Abstract
We report here the results of a systematic high-resolution X-ray crystallographic analysis of complexes of the hepatitis C virus (HCV) RNA polymerase with ribonucleoside triphosphates (rNTPs) and divalent metal ions. An unexpected observation revealed by this study is the existence of a specific rGTP binding site in a shallow pocket at the molecular surface of the enzyme, 30 Å away from the catalytic site. This previously unidentified rGTP pocket, which lies at the interface between fingers and thumb, may be an allosteric regulatory site and could play a role in allowing alternative interactions between the two domains during a possible conformational change of the enzyme required for efficient initiation. The electron density map at 1.7-Å resolution clearly shows the mode of binding of the guanosine moiety to the enzyme. In the catalytic site, density corresponding to the triphosphates of nucleotides bound to the catalytic metals was apparent in each complex with nucleotides. Moreover, a network of triphosphate densities was detected; these densities superpose to the corresponding moieties of the nucleotides observed in the initiation complex reported for the polymerase of bacteriophage φ6, strengthening the proposal that the two enzymes initiate replication de novo by similar mechanisms. No equivalent of the protein stacking platform observed for the priming nucleotide in the φ6 enzyme is present in HCV polymerase, however, again suggesting that a change in conformation of the thumb domain takes place upon template binding to allow for efficient de novo initiation of RNA synthesis.
About 3% of the world population is chronically infected with hepatitis C virus (HCV), an infection that leads to liver cancer in many patients. There is neither vaccine nor effective therapy available against this pathogen, and, therefore, there is an urgent need for efficient means of combating and, ultimately, curing this viral disease. The RNA-dependent RNA polymerase (RdRp) activity of HCV protein NS5B is an absolute requirement for replication of the virus. This enzyme exhibits important differences with cellular polymerases and is therefore a good target for developing specific anti-HCV therapies. The crystal structure of unliganded NS5B has been determined by three groups independently (2, 8, 27); this work shows that NS5B contains the classic structural domains, denoted fingers, palm, and thumb, of other single-chain polynucleotide polymerases (21). The strictly conserved aspartic acids that chelate the catalytic Mg2+ ions are found in the palm, the fold of which is conserved among all members of the polymerase I and α families (43). In contrast to most other single-chain polymerases of known structure, the finger and thumb domains are connected in the HCV enzyme and are therefore not free to change conformation independently of each other. This is also the case for other RdRps, as confirmed by the recently reported structure of the double-stranded RNA bacteriophage φ6 polymerase (12). The overall similarity between the phage φ6 and the HCV polymerases indicates an unexpected evolutionary link between the corresponding genes in these otherwise unrelated viruses. An additional distinctive feature of these two polymerases is their ability to initiate replication from an RNA template without the need of a primer (de novo initiation), as reported for HCV (30, 34, 46) and φ6 (31). In addition, it has been shown that high concentrations of riboguanosine triphosphate (rGTP) stimulate RNA synthesis for both the φ6 (35) and HCV (29, 30) enzymes, presumably by enhancing the efficiency of the initiation process. The cocrystals of the φ6 polymerase in complex with a DNA oligonucleotide and rGTP provided the first snapshot of an initiation complex formed by these enzymes (12). Understanding the details of such a process, which is specific to a particular subset of RNA viruses (11), can provide a handle for devising specific drugs that block initiation of viral replication. To obtain more structural information about the initiation of replication of HCV and its rGTP activation, we undertook the determination of structures of complexes of NS5B with all four ribonucleoside triphosphates (rNTPs) and divalent cations (essentially Mn2+ but also Mg2+) to high resolution. For rGTP we observed electron density for a nucleotide bound to a surface pocket at a distance of about 30 Å from the catalytic site. For all four rNTPs (rATP, rGTP, rCTP, and rUTP), we observed electron density at and near the active site, readily interpretable as metal ions and the triphosphate (tP) moieties of three rNTPs. The latter superpose remarkably well to the tPs of the bound nucleotides observed in the initiation complex of the φ6 polymerase at the C (catalytic), P (priming), and I (interrogation) sites (12). In the absence of template, the corresponding nucleoside moieties are disordered in our crystals. Furthermore, the location of the tP corresponding to the priming nucleotide indicates that its nucleoside moiety lies within a large water-filled cavity of the enzyme. This observation suggests that a conformational change of the protein needs to take place to provide for a stacking platform for initiation, such as the platform provided by Tyr630 in the φ6 polymerase. We describe here the results obtained in each case and discuss the implications in the light of the superposition to the initiation complex observed for the φ6 polymerase.
MATERIALS AND METHODS
Crystallization and diffraction data collection.
Crystals of a form of the HCV RdRp lacking the 55 C-terminal amino acids (HCV-NS5B-ΔC55; either as a native or selenomethionine-derivatized protein) were grown as described previously (8). This form ends at the C terminus of the thumb domain and therefore lacks the 21-residue membrane-anchoring region and the 34-amino-acid linker connecting thumb and membrane anchor. Crystals were soaked for 4 to 15 h at 4°C in a solution containing either rGTP, rUTP, rCTP, or rATP and either MnCl2 or MgCl2 (see Table 1 for concentrations) in 0.09 M MES (morpholineethanesulfonic acid) buffer (pH 6.8)-7% (vol/vol) 2-propanol-7.2% (wt/vol) polyethylene glycol 4000. Crystals were briefly transferred to a cryoprotectant solution containing 18% (vol/vol) 2-methyl-2,4-pentanediol added to the other components of the soaking solution and then were flash-cooled by plunging them into liquid ethane. For rATP and rGTP, a slight precipitate was observed in the cryoprotectant solution when the nucleotide concentration used was 10 mM, indicating that the actual concentrations seen by the crystal in these cases were somewhat lower than those reported in Table 1. Diffraction data were collected at beam line ID14 of the European Synchrotron Radiation Facility or in-house on a rotating-anode X-ray generator with focusing mirrors and a Mar345 image plate detector. Data were processed and reduced with the HKL suite of programs (36). Analysis of the diffraction data showed that the soaking procedure induced a change in the crystal space group. The crystals grew in space group P212121 with cell parameters a = 67 Å, b = 97 Å, and c = 194 Å and two molecules in the asymmetric unit and shifted to P21212 with cell parameters a = 67.1 Å, b = 96.5 Å, and c = 96.2 Å and one molecule in the asymmetric unit upon being soaked with nucleotides.
TABLE 1.
