequence and subsequent annotation suggest that there are as many as seven genes that are related in sequence to EBP1, with OYE21 sharing the most sequence identity (86.7%) (12, 27). Real-time RT-PCR primers were designed based on sequence differences between EBP1 and OYE21 to detect expression of EBP1. Real-time RT-PCR analysis was conducted on some of the RNA samples that were used for the microarray analysis to determine if EBP1 was differentially expressed under any of these growth conditions. Results from analysis of EBP1 expression are included in Table A2 in the supplemental material. Under the conditions tested, EBP1 was not differentially expressed in response to estrogen exposure. Therefore, if C. albicans uses Ebp1p in its response to estrogen exposure, it does so without an increase in transcription of the EBP1 gene.
DISCUSSION
The effect of estrogen on the cellular and molecular biology of C. albicans was studied using recently collected vaginal isolates (GC15 and GC29) and a laboratory-maintained strain (SC5314). These strains were grown in the presence of physiologically relevant estrogenic compounds at concentrations ranging from physiological (10−10 M) to supraphysiological (10−5 M). The addition of 17-β-estradiol to SC5314 cultures caused increased germ tube formation at the 10−5 and 10−7 M concentrations in a dose-dependent manner (Fig. 3). Strain GC15 also formed more germ tubes in the presence of 10−5 M 17-β-estradiol, while the addition of 17-β-estradiol did not increase germ tube formation for strain GC29.
Both GC15 and GC29 are recent vaginal isolates from women with clinical signs of VVC. The effect of 17-β-estradiol was so marked at the 10−5 M concentration that germ tube length was visibly longer for strains SC5314 and GC15 compared to the untreated control cells (Fig. 3b). The addition of ethynyl estradiol to GC15 cells also resulted in increased germ tube formation and length. The results of our studies compare favorably with those of Kinsman et al. (30), who showed that the addition of 10−7 M 17-β-estradiol to C. albicans during incubation in 10% serum resulted in increased germ tube formation and length. The results from this group were dose dependent in the range of 10−6 to 10−8 M 17-β-estradiol. The disparity between 17-β-estradiol concentration and measured effects for our study and that of Kinsman et al. (30) might be due to the presence of estrogen in their serum preparation. White and Larsen (49) also noted changes in germ tube formation when 17-β-estradiol was added to C. albicans cells in serum, however, in their study, germ tube formation decreased as 17-β-estradiol concentration increased between 10−8 and 10−6 M. It is possible that strain effects play a role in generating different results since the strain effects we noted within our collection of clinical vaginal isolates were large (see Materials and Methods).
The data presented here suggest that addition of 17-β-estradiol to C. albicans cells increases germ tube formation and length, although at physiological concentrations of estrogen, these effects are likely to be slight. However, even slight increases in the formation and length of C. albicans germ tubes may help the organism withstand sloughing of the vaginal epithelium and maintain colonization of the host.
Biomass analysis of the effect of 17-β-estradiol addition to cultured C. albicans cells showed no statistical difference between estrogen-treated and control cultures for any of the C. albicans strains. In a previous study, Gujjar et al. (22) observed a significant increase in culture biomass when 10−6 M 17-β-estradiol was added to C. albicans cells cultured for 7 days in a chemically defined medium. The increased germ tube length that we noted in the experiments described above suggest that a concomitant increase in biomass should also occur. It is likely that the method used to measure differences in biomass (weighing of dried, filtered cells) was not sensitive enough to detect this change. The effect of slight increases in biomass of C. albicans cells in the host are not known, but intuitively, increased biomass would likely promote colonization of the vaginal epithelium.