Statistics of data collection and refinement
| Soaking conditionsa (mM) | Resolution rangeb (Å) | Rsymc (%) | I/σIe | Completeness (%) | No. of unique reflections measured | Avg redun- dancy of measure- ments | No. of atoms of protein/ligand/water molecule | rmsdf bonds (Å)/ angles (°) | R/Rfree (%)d |
|---|---|---|---|---|---|---|---|---|---|
| GTP, 10; MnCl2, 11 | 25-1.7 (1.73-1.70) | 6.9 (42.3) | 12.3 (2.9) | 99.3 (98.5) | 68,572 | 3.2 | 4,127/67/655 | 0.010/1.5 | 19.3/22.2 |
| UTP, 10; MnCl2, 11 | 25-1.85 (1.88-1.85) | 7.5 (38.4) | 12.3 (3.3) | 99.5 (99.9) | 53,875 | 3.5 | 4,143/69/484 | 0.010/1.5 | 19.7/22.9 |
| ATP, 10; MnCl2, 11 | 20-2.0 (2.05-2.00) | 8.1 (32.6) | 10.2 (2.4) | 94.0 (94.8) | 40,377 | 2.6 | 4,133/2/481 | 0.007/1.3 | 20.2/23.7 |
| CTP, 10; MnCl2, 11 | 20-1.65 (1.68-1.65) | 8.6 (31.2) | 13.5 (3.9) | 99.0 (95.1) | 74,850 | 3.7 | 4,133/27/591 | 0.010/1.5 | 20.0/22.5 |
| GTP, 10; MgCl2, 9 | 30-1.85 (1.88-1.85) | 8.8 (40.1) | 14.3 (3.7) | 99.6 (98.9) | 53,606 | 4.5 | 4,133/10/426 | 0.010/1.5 | 18.8/21.5 |
| GTP, 1; MgCl2, 4 | 20-2.3 (2.38-2.30) | 6.3 (19.0) | 15.2 (4.2) | 100.0 (100.0) | 28,205 | 4.6 | 4,133/67/341 | 0.010/1.6 | 20.9/24.7 |
A slight precipitate in the solutions indicated that the concentrations of nucleotides and/or divalent ions were less than those initially added.
In parentheses, statistics for the highest-resolution shell.
Rsym was determined by the equation 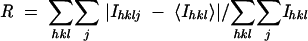 where I is measured intensities, h, k, and l are the unique indices of all reflections measured more than once, and j is the index for symmetry-redundant reflections.
where I is measured intensities, h, k, and l are the unique indices of all reflections measured more than once, and j is the index for symmetry-redundant reflections.
R and Rfree were determined by the equation 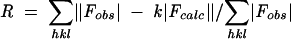 where h, k, and l are the indices of the reflections used in refinement (R) or of 5% of the reflections set aside and not used in refinement (Rfree). The same set of reflections was used for Rfree in all structures. Fobs are the structure factors deduced from the measured intensities, and Fcalc are the structure factors calculated from the model. k is a scale factor to put Fcalc on the same scale as the Fobs.
where h, k, and l are the indices of the reflections used in refinement (R) or of 5% of the reflections set aside and not used in refinement (Rfree). The same set of reflections was used for Rfree in all structures. Fobs are the structure factors deduced from the measured intensities, and Fcalc are the structure factors calculated from the model. k is a scale factor to put Fcalc on the same scale as the Fobs.
σ1, root mean square error of measurement of intensities.
rmsd, root mean square deviation.
Structure determination.
Program AMoRe (33) was used for molecular replacement (MR) with the structure of unliganded NS5B as a search model (8). Refinement was carried out with the CNS package (10). An initial positional and B-factor refinement of the MR solution led to an Rfree (9) value below 28%. Electron density was apparent at this stage in an Fo-Fc difference map in the region of the catalytic site. Two peaks near Asp220, -318, and -319 were clearly attributable to two catalytic metal ions when the metal ion used was Mn2+. For the rGTP complexes, strong density was also observed near residues 495 to 499, independently of the metal ion used (Mn2+ or Mg2+). After the two manganese ions were built at the catalytic site and after the amino acid side chains surrounding these ions were manually adjusted, three cycles of refinement and automatic water molecule picking were performed. Care was taken to avoid introduction of spurious water molecules either in the region of the catalytic site or near residues 495 to 499. After addition of ∼300 water molecules, the Rfree had dropped below 25% and the electron density maps improved to the extent that the nucleotides and/or tP moieties could be readily built. Statistics of data collection and refinement for complexes with each of the four ribonucleotides used are provided in Table 1.
Sequence alignments.
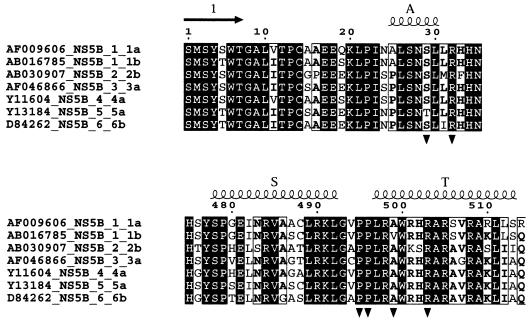
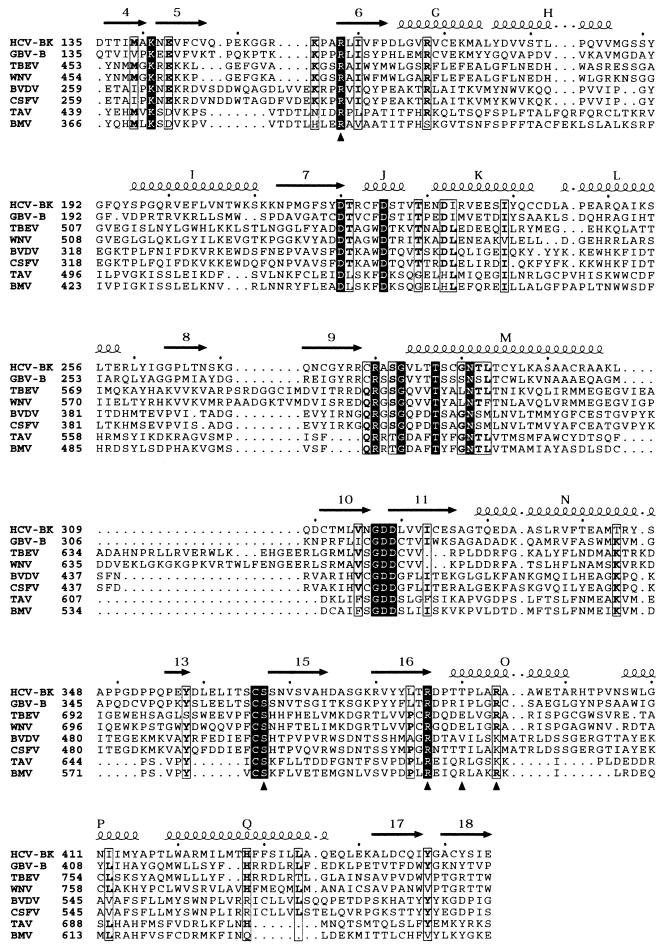
The alignments displayed in Fig. 4 and 5 were made using the facilities of the Réseau National Hépatites web server (http://hcvpub.ibcp.fr). For Fig. 4, one sequence each of HCV-NS5B proteins from genotypes 1a, 1b, 2, 3, 4, 5, and 6 was retrieved from the server and the seven sequences were aligned with CLUSTALW (45). To get a reliable alignment between HCV-NS5B and more-distant polymerases (see Fig. 5), a PSI-BLAST (3) search was performed with the sequence of the HCV BK strain used in this study and all polymerase sequences from the SWISSPROT database. Five sequences of Flaviviridae polymerases and two from Bromoviridae were kept and the PSI-BLAST alignment was then manually adjusted by using the secondary structure of HCV-NS5B as a guide.
FIG. 4.
Partial sequence alignment of seven polymerase sequences from HCV with the six major genotypes, with two representatives of HCV with genotype 1 (1a and 1b). Strictly conserved residues are represented as white on a black background. ▾, residues involved in binding the nucleotide at the surface site. A, S, and T, α-helices; arrow (1), β-strand.
FIG. 5.
Partial sequence alignment of polymerases of HCV (strain BK), GBV-B (the closest known relative of HCV), flaviviruses (tick-borne encephalitis virus [TBEV] and West Nile virus [WNV]), pestiviruses (BVDV and classical swine fever virus [CSFV]), and bromoviruses (BMV and tomato aspermy virus [TAV]). Secondary structure elements are indicated for the HCV polymerase, with α-helices labeled with capital letters and β-strands labeled with numbers. ▴, residues involved in binding the nucleotide at the priming site. Strictly conserved residues are represented as white on a black background. Residues more than 50% conserved are boxed, with the most commonly found amino acids in bold.
Graphic representation for figures.
Figures 1 to 3 were made with Bobscript (14) and Molscript (26) and were rendered with Raster3D (32). Figures 4 and 5 were made with ESPript (15) by using the web server interface (http://prodes.toulouse.inra.fr/ESPript).
FIG. 1.
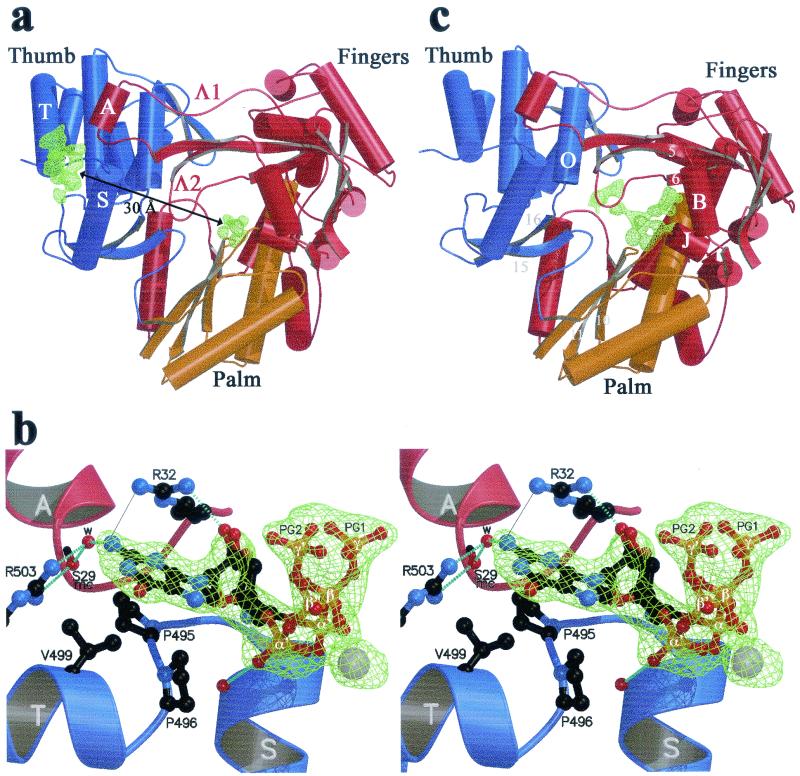
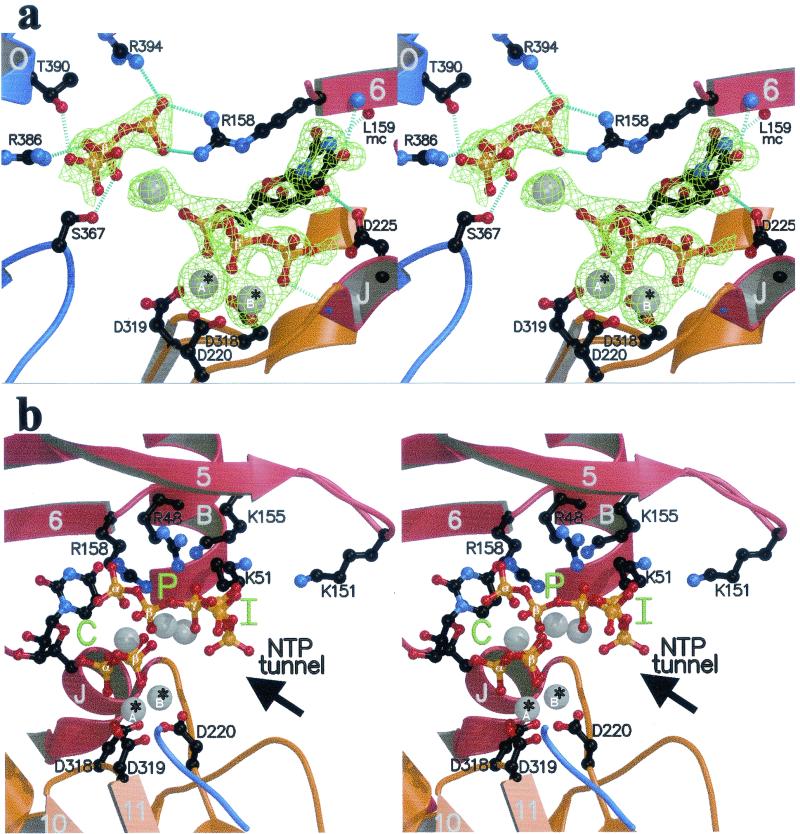
(a and b) The rGTP complex. (a) Overall view, with the finger, palm, and thumb domains colored red, yellow, and blue, respectively. Arrows, β-strands; cylinders, α-helices; thin tubes, connecting loops. The Fo-Fc simulated annealing electron density omit map is displayed at a contour level of 3.5σ as a green net at the surface and C sites. Secondary structure elements next to the surface site (α-helices A, S, and T and loops Λ1 and Λ2) are labeled as described previously (8). This view is from the back of the enzyme, with respect to the views defined in reference 8. (b) Stereo view of the putative rGTP activator site. The complex has been rotated 90° around a vertical axis with respect to the view shown in panel a, so that Arg32 (at the base of helix A) now lies in the background. The residues involved in contacts with rGTP are displayed in ball-and-stick representation. Two alternative conformations for the sugar and tP of the nucleotide exhibiting about 50% occupancy are represented (with phosphates labeled α, β, and γ and α′, β′, and γ′ denoting the two conformations). Atoms are color coded as follows: carbon, black; nitrogen, blue; oxygen, red; phosphorus, orange; manganese, grey. Broken light blue lines, hydrogen bonds. A thin grey continuous line between one of the guanidinium nitrogens of Arg32 and the N2 position of the guanine base denotes a water-mediated hydrogen bond between the two donors (see text). mc, main chain; w, water molecule. (c) rUTP complex, overall view. The difference electron density with respect to the unliganded enzyme (Fo-Fc simulated annealing omit map contoured at 3σ) is displayed as a green net in the region of the C and P sites. Secondary structure elements relevant to the binding of nucleotides at the C and P sites are labeled.