Knowledge of the gene expression changes that occur in C. albicans cells following exposure to estrogenic compounds are mainly from studies of the effects of antifungal compounds on C. albicans (16, 28, 31, 50). Our analysis used microarray technology to gain a larger picture of the effects of estrogen exposure on C. albicans cells. Cell biology experiments were followed by microarray analysis that emphasized understanding the gene expression changes observed under growth conditions that showed increased germ tube formation and length. One general conclusion from the entirety of our microarray work is that exposure of cultured C. albicans cells to exogenous estrogen affects the expression of a relatively small number of genes. The lists of differentially expressed genes mainly differed depending on the combination of strain/estrogen/incubation time analyzed. A limited number of genes were differentially expressed in more than one growth condition (Table 3), with most genes entered on this list because of their differential expression in strain GC15 treated with ethynyl estradiol for 10 min and in strain DSY654 treated with 17-β-estradiol for 85 min.
One gene that was differentially expressed under many growth conditions, and often with the highest magnitude of change, was CDR1. This result matches previous publications that showed increased expression of CDR1 in the presence of estrogenic compounds (16, 28, 31, 50). On our PCR product-based microarray, the CDR1 probe also hybridizes to cDNA derived from CDR2 because of the large degree of sequence identity between the genes (41). To distinguish between transcript derived from CDR1 and those from CDR2, real-time RT-PCR primers were designed to specifically recognize each gene (Table 1). The results of real-time RT-PCR analysis showed that the signal observed on the microarray was a combination of increased transcription from both CDR1 and CDR2 (see Table A2 in the supplemental material). These results suggest that, in the presence of exogenous estrogen, cultured C. albicans cells respond as they would in the presence of antifungal drugs and up-regulate production of efflux pumps to eliminate the estrogen from the cell. This logic is consistent with the work of de Micheli et al. (16) who showed the presence of drug responsive element sequences in the promoter of CDR1 and CDR2.
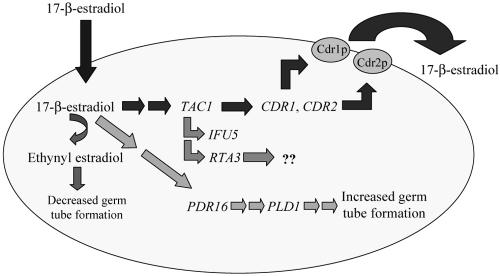
The extensive microarray analysis performed in the context of our studies led to a model to explain the molecular basis for increased germ tube formation and length that occurs for estrogen-exposed C. albicans cells (Fig. 6). The method of entry of 17-β-estradiol into the C. albicans cell is not known, but it is assumed to occur by diffusion (32). Once inside the cell, 17-β-estradiol increases transcription of TAC1, which then increases transcription of CDR1, CDR2, IFU5, and RTA3 (13). Increased transcription of CDR1 and CDR2 likely increases the production of efflux pumps, which remove 17-β-estradiol from the C. albicans cell (32). These interactions are supported by data published previously by other groups and also by our microarray data. Since Cdr1p and Cdr2p pump 17-β-estradiol out of the C. albicans cell, the intracellular levels of 17-β-estradiol should be relatively low.
FIG. 6.
Model to reconcile cell biology effects of estrogen exposure on cultured C. albicans cells with microarray observations of gene expression patterns.
Our microarray data show that exposure to 17-β-estradiol also increases transcription of PDR16, which is depicted as an alternative pathway in Fig. 6. In S. cerevisiae, PDR16 (SFH3) is a nonclassical phosphatidylinositol transfer protein that belongs to a family of SEC14 homologues (SFHs) (33). PDR16 was recently cloned and characterized in C. albicans and is associated with drug resistance (15). The basal transcriptional activity of PDR16 increased in a clinical azole-resistant strain compared to a susceptible strain (15). In fluconazole-resistant Candida glabrata, a PDR16 mutation led to fluconazole susceptibility, suggesting PDR16 involvement in fluconazole resistance (29). In addition to azole resistance, the down-regulated expression of PDR16 is suggested in phospholipid metabolism (8).