FIG. 3.
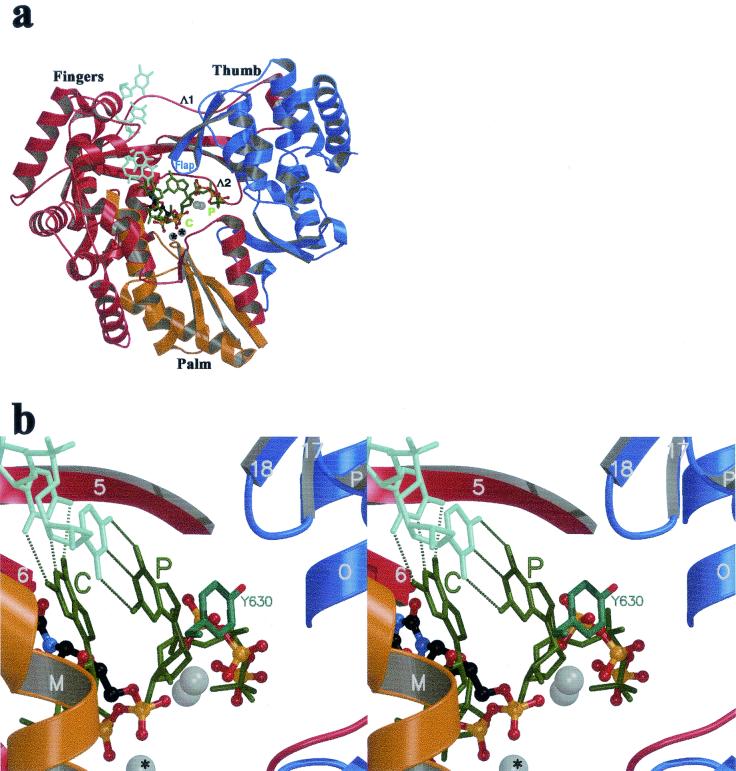
Model for an HCV initiation complex derived from the φ6 polymerase quaternary complex. The images were made after superposition of the equivalent atoms of the HCV and the phage φ6 polymerase (using the equivalent atoms in the fingers and palm defined previously [12]). (a) Stick representation of the template strand (light green) and the two rGTPs (dark green) of the φ6 polymerase initiation complex, along with the rUTP complex (the I site has been removed for clarity). The color code by domains of the protein is the same as that of Fig. 1a; the orientation is the standard front view of the enzyme. The Mn2+ ions are displayed as for Fig. 1 and 2. The C and P sites are labeled in green, with the atoms observed in the rUTP complex displayed as described in Fig. 1a. Flap, β-hairpin 17-18 of the thumb (8). Λ1 and Λ2, loops connecting fingers and thumb at the back of the enzyme. (b) Stereo diagram displaying a close-up view of the P site. Tyr630 of the φ6 polymerase, stacking against the priming base, is displayed in stick representation. Atoms of the rUTP complex are displayed as in Fig. 2a.
Data deposition.
Coordinates and structure factors for the first two structures described in Table 1 have been deposited with the Protein Data Bank. Coordinates and structure factors for the other structures in Table 1 will be made available on our website (http://www.cnrs-gif.fr/gv).
RESULTS
The surface rGTP site.
The surface binding site observed in crystals soaked with rGTP lies at about 30 Å from the catalytic site, next to the region of interaction between fingertips and thumb (Fig. 1a). The nucleotide contacts residues from the loop between α-helices S and T of the thumb and from α-helix A, at the tip of the Λ1 loop of the fingertips (we use the secondary structure nomenclature defined in reference 8 throughout). Six amino acids of NS5B make direct or water-mediated contacts to the nucleotide (Table 2; see Fig. 4), four from the thumb domain (Pro495 and -496 from the αS-αT loop and Val499 and Arg503 from α-helix T) and two from the fingertips (Arg32 and Ser29 in α-helix A). The polymerase makes direct contacts only with the nucleoside moiety of the bound nucleotide. The tP moiety is less well ordered and exhibits two alternative conformations (which have been modeled as shown in Fig. 1b). The ring of Pro495 and the side chain of Val499 provide a hydrophobic platform against which the guanine base lies flat, with lateral contacts to the side chain ring of Pro496, which is roughly perpendicular to the plane of the base. The side chain of Arg32 is in position to make a bidentate hydrogen bond to the 2′ OH group of the ribose and to the N2 position of the guanine base (Fig. 1b). In the latter case, the distance between the two nitrogens is 3.6 Å, indicating that the second hydrogen bond is water mediated, as expected since both the guanidinium and the guanine nitrogens are proton donors. The side chain atoms of Arg32 exhibit fairly high temperature factors in all of our structures, suggesting that this surface residue is mobile. Our maps show density for its guanidinium group only in the complexes with rGTP, indicating that the binding of the nucleotide restricts the mobility of the Arg32 side chain. In contrast, the electron density for the side chain of Arg503 is very well defined in all structures. Its guanidinium group makes a bidentate hydrogen bond to the N2 of the base (again, via a water molecule) and to the main-chain carbonyl of Ser 29 (at the C terminus of helix A). The water molecule mediating the bond to the N2 of the guanine is also hydrogen bonded to the main-chain carbonyl of residue 29, giving rise to a network of interactions involving fingers, thumb, and rGTP. This nucleotide-binding site is fully occupied at 10 mM rGTP, and its occupancy is still 0.7 to 0.8 at 1 mM rGTP, whether the metal ion is Mn2+ or Mg2+ (see Table 1 for three of the five complexes made at various rGTP and Mn2+ or Mg2+ concentrations). Furthermore, this site is specific for rGTP: soaking the crystals with rATP, rCTP, or rUTP, even at 10 mM concentration (Table 1), revealed no binding at this site. These results were reproduced three times with rATP and verified once with rCTP and rUTP. Arg32 seems to be an important specificity determinant. Its bidentate hydrogen bond to the ribose and the base can only exist with rGTP, since the orientation of the base is defined by its interactions with the two proline rings (P495 and P496) and Val499. The side chains lining this surface rGTP site appear to be strictly conserved among HCV genotypes (Table 2; see Fig. 4), with the notable exception of position 499, which displays restricted variability (see Discussion).
TABLE 2.