In S. cerevisiae, PDR16 (SFH3) demonstrates an effect on sterols and phospholipids (48). Strains lacking PDR16 show a decreased percentage of ergosterol and an increased percentage of episterol and fecosterol, suggesting an effect of PDR16 on the intermediate steps between episterol and ergosterol. van den Hazel et al. (48) hypothesized that PDR16 is involved in the regulation of sterol concentrations by either affecting sterol enzymes or altering the sterol availability. Pdr16p localizes to microsomes and the plasma membrane (42). Upon the deletion of PDR16, the phospholipid composition changed and was further reduced when the gene was deleted in combination with PDR17 (48). In pdr16 pdr17 deletion mutants, membrane-bound lipid transfer was affected (48). Furthermore, in strains lacking SFHs, the products of phospholipid D (PLD), choline and phosphatidic acid, are reduced, indicating that SFH proteins act on PLD activity. Routt and Bankaitis (40) hypothesized that SFH proteins are involved in the activation of PLD, by acting on phosphatidylinositol 4,5-bisphosphate (PIP2) synthesis.
In C. albicans, PLD activity is increased during the yeast-to- hypha transition (36). Phosphatidic acid, a product of PLD activity, stimulates phosphatidylinositiol-4-phosphate 5-kinase (PI4P5K) (23). PI4P5K activity is increased during temperature-induced hypha formation (23). Phosphatidic acid is suggested as an effector molecule involved in the yeast to hypha transition (2), by acting as a positive feedback regulator of PI4P5K (23). Additionally, PLD mutants show reduced invasion on solid medium and reduced virulence in a mouse model (17, 26). PLD is suggested to be involved in a hypha signaling pathway linked with tissue invasion. As suggested in S. cerevisiae (33, 40), PDR16 in C. albicans could be acting on PI4P5K to synthesize more PIP2. PIP2 then stimulates PLD, which is involved in the yeast-to-hypha transition. The potential interrelationships between 17-β-estradiol, transcription of PDR16 and PLD1, and the link between Pld1p activity and hypha formation suggest a model by which increased hypha formation is promoted in C. albicans in the presence of estrogen.
Another intriguing component to this model is the fact that 17-β-estradiol could be modified in C. albicans to form other compounds that contribute to the altered ability of the cell to form germ tubes. In S. cerevisiae, steroids such as estradiol are esterified by an alcohol acetyltransferase (ATF2) (9). The preferred substrate for Aft2p is pregnenalone. In a mutant deficient for Atf2p activity, pregnenalone slows the growth rate, indicating a toxic effect. Cauet et al. (9) hypothesized that the cellular target of pregnenalone is ERG2 and efflux pumps are responding to a potential growth inhibition of ergosterol biosynthesis. They suggest that pregnenolone's toxicity is tempered by esterification with Atf2p and by efflux from the cell through Snq2p and Pdr5p. This example demonstrates how steroid molecules might interfere with growth rate of a fungal cell. Alterations in growth rate would affect germ tube formation.
Despite the fact that many pieces of key evidence still need to be deduced to support it, the model proposed in Fig. 6 reconciles our cell biological and microarray data and provides a starting point for explaining how exposure to exogenous estrogens increases germ tube formation and length. In the presence of supraphysiological levels of 17-β-estradiol, C. albicans up-regulates CDR1 and CDR2 to pump the estrogen from the cell producing relatively low intracellular levels of 17-β-estradiol, despite the supraphysiological levels of estrogen in the growth medium. The addition of 17-β-estradiol to C. albicans cells also up-regulates expression of PDR16. The potential mechanism for how increased PDR16 expression might result in increased germ tube formation was provided above and involves the action of Pld1p.
The potential for 17-β-estradiol to be converted to another biologically active compound was also discussed above. We propose that this conversion step is far less favored than the steps already described and that it occurs only in the presence of relatively high intracellular estrogen concentrations, such as those that are likely to occur in the absence of efflux pump activity in strain DSY654. At high intracellular levels of 17-β-estradiol, the less-efficient conversion of 17-β-estradiol could occur. We propose that the end product of this reaction might be ethynyl estradiol in order to explain the unexpectedly similar gene expression results observed for strain GC15 in the presence of ethynyl estradiol (10 min; efflux pumps functional) and strain DSY654 in the presence of 17-β-estradiol (85 min; no efflux pump activity). Gene expression patterns at the early time point for the GC15/ethynyl estradiol experiment could resemble those for the DSY654 analysis if the efflux pumps were slower to remove intracellular ethynyl estradiol and higher intracellular concentrations were achieved, causing the cell to favor the pathway where decreased germ tube formation occurs. By the 85-min time point, the efflux pumps may have reduced the intracellular estrogen levels to the point where the system functions much as it does in the presence of 17-β-estradiol, perhaps through up-regulation of the CDR1 and CDR2 genes. In the mutant strain DSY654, the effect on gene expression patterns may be slowed by the need to convert 17-β-estradiol to ethynyl estradiol and produce the concomitant effects. Our gene expression data suggest that sufficient conversion had occurred by 85 min.
The model in Fig. 6 describes the effect of estrogen on germ tube formation and length as a balance between competing reactions and interactions. Our data also imply the presence of a strain-specific effect that alters the balance between the competing reactions. However, the model explains the concentration dependent-effects on cellular morphology that we observed and suggests that a concentration of 17-β-estradiol of <10−5 M and >10−7 M will result in increased germ tube formation that can be measured using the experimental methods presented here. More precise measurements would be required to determine if these effects occur at physiological estrogen concentrations and contribute to the physiological balance between C. albicans and its host.
Supplementary Material
Acknowledgments
We thank Suzanne Trupin for collecting the clinical isolates that were used in this work and Dominque Sanglard for strain DSY654.
This work was supported by a Minority Predoctoral Fellowship (F31 AI54328) from the National Institute of Allergy and Infectious Diseases and by Public Health Service grant DE14158 from the National Institute of Dental and Craniofacial Research of the National Institutes of Health. This investigation was conducted in a facility constructed with support from Research Facilities Improvement Program Grant Number C06 RR16515-01 from the National Center for Research Resources, National Institutes of Health.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Bailey, D. A., P. J. Feldmann, M. Bovey, N. A. Gow, and A. J. Brown. 1996. The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J. Bacteriol. 178:5353-5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, C. A., K. Desrosiers, and J. W. Dolan. 2002. Propranolol inhibits hyphal development in Candida albicans. Antimicrob. Agents Chemother. 46:3617-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauters, T. G., M. A. Dhont, M. I. L. Temmerman, and H. J. Nelis. 2002. Prevalence of vulvovaginal candidiasis and susceptibility to fluconazole in women. Am. J. Obstet. Gynecol. 187:569-574. [DOI] [PubMed] [Google Scholar]
- 4.Bindal, R. D., K. E. Carlson, B. S. Katzenellenbogen, and J. A. Katzenellenbogen. 1988. Lipophilic impurities, not phenolsulfonpthalein, account for the estrogenic activity in commercial preparations of phenol red. J. Steroid Biochem. 31:287-293. [DOI] [PubMed] [Google Scholar]