Summary of all interactions observed between HCV polymerase and nucleotides
| Nucleotide site | Residue | Nature of interactions |
|---|---|---|
| Surface | Ser29 | 1 water-mediated hydrogen bond to ribose; main-chain interaction |
| Arg32 | 1 hydrogen bond to ribose, 1 water-mediated hydrogen bond to base | |
| Pro495 | van der Waals contacts with base | |
| Pro496 | van der Waals contacts with base | |
| Val499 | van der Waals contacts with base | |
| Arg503 | 1 water-mediated hydrogen bond to base | |
| C | Asp220 | Coordination of metals A and B |
| Asp318 | Coordination of metals A and B | |
| Asp319 | Coordination of metal A | |
| P | Arg158 | 2 hydrogen bonds to α phosphatea |
| Ser367 | 1 hydrogen bond to β phosphate | |
| Arg386 | 2 hydrogen bonds to β phosphate | |
| Thr390 | 1 hydrogen bond to γ phosphatea | |
| Arg394 | 1 hydrogen bond to α phosphate αa | |
| I | Arg48 | Electrostaticb |
| Lys51 | Electrostaticb | |
| Lys151 | Electrostaticc | |
| Lys155 | Electrostaticb | |
| Arg158 | Electrostaticb | |
| rUTP Cd | Leu159 | 2 hydrogen bonds to base; main-chain interaction |
| Asp225 | 1 hydrogen bond to ribose |
α and γ phosphates were assigned with respect to the expected position of the nucleoside moiety, which is not ordered in our structures.
Distance between the charged residue and the tP is in the range 3.5 to 3.8 Å, indicating a strong electrostatic interaction but no hydrogen bonding.
Distance between the charged residue and the tP is in the range 4 to 5 Å, indicating a weak electrostatic interaction.
The nucleoside moiety, and these interactions, were observed at the C site only with rUTP. In this case, the tP moiety is also shifted compared to the other structures (see text for details).
The C and I rNTP sites.
X-ray diffraction analysis of crystals soaked with rNTPs and Mn2+ or Mg2+ ions under the conditions described in Materials and Methods revealed extra density at and near the catalytic site of the enzyme (Fig. 1c). Except for rUTP (see below), only densities corresponding to the tP moieties of rNTPs were present in the difference maps, indicating that the nucleoside moieties are disordered in the absence of template. In addition to the tP of a nucleotide at the catalytic site, densities for two other tPs were present in each complex. The refined occupancies were between 0.6 and 0.85, depending on the particular complex. While the occupancies of the tPs observed at the P and I sites were the same independently of the divalent cation, that of the C site was much lower when Mg2+ was used instead of Mn2+. Crystallographic refinement was therefore pursued first for the models corresponding to complexes obtained in the presence of Mn2+, which are described in detail below. It was verified that the refined models obtained superpose well to electron density maps obtained from crystals soaked with rNTP and Mg2+, in particular concerning the weaker density at the C site.
The C site.
The electron density maps obtained from crystals soaked with rCTP, rGTP, and rATP in the presence of Mn2+ displayed clear density for the tP moiety of a nucleotide bound to two metal ions (labeled A and B in all the figures). The metals lie 3.6 Å apart and are coordinated by Asp220, -318, and -319 (Table 2) from conserved polymerase motifs A and C (39). Metal B is coordinated by all three phosphates of the nucleotide, and metal A interacts with the α-phosphate only, likely in position to contact the 3′ OH of the ultimate nucleotide of the nascent RNA chain during polymerization. The side chains of Asp220 and Asp318 bridge the two metals, whereas that of Asp319 interacts only with metal A. Superimposing the crystal structures of NS5B with that of human immunodeficiency virus type 1 reverse transcriptase (RT), based on the palm and fingertips, brings the tP density observed at the C site into superposition with the corresponding portion of the dNTP of the RT-template-primer-dTTP quaternary complex (20). The corresponding superposition to the φ6 polymerase results in an almost perfect match of the tP density observed in our structures with that of the nucleotide at the C site of the φ6 polymerase initiation complex (12). In addition, the latter superposition also identifies sites P and I, as discussed below.
Surprisingly, soaking the crystals with rUTP revealed density for the complete nucleotide bound at the active site, as displayed in the stereo diagram of Fig. 2a. The base is ordered because it interacts with the polypeptide main chain, with its N3 and O4 positions hydrogen bonded to the main-chain carbonyl and nitrogen atoms, respectively, of Leu159. This interaction is not expected to take place in the presence of template, where N3 an O4 would be involved in Watson-Crick base pairing to the matching base facing the catalytic site. On the other hand, the 2′ OH of the ribose hydrogen bonds to the side chain of Asp225 in motif A, an interaction which is expected to remain valid in the presence of template and which is likely to confer specificity for ribonucleotides. The tP moiety is also shifted with respect to the normal nucleotide-binding mode at the catalytic site, in such a way that the β and γ phosphates occupy the former positions of the α and β phosphates, respectively (Fig. 3b).
FIG. 2.
The rUTP complex. Secondary structure elements are color coded as in Fig. 1a and labeled as in Fig. 1c. (a) Close-up view of the region around the catalytic site of the enzyme (stereo view). The orientation is as in Fig. 1c, and the representation of atoms is as in Fig. 1b. Amino acids involved in binding the nucleotides are labeled. To the right is the nucleotide bound to the C site, with the uracil base hydrogen bonded to the polypeptide main chain. To the left is the tP moiety of the nucleotide bound at the P site. Hydrogen bonds are represented as in Fig. 1b. Divalent metals (Mn2+) are displayed as gray spheres; *A and *B identify the catalytic ions (see text). mc, main chain. (b) Stereo view displaying the C, P, and I sites (green) in the enzyme's active-site region in the rUTP complex. Shown is a view from the thumb. Labeling is as described in the legend to panel a. Arrow, access route of nucleotides through the NTP tunnel defined previously (8).
In summary, nucleotides can bind to the catalytic site of NS5B through their tP moieties even in the absence of template. For rUTP, fortuitous interactions of the base with the polypeptide main chain combine with interactions that are biologically relevant (the 2′ OH of the ribose with the side chain of Asp225). These interactions lead to a binding mode in which the tP moiety is shifted with respect to the expected binding in the productive nucleotide incorporation mode.
The P site.
The stereo plot of Fig. 2a displays all the ligands (listed in Table 2) to the nucleotide in the P site. Arg158 (from the Λ2 loop of the fingers, in motif F identified recently [27]) makes a bidentate hydrogen bond to the α phosphate, which is also contacted by Arg394 (helix αO, thumb). Ser367 in motif E and Arg386, which lie at the base of β-hairpin 15-16 (Fig. 1c), hydrogen bond to the β phosphate, and Thr390 hydrogen bonds to the γ phosphate. A Mn2+ ion was detected between the tPs from the P and C sites, likely screening electrostatic repulsion between the two adjacent nucleotides. Of the five residues interacting with the tP at the P site, three (R158, S367, and R386) appear strictly conserved among all HCV genotypes (not shown). Interestingly, these amino acids are also conserved throughout the Flaviviridae family (with the notable exception of GB virus A [GBV-A] and GBV-C), and this conservation extends to the polymerases of other positive-stranded RNA viruses known to initiate replication de novo, such as brome mosaic virus (BMV) (23), as shown in Fig. 5.
In the initiation complex of the φ6 polymerase, the base of the nucleotide at the P site stacks against a tyrosine residue (φ6-Tyr630). This side chain acts as a protein platform to hold the base in the right orientation so that it can make a Watson-Crick base pair with the 3′ base of the template (Fig. 3b). The polymerization reaction is thus primed by a single nucleotide. In NS5B, there is a water-filled cavity in this region and the nucleoside moiety is disordered in the absence of both template and platform.
The I site.
The stereo plot shown in Fig. 2b displays all three observed tPs in the case of the rUTP complex. The I site is found further toward the NTP tunnel, and the tP of this nucleotide exhibits no direct hydrogen bonding to side chains, in contrast to the P site (Table 2). It is held in place instead by electrostatic interactions only, in keeping with the notion that this site holds the nucleotides transiently, allowing template interrogation. A rapid turnover in this region and across the NTP tunnel during elongation is indeed expected to occur. The five side chains interacting with this nucleotide, Arg48, Lys51, Lys151, Lys155, and Arg158, appear strictly conserved in all HCV genotypes (not shown).
DISCUSSION
The results reported here provide a detailed high-resolution picture of the binding of rNTPs to the HCV polymerase in the absence of template. For the crystallographic analyses, we mostly used saturating conditions of ligands (Table 1) in order to maximize the occupancy and facilitate the interpretation of the electron density maps. Under these conditions, we were able to make the following important and unexpected observations: (i) the presence of a specific binding site for rGTP at the surface of the enzyme (a site occupied also at nucleotide and metal concentrations closer to the physiological conditions); (ii) an alternative rUTP binding site, in the absence of template, superposing partially with the binding site at the catalytic center; and (iii) the presence in the active site area of electron density for the tP moieties of three nucleotides that match precisely the locations of the nucleotides observed in the initiation complex of the bacteriophage φ6 polymerase.
The biological significance of the low-affinity surface rGTP site is currently not understood. From our crystallographic analysis we know that this site is 70 to 80% occupied at 1 mM rGTP. This observation suggests that the dissociation constant of rGTP is in the 0.25-to-0.45 mM range under our experimental conditions, that is, conditions compatible with the stability and cryoprotection of crystals, as indicated in Materials and Methods. In the cytoplasm of an infected cell, different environmental conditions may well affect the binding, but the affinity of rGTP for this site is likely to be of the same order of magnitude, given that the physiological ionic strength and pH are similar to the ones used for stabilizing the crystals. In sharp contrast, soaking the crystals with rATP, rCTP, or rUTP, even at a 10 mM concentration, does not lead to binding at the surface site, demonstrating the specificity for rGTP. Several studies have shown a specific effect of high rGTP concentrations on the efficiency of RNA synthesis by NS5B (29, 30). Luo et al. (30) have concluded that the rGTP effect is likely to be related to its incorporation as an initiating nucleotide, which they interpret as being rate limiting for RNA synthesis. We have reassessed the reported data both on stimulation of NS5B and de novo synthesis (24, 29, 30, 34, 46), bearing in mind two possibilities: first, that the two phenomena may be linked and, second, that the remote surface site might play a role in activating de novo initiation, with an overall stimulation of RNA synthesis as a result. No definite conclusions can be drawn at the moment, however, as different studies are not directly comparable due to major differences both in biochemical assay conditions and ways of analyzing the products. However, a possibly important observation is that amino acid 499, an integral part of the surface rGTP binding site, is variable among HCV isolates. NS5B residue 499 is a valine in most isolates of genotype 1b, as in the BK isolate used in this study and in those of Lohmann et al. (29) and Zhong et al. (46); it is an alanine in most isolates with the other genotypes (Fig. 4). However, some isolates of genotype 1b sport an alanine or a threonine at this position, as with the enzymes used by Luo et al. (30) and Oh et al. (34), respectively. These differences might explain some of the discrepancies reported in different studies, which suggest that variants with Ala at position 499 are more easily activated by rGTP. However, given that the polymerases used in the reported experiments differed at other positions besides position 499, it is clear that parallel biochemical studies with mutants differing only in amino acids observed as binding to the surface nucleotide are needed to clarify the possible effects of rGTP binding to this surface site.
If the involvement of the surface site in de novo initiation is established by biochemical data, the question of the underlying mechanism remains to be addressed. One possibility is an allosteric regulation, with rGTP binding at this site helping to induce a conformational change of the enzyme that would be important for initiation. However, we see no change of conformation in the crystal of the enzyme with bound rGTP. It is possible that the presence of both template in the RNA binding groove (2, 8, 27) and rGTP at the surface site is necessary for such a conformational change, and more structural studies are obviously needed in order to test this hypothesis. An alternative possibility is that the surface site may provide an oligomerization surface for NS5B and that the oligomeric form would adopt the correct conformation required for efficient initiation. Interestingly, a very recent report (40) points to a possible dimerization of NS5B needed for RdRp. This report identifies two surface amino acids, Glu18 and His502, as being essential for dimerization and activity. His502, in α-helix T, comes immediately before Arg503, which is part of the surface rGTP site (Fig. 1b and 4). A possible role for rGTP could be, therefore, the stabilization of a putative catalytic NS5B dimer. For the poliovirus polymerase (protein 3DPOL), it has been proposed that multimerization of the enzyme might be important for polymerase function (17, 18, 37). Furthermore, it has also been reported that rGTP binding to 3DPOL results in a dramatic protection of the enzyme from heat denaturation and activity loss (42) and that rGTP concentrations of about 2 mM are needed for maximum protection. A separate nucleotide-binding site, not implicated in the polymerase activity of 3DPOL, indeed maps to the surface of the enzyme and involves either 3DPOL-Lys278 or -283 (41), which align with amino acids 272 and 278, respectively, of NS5B, along β-strand 9 in the fingertips. Mutations in 3DPOL that eliminate this binding site result in an enzyme that is as active as the wild type when tested in vitro but, when introduced into an infectious clone, lead to a virus with extremely poor growth which quickly reverts to the wild-type nucleotide-binding phenotype (41). The location of this nucleotide-binding site, in a region that was disordered in the crystal structure of 3DPOL (17) (determined in the absence of bound rGTP), is away, however, from the finger-thumb connection in which the observed surface rGTP site of NS5B lies. The surface sites in the two enzymes are therefore not equivalent and may play different roles, in keeping with the different mechanism of initiation of poliovirus replication (38).
In addition to revealing the rGTP binding site at the surface of the enzyme, our data demonstrate that nucleotides bind at the active site in the absence of RNA template. The affinity of the HCV polymerase for nucleotides is therefore sufficiently high, in particular when the divalent cation is Mn2+, to allow for detailed crystallographic analyses. In contrast to most other polymerases, in which a large rearrangement of the fingers domain is necessary to bring together the amino acid side chains that are needed to contact the nucleotides (13, 20), NS5B has a preformed active site (2, 8, 27). Comparison of our results with the published structures of catalytically competent complexes of homologous polymerases, therefore, becomes particularly informative.