- 5.Birse, C. E., M. Y. Irwin, W. A. Fonzi, and P. S. Sypherd. 1993. Cloning and characterization of ECE1, a gene expressed in association with cell elongation of the dimorphic pathogen Candida albicans. Infect. Immun. 61:3648-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradshaw, C. S., A. N. Morton, S. M. Garland, M. B. Morris, L. M. Moss, and C. K. Fairley. 2005. Higher-risk behavioral practices associated with bacterial vaginosis compared with vaginal candidiasis. Obstet. Gynecol. 106:105-114. [DOI] [PubMed] [Google Scholar]
- 7.Buckman, J., and S. M. Miller. 1998. Binding and reactivity of Candida albicans estrogen binding protein with steroid and other substrates. Biochemistry 37:14326-14336. [DOI] [PubMed] [Google Scholar]
- 8.Cao, Y. Y., Y. B. Cao, Z. Xu, K. Ying, Y. Li, Y. Xie, Z. Y. Zhu, W. S. Chen, and Y. Y. Jiang. 2005. cDNA microarray analysis of differential gene expression in Candida albicans biofilm exposed to farnesol. Antimicrob. Agents Chemother. 49:584-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cauet, G., E. Degryse, C. Ledoux, R. Spagnoli, and T. Achstetter. 1999. Pregnenolone esterification in Saccharomyces cerevisiae. A potential detoxification mechanism. Eur. J. Biochem. 261:317-324. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, G., K. Wozniak, M. A. Wallig, P. L. Fidel, Jr., S. R. Trupin, and L. L. Hoyer. 2005. Comparison between Candida albicans agglutinin-like sequence gene expression patterns in human clinical specimens and models of vaginal candidiasis. Infect. Immun. 73:1656-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collart, M. A., and S. Oliviero. 1993. Preparation of yeast RNA, p. 13.12.1-13.12.5. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 2. John Wiley and Sons, Inc., New York, N.Y. [DOI] [PubMed] [Google Scholar]
- 12.Combet, C., C. Blanchet, C. Geourjon, and G. Deleage. 2000. NPS@: network protein sequence analysis. Trends Biochem. Sci. 25:147-150. [DOI] [PubMed] [Google Scholar]
- 13.Coste, A. T., M. Karababa, F. Ischer, J. Bille, and D. Sanglard. 2004. TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot. Cell 3:1639-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotch, M. F., S. L. Hillier, R. S. Gibbs, and D. A. Eschenbach. 1998. Epidemiology and outcomes associated with moderate to heavy Candida colonization during pregnancy. Am. J. Obstet. Gynecol. 178:374-380. [DOI] [PubMed] [Google Scholar]
- 15.De Deken, X., and M. Raymond. 2004. Constitutive activation of the PDR16 promoter in a Candida albicans azole-resistant clinical isolate overexpressing CDR1 and CDR2. Antimicrob. Agents Chemother. 48:2700-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Micheli, M., J. Bille, C. Schueller, and D. Sanglard. 2002. A common drug-responsive element mediates the upregulation of the Candida albicans ABC transporters CDR1 and CDR2, two genes involved in antifungal drug resistance. Mol. Microbiol. 43:1197-1214. [DOI] [PubMed] [Google Scholar]
- 17.Dolan, J. W., A. C. Bell, B. Hube, M. Schaller, T. F. Warner, and E. Balish. 2004. Candida albicans PLD1 activity is required for full virulence. Med. Mycol. 42:439-447. [DOI] [PubMed] [Google Scholar]
- 18.Felk, A., M. Kretschmar, A. Albrecht, M. Schaller, S. Beinhauer, T. Nichterlein, D. Sanglard, H. C. Korting, W. Schafer, and B. Hube. 2002. Candida albicans hyphal formation and the expression of the Efg1-regulated proteinases Sap4 to Sap6 are required for the invasion of parenchymal organs. Infect. Immun. 70:3689-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geiger, A. M., and B. Foxman. 1996. Risk factors for vulvovaginal candidiasis: a case-control study among university students. Epidemiology 7:182-187. [DOI] [PubMed] [Google Scholar]
- 20.Gillum, A. M., E. Y. H. Tsay, and D. R. Kirsch. 1984. Isolation of the Candida albicans genes for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179-182. [DOI] [PubMed] [Google Scholar]
- 21.Green, C. B., X. Zhao, K. M. Yeater, and L. L. Hoyer. 2005. Construction and real-time RT-PCR validation of Candida albicans PALS-GFP reporter strains and their use in flow cytometry analysis of ALS gene expression in budding and filamenting cells. Microbiology 151:1051-1060. [DOI] [PubMed] [Google Scholar]
- 22.Gujjar, P., M. Finucane, and B. Larsen. 1997. The effect of estradiol on Candida albicans growth. Ann. Clin. Lab. Sci. 27:151-156. [PubMed] [Google Scholar]
- 23.Hairfield, M. L., C. Westwater, and J. W. Dolan. 2002. Phosphatidylinositol-4-phosphate 5-kinase activity is stimulated during temperature-induced morphogenesis in Candida albicans. Microbiology 148:1737-1746. [DOI] [PubMed] [Google Scholar]
- 24.Hicks, J. B., and I. Herskowitz. 1976. Interconversion of yeast mating types I. Direct observations of the action of the homothallism (HO) gene. Genetics 83:245-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoyer, L. L., T. L. Payne, M. Bell, A. M. Myers, and S. Scherer. 1998. Candida albicans ALS3 and insights into the nature of the ALS gene family. Curr. Genet. 33:451-459. [DOI] [PubMed] [Google Scholar]
- 26.Hube, B., D. Hess, C. A. Baker, M. Schaller, W. Schafer, and J. W. Dolan. 2001. The role and relevance of phospholipase D1 during growth and dimorphism of Candida albicans. Microbiology 147:879-889. [DOI] [PubMed] [Google Scholar]
- 27.Jones, T., N. A. Federspiel, H. Chibana, J. Dungan, S. Kalman, B. B. Magee, G. Newport, Y. R. Thorstenson, N. Agabian, P. T. Magee, R. W. Davis, and S. Scherer. 2004. The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. USA 101:7329-7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karnani, N., N. A. Gaur, S. Jha, N. Puri, S. Krishnamurthy, S. K. Goswami, G. Mukhopadhyay, and R. Prasad. 2004. SRE1 and SRE2 are two specific steroid-responsive modules of Candida drug resistance gene 1 (CDR1) promoter. Yeast 21:219-239. [DOI] [PubMed] [Google Scholar]
- 29.Kaur, R., I. Castano, and B. P. Cormack. 2004. Functional genomic analysis of fluconazole susceptibility in the pathogenic yeast Candida glabrata: roles of calcium signaling and mitochondria. Antimicrob. Agents Chemother. 48:1600-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kinsman, O. S., K. Pitblado, and C. J. Coulson. 1988. Effect of mammalian steroid hormones and luteinizing hormone on the germination of Candida albicans and implication for vaginal candidosis. Mycoses 31:617-626. [DOI] [PubMed] [Google Scholar]
- 31.Krishnamurthy, S., V. Gupta, R. Prasad, S. Panwar, and R. Prasad. 1998. Expression of CDR1, a multidrug resistance gene of Candida albicans: transcriptional activation by heat shock, drugs and human steroid hormones. FEMS Microbiol. Lett. 160:191-197. [DOI] [PubMed] [Google Scholar]
- 32.Krishnamurthy, S., V. Gupta, P. Snehlata, and R. Prasad. 1998. Characterization of human steroid hormone transport mediated by Cdrlp, a multidrug transporter of Candida albicans, belonging to the ATP binding cassette super family. FEMS Microbiol. Lett. 158:69-74. [DOI] [PubMed] [Google Scholar]
- 33.Li, X., S. M. Routt, Z. Xie, X. Cui, M. Fang, M. A. Kearns, M. Bard, D. R. Kirsch, and V. A. Bankaitis. 2000. Identification of a novel family of nonclassic yeast phosphatidylinositol transfer proteins whose function modulates phospholipase D activity and Sec14p-independent cell growth. Mol. Biol. Cell 11:1989-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 35.Madani, N. D., P. J. Malloy, P. Rodriquez-Pombo, A. Krishnan, and D. Feldman. 1994. Candida albicans estrogen-binding protein gene encodes an oxidoreductase that is inhibited by estradiol. Proc. Natl. Acad. Sci. USA 91:922-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McLain, N., and J. W. Dolan. 1997. Phospholipase D activity is required for dimorphic transition in Candida albicans. Microbiology 143:3521-3526. [DOI] [PubMed] [Google Scholar]
- 37.Odds, F. 1988. Candida and candidosis, 2nd ed. Balliere Tindall, London, England.