The finding that rUTP binds at the catalytic site with the base hydrogen bonded to the polypeptide main chain is, although artificial in the sense that rUTP cannot bind in such a way in the presence of template, important for the conception of new specific antiviral drugs. It is conceivable that rUTP analogs that bind to the C site in this alternative mode with higher affinity, acting as competitive inhibitors of the enzyme, can be obtained. Because the relevant interactions involve only the polypeptide main chain, the catalytic metal ions, and the strictly conserved Asp225 it is conceivable that the virus would not easily find mutations to escape inhibition by such compounds. Indeed, mutations of Asp225 have been shown to completely inactivate the enzyme (28).
A final important observation is the presence of three distinct nucleotide-binding sites in the catalytic center of the enzyme, albeit with the sugars and bases disordered (except for rUTP, as explained above). The superposition of the observed densities of the tP moieties of rCTP, rGTP, and rATP to the nucleotides that were observed in the initiation complex of the φ6 polymerase is remarkable. Particularly interesting is the fact that three of the amino acids in NS5B that bind the tP moiety of the nucleotide at the P site are strictly conserved among HCV with different genotypes (not shown) and in most other members of the Flaviviridae (Fig. 5). Arg158 lies in the previously identified motif E of RdRp (27), in the fingers. The other conserved residues involved, Ser367 and Arg386, are in the thumb, at the base of β-hairpin 15-16 (Fig. 1c, 2a, and 5). Arg386 has been shown by Hong and collaborators to be critical for extension of a dinucleotide primer by NS5B (19), which led these authors to suggest that it might also be involved in de novo initiation. The conservation of the three above-mentioned amino acids among polymerases of positive-stranded RNA viruses was recognized by Koonin (25), who labeled the conserved RdRp motifs I to VIII. Arg158 lies in motif II, and Ser367 and Arg386 lie in motifs VII and VIII, respectively. Inspection of the alignment presented by Koonin shows that the combination of Ser and Arg in motifs VII and VIII separated by about 19 amino acids, as is the case in NS5B, is a conserved feature of polymerases of many other viruses, including members of the three supergroups of RdRp identified in that work (25). Enzymes such as poliovirus 3DPOL, which is known to use a genome-linked protein primer for initiation (38), and coliphage Qβ RdRp, which needs a host protein cofactor for initiation (6, 7), do not display the conserved Ser and Arg residues. On the other hand, the polymerase of φ6, which is a more distant, double-stranded RNA phage, does not display the conservation in motifs VII and VIII either, even though it directs de novo initiation. The structure of the initiation complex of the φ6 enzyme indeed shows that, in spite of the remarkable spatial superposition of the nucleotides, the tP moiety of the priming nucleotide is contacted by side chains from regions of the protein that do not have a counterpart in NS5B. The conservation pattern detected in motifs VII and VIII is therefore likely to be related to the particular structure used in binding the priming nucleotide by RdRps from a subset of positive-stranded RNA viruses and may be used to define a new class of enzymes that direct de novo initiation. The alignment presented by Koonin suggests that polymerases from the corona- and toroviruses, as well as plant viruses such as poty- and luteoviruses, may also belong to this new class. Indeed, the 19 intervening amino acids between the two conserved residues make a β-hairpin in NS5B, (one of two β-hairpins in an otherwise α-helical thumb domain [2, 8, 27]). Both β-hairpins are absent in the poliovirus thumb, which is a smaller, exclusively α-helical domain (17). It is worth noting, however, that even the poliovirus enzyme, which is known to initiate replication in vivo with protein template VPg (38), has been shown to be capable of de novo initiation under special in vitro conditions (requiring 5 mM Mn2+ [4, 5]). This observation is a warning that detecting de novo initiation in vitro does not by itself prove that this process is biologically relevant. Incidentally, our observation that the C site is more occupied when the divalent ion is Mn2+ is suggestive of a stabilizing effect of this ion, favoring the binding of both initiating nucleotides in the active-site region. This may lead, for HCV-NS5B also, to de novo initiation via a nonphysiological mechanism in some in vitro conditions. Nevertheless, our observations point to a conserved motif involved in binding the “priming” nucleotide. The sequence alignment of Fig. 5 shows that this R-19X-S motif is also present in the polymerase of BMV, which was shown to efficiently initiate RNA replication de novo (23, 44). The fact that this motif is shared not only by RdRps from the Flaviviridae family but also by those from viruses from other viral families, including BMV, the properties of which have been very well characterized biochemically, is a strong argument in favor of the relevance of the NS5B P site in vivo. Furthermore, the conservation of the R-19X-S motif suggests that polymerases from the Flaviviridae family, including pestiviruses and flaviviruses, are likely to initiate RNA replication de novo with properties similar to those of the BMV enzyme. Indeed, the polymerase of the pestivirus bovine viral diarrhea virus (BVDV) has been shown to direct primer-independent synthesis of RNA (22), and two very recent reports show that the polymerases of the flaviviruses Kunjin (16) and dengue 2 (1) also direct de novo initiation of RNA synthesis.
An important piece of information provided by the superposition of our structures to the initiation complex of the φ6 polymerase is the identification of the orientation of the nucleotides in the active site, i.e., which one is the α phosphate and which is the γ phosphate (Fig. 3). Concerning the nucleotide at site P, this information also reveals the presence of a big solvent cavity in the region where the nucleoside moiety should lie. This observation strongly suggests that a different conformation of the NS5B thumb is needed for efficient initiation, in which the protein, through an element similar to the protein platform provided by residue Tyr630 of the φ6 polymerase, would hold the base in place to make Watson-Crick interactions with the 3′ base of the template. Since fingers and thumb are connected via the Λ1 loop, a change of conformation of the thumb will also affect the fingers. In this context, the presence of a surface rGTP site precisely at the finger-thumb connection is suggestive of a possible role of rGTP either in triggering the conformational change or in stabilizing the active conformation. These results thus point to specific biochemical and structural studies to tackle the determination of the precise mechanism of initiation of RNA synthesis by the HCV polymerase and to help clarify the role of rGTP in this process.
Acknowledgments
We thank J. Lescar and the staff of ESRF beam line ID14 for help with diffraction data collection and J. Janin for helpful discussions.
This work was funded in part by grants from CNRS, ARC, FRM, the SESAME program from “Région Ile-de-France,” and BIOCRYST Pharmaceuticals and a PRFMMIP (“Réseau Hépatite C”) grant from the French Ministry of Research to F.A.R.