- 38.Powell, B. L., C. L. Frey, and D. J. Drutz. 1984. Identification of a 17β-estradiol binding protein in Candida albicans and Candida (Torulopsis) glabrata. Exp. Mycol. 8:304-313. [Google Scholar]
- 39.Puri, N., S. Krishnamurthy, S. Habib, S. Hasnain, S. Goswami, and R. Prasad. 1999. CDR1, a multidrug resistance gene from Candida albicans, contains multiple regulatory domains in its promoter and the distal AP-1 element mediates its induction by miconazole. FEMS Microbiol. Lett. 180:213-219. [DOI] [PubMed] [Google Scholar]
- 40.Routt, S. M., and V. A. Bankaitis. 2004. Biological functions of phosphatidylinositol transfer proteins. Biochem. Cell Biol. 82:254-262. [DOI] [PubMed] [Google Scholar]
- 41.Sanglard, D., F. Ischer, M. Monod, and J. Bille. 1997. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology 143:405-416. [DOI] [PubMed] [Google Scholar]
- 42.Schnabl, M., O. V. Oskolkova, R. Holic, B. Brezna, H. Pichler, M. Zagorsek, S. D. Kohlwein, F. Paltauf, G. Daum, and P. Griac. 2003. Subcellular localization of yeast Sec14 homologues and their involvement in regulation of phospholipid turnover. Eur. J. Biochem. 270:3133-3145. [DOI] [PubMed] [Google Scholar]
- 43.Skowronski, R., and D. Feldman. 1989. Characterization of an estrogen-binding protein in the yeast Candida albicans. Endocrinology 124:1965-1972. [DOI] [PubMed] [Google Scholar]
- 44.Sobel, J. D., S. Faro, R. W. Force, B. Foxman, W. J. Ledger, P. R. Nyirjesy, B. D. Reed, and P. R. Summers. 1998. Vulvovaginal candidiasis: epidemiologic, diagnostic, and therapeutic considerations. Am. J. Obstet. Gynecol. 178:203-211. [DOI] [PubMed] [Google Scholar]
- 45.Spinillo, A., A. Bernuzzi, C. Cevini, R. Gulminetti, S. Luzi, and A. D. Santolo. 1997. The relationship of bacterial vaginosis, Candida and Trichomonas infection to symptomatic vaginitis in postmenopausal women attending a vaginitis clinic. Maturitas 27:253-260. [DOI] [PubMed] [Google Scholar]
- 46.Spinillo, A., E. Capuzzo, S. Nicola, F. Baltaro, A. Ferrari, and A. Monaco. 1995. The impact of oral contraception on vulvovaginal candidiasis. Contraception 51:293-297. [DOI] [PubMed] [Google Scholar]
- 47.Staab, J. F., and P. Sundstrom. 1998. Genetic organization and sequence analysis of the hypha-specific cell wall protein gene HWP1 of Candida albicans. Yeast 14:681-686. [DOI] [PubMed] [Google Scholar]
- 48.van den Hazel, H. B., H. Pichler, M. A. do Valle Matta, E. Leitner, A. Goffeau, and G. Daum. 1999. PDR16 and PDR17, two homologous genes of Saccharomyces cerevisiae, affect lipid biosynthesis and resistance to multiple drugs. J. Biol. Chem. 274:1934-1941. [DOI] [PubMed] [Google Scholar]
- 49.White, S., and B. Larsen. 1997. Candida albicans morphogenesis is influenced by estrogen. Cell. Mol. Life. Sci. 53:744-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, X., M. Essmann, E. T. Burt, and B. Larsen. 2000. Estrogen effects on Candida albicans: a potential virulence-regulating mechanism. J. Infect. Dis. 181:1441-1446. [DOI] [PubMed] [Google Scholar]
- 51.Zhao, X., P. J. Malloy, C. Ardies, and D. Feldman. 1995. Oestrogen-binding protein in Candida albicans: antibody development and cellular localization by electron immunocytochemistry. Microbiology 141:2685-2692. [DOI] [PubMed] [Google Scholar]
- 52.Zhao, X., S.-H. Oh, K. M. Yeater, and L. L. Hoyer. 2005. Analysis of the Candida albicans Als2p and Als4p adhesins suggests the potential for compensatory function within the Als family. Microbiology 151:1619-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.