REFERENCES
- 1.Ackermann, M., and R. Padmanabhan. 2001. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 276:39926-39937. [DOI] [PubMed] [Google Scholar]
- 2.Ago, H., T. Adashi, A. Yoshida, M. Yamamoto, N. Habuka, K. Yatsunami, and M. Miyano. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure 7:1417-1426. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnold, J. J., and C. E. Cameron. 1999. Poliovirus RNA-dependent RNA polymerase (3Dpol) is sufficient for template switching in vitro. J. Biol. Chem. 274:2706-2716. [DOI] [PubMed] [Google Scholar]
- 5.Arnold, J. J., S. K. Ghosh, and C. E. Cameron. 1999. Poliovirus RNA-dependent RNA polymerase (3D(pol)). Divalent cation modulation of primer, template, and nucleotide selection. J. Biol. Chem. 274:37060-37069. [DOI] [PubMed] [Google Scholar]
- 6.Blumenthal, T. 1980. Interaction of host-coded and virus-coded polypeptides in RNA phage replication. Proc. R. Soc. London B Biol. Sci. 210:321-335. [DOI] [PubMed] [Google Scholar]
- 7.Blumenthal, T., and D. Hill. 1980. Roles of the host polypeptides in Q beta RNA replication. Host factor and ribosomal protein S1 allow initiation at reduced GTP concentration. J. Biol. Chem. 255:11713-11716. [PubMed] [Google Scholar]
- 8.Bressanelli, S., L. Tomei, A. Roussel, I. Incitti, R. L. Vitale, M. Mathieu, R. De Francesco, and F. A. Rey. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. USA 96:13034-13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brünger, A. T. 1997. Free R value: cross-validation in crystallography. Methods Enzymol. 277:366-396. [DOI] [PubMed] [Google Scholar]
- 10.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 11.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butcher, S. J., J. M. Grimes, E. V. Makeyev, D. H. Bamford, and D. I. Stuart. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410:235-240. [DOI] [PubMed] [Google Scholar]
- 13.Doublie, S., S. Tabor, A. M. Long, C. C. Richardson, and T. Ellenberger. 1998. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature 391:251-258. [DOI] [PubMed] [Google Scholar]
- 14.Esnouf, R. M. 1997. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph. Model. 15:132-134, 112-113. [DOI] [PubMed] [Google Scholar]
- 15.Gouet, P., E. Courcelle, D. I. Stuart, and F. Metoz. 1999. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15:305-308. [DOI] [PubMed] [Google Scholar]
- 16.Guyatt, K. J., E. G. Westaway, and A. A. Khromykh. 2001. Expression and purification of enzymatically active recombinant RNA-dependent RNA polymerase (NS5) of the flavivirus Kunjin. J. Virol. Methods 92:37-44. [DOI] [PubMed] [Google Scholar]
- 17.Hansen, J. L., A. M. Long, and S. C. Schultz. 1997. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure 5:1109-1122. [DOI] [PubMed] [Google Scholar]
- 18.Hobson, S. D., E. S. Rosenblum, O. C. Richards, K. Richmond, K. Kirkegaard, and S. C. Schultz. 2001. Oligomeric structures of poliovirus polymerase are important for function. EMBO J. 20:1153-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong, Z., C. E. Cameron, M. P. Walker, C. Castro, N. Yao, J. Y. Lau, and W. Zhong. 2001. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology 285:6-11. [DOI] [PubMed] [Google Scholar]
- 20.Huang, H., R. Chopra, G. L. Verdine, and S. C. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669-1675. [DOI] [PubMed] [Google Scholar]
- 21.Joyce, C. M., and T. A. Steitz. 1995. Polymerase structures and function: variations on a theme? J. Bacteriol. 177:6321-6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kao, C. C., A. M. Del Vecchio, and W. Zhong. 1999. De novo initiation of RNA synthesis by a recombinant flaviviridae RNA-dependent RNA polymerase. Virology 253:1-7. [DOI] [PubMed] [Google Scholar]
- 23.Kao, C. C., and J. H. Sun. 1996. Initiation of minus-strand RNA synthesis by the brome mosaic virus RNA-dependent RNA polymerase: use of oligoribonucleotide primers. J. Virol. 70:6826-6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kao, C. C., X. Yang, A. Kline, Q. M. Wang, D. Barket, and B. A. Heinz. 2000. Template requirements for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 74:11121-11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koonin, E. V. 1991. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 72:2197-2206. [DOI] [PubMed] [Google Scholar]
- 26.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 27.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 28.Lohmann, V., F. Korner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lohmann, V., H. Overton, and R. Bartenschlager. 1999. Selective stimulation of hepatitis C virus and pestivirus NS5B RNA polymerase activity by GTP. J. Biol. Chem. 274:10807-10815. [DOI] [PubMed] [Google Scholar]
- 30.Luo, G., R. K. Hamatake, D. M. Mathis, J. Racela, K. L. Rigat, J. Lemm, and R. J. Colonno. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makeyev, E. V., and D. H. Bamford. 2000. Replicase activity of purified recombinant protein P2 of double-stranded RNA bacteriophage phi6. EMBO J. 19:124-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merritt, E. A., and D. J. Bacon. 1997. Raster3D photorealistic molecular graphics. Methods Enzymol. 277:505-524. [DOI] [PubMed] [Google Scholar]
- 33.Navaza, J. 1987. On the fast rotation function. Acta Crystallogr. A 43:645-653. [DOI] [PubMed] [Google Scholar]
- 34.Oh, J. W., T. Ito, and M. M. Lai. 1999. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J. Virol. 73:7694-7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ojala, P. M., and D. H. Bamford. 1995. In vitro transcription of the double-stranded RNA bacteriophage phi 6 is influenced by purine NTPs and calcium. Virology 207:400-408. [DOI] [PubMed] [Google Scholar]
- 36.Otwinowski, Z., and W. Minor. 1996. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 37.Pata, J. D., S. C. Schultz, and K. Kirkegaard. 1995. Functional oligomerization of poliovirus RNA-dependent RNA polymerase. RNA 1:466-477. [PMC free article] [PubMed] [Google Scholar]
- 38.Paul, A. V., J. H. van Boom, D. Filippov, and E. Wimmer. 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393:280-284. [DOI] [PubMed] [Google Scholar]
- 39.Poch, O., I. Sauvaget, M. Delarue, and N. Tordo. 1989. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 8:3867-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qin, W., H. Luo, T. Nomura, N. Hayashi, T. Yamashita, and S. Murakami. 2002. Oligomeric interaction of hepatitis C virus NS5B is critical for catalytic activity of RNA dependent RNA polymerase. J. Biol. Chem. 277:2132-2137. [DOI] [PubMed] [Google Scholar]
- 41.Richards, O. C., and E. Ehrenfeld. 1997. One of two NTP binding sites in poliovirus RNA polymerase required for RNA replication. J. Biol. Chem. 272:23261-23264. [DOI] [PubMed] [Google Scholar]
- 42.Richards, O. C., P. Yu, K. L. Neufeld, and E. Ehrenfeld. 1992. Nucleotide binding by the poliovirus RNA polymerase. J. Biol. Chem. 267:17141-17146. [PubMed] [Google Scholar]
- 43.Steitz, T. A., S. J. Smerdon, J. Jager, and C. M. Joyce. 1994. A unified polymerase mechanism for nonhomologous DNA and RNA polymerases. Science 266:2022-2025. [DOI] [PubMed] [Google Scholar]
- 44.Sun, J. H., and C. C. Kao. 1997. RNA synthesis by the brome mosaic virus RNA-dependent RNA polymerase: transition from initiation to elongation. Virology 233:63-73. [DOI] [PubMed] [Google Scholar]
- 45.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhong, W., A. S. Uss, E. Ferrari, J. Y. Lau, and Z. Hong. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]