Abstract
Methylation of cytosine residues in DNA plays a critical role in the silencing of gene expression, organization of chromatin structure, and cellular differentiation of eukaryotes. Previous studies failed to detect 5-methylcytosine in Dictyostelium genomic DNA, but the recent sequencing of the Dictyostelium genome revealed a candidate DNA methyltransferase gene (dnmA). The genome sequence also uncovered an unusual distribution of potential methylation sites, CpG islands, throughout the genome. DnmA belongs to the Dnmt2 subfamily and contains all the catalytic motifs necessary for cytosine methyltransferases. Dnmt2 activity is typically weak in Drosophila melanogaster, mouse, and human cells and the gene function in these systems is unknown. We have investigated the methylation status of Dictyostelium genomic DNA with antibodies raised against 5-methylcytosine and detected low levels of the modified nucleotide. We also found that DNA methylation increased during development. We searched the genome for potential methylation sites and found them in retrotransposable elements and in several other genes. Using Southern blot analysis with methylation-sensitive and -insensitive restriction endonucleases, we found that the DIRS retrotransposon and the guaB gene were indeed methylated. We then mutated the dnmA gene and found that DNA methylation was reduced to about 50% of the wild-type level. The mutant cells exhibited morphological defects in late development, indicating that DNA methylation has a regulatory role in Dictyostelium development. Our findings establish a role for a Dnmt2 methyltransferase in eukaryotic development.
DNA methylation is linked to various aspects of epigenetic regulation, including silencing of gene expression, organization of chromatin structure, and cellular differentiation (16, 27, 35, 39). DNA methyltransferases add a methyl group to the C-5 position of cytosine in genomic DNA. These epigenetic modifications can be replicated by the maintenance methyltransferase, Dnmt1, during DNA replication (4). Methylation of CpG dinucleotides in promoter regions usually leads to reduced gene expression (12, 15). DNA methylation contributes to stable and efficient repression by blocking transcription factors from binding to promoters and by recruiting 5-methylcytosine (5mC) binding proteins that act as repressors. DNA methylation also induces histone deacetylation, which results in chromatin condensation, such as in the silencing of the inactive X chromosome, imprinted genes, and parasitic DNAs (4, 15, 44). Retrotransposable elements (RTEs) are also heavily methylated in mammalian and plant cells (18, 23). Although numerous studies have revealed a negative correlation between DNA methylation of promoter regions and gene expression, the precise role of tissue-specific DNA methylation patterns in development is still controversial (16, 26, 31).
In the past 15 years, it has been accepted that DNA methylation does not occur in Dictyostelium. Smith and Ratner (40) reported that methylation was absent from CCGG sites in repetitive DNA and in the actin multigene family. Drosophila melanogaster was also thought to be an exception for a long time, but recent evidence demonstrated a functional DNA methylation system in Drosophila (14, 25). A small amount of 5mC, consisting of 0.1 to 0.2% of the total cytosine residues, has been detected by methylcytosine antibodies and by high-performance liquid chromatography (HPLC) analysis.
DNA methylation in Drosophila is mediated by the DNA methyltransferase Dnmt2 (32). The Dnmt2 methyltransferase family is highly conserved from yeast to humans, but its trans-methyl activity is rather weak. The biological function of this family is unknown because disruption of the genes caused no readily observable phenotypes in the mouse, yeast, or fly (20, 28, 45). Nevertheless, the evolutionary conservation of this gene family suggests an ancestral origin and an essential function in eukaryotes.
The Dictyostelium genome-sequencing consortium reported an unusual distribution of G+C-rich regions throughout the genome and an underrepresentation of CpG dinucleotides relative to the isomer GpC (9). Such a bias is believed to reflect methylation of cytosine in CpGs, probably because methylated cytosine promotes the mutagenic transition from CpG to TpG. In addition, the Dictyostelium genome sequence revealed the existence of a DNA methyltransferase for 5mC (30). The Dictyostelium DnmA (dictyBase identification no. DDB0231095) is highly similar to other members of the Dnmt2 subfamily. These observations suggested that methylation of cytosine may occur in Dictyostelium and that it may serve as a useful model system for the study of Dnmt2 transmethylases.
We show here that the Dictyostelium genome does contain 5mC, albeit at very low levels. We also show that DNA methylation is developmentally regulated and that deletion of the dnmA gene results in reduced methylation and in developmental defects. We found that CpG dinucleotides have a unique distribution in the Dictyostelium genome and that 5mC residues are found around some of the DIRS transposable elements and in the guaB gene.
MATERIALS AND METHODS
Growth, development, and generation of mutants.
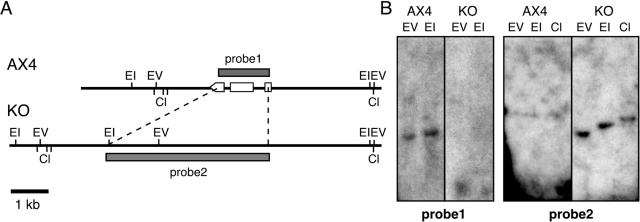
Wild-type Dictyostelium strain AX4 (19) and the dnmA− mutant were grown in HL-5 liquid medium (41) and developed as described previously (36). The dnmA knockout strain was generated in AX4 by substituting a 1.2-kb fragment of the Dictyostelium dnmA gene (nucleotides 80 to 1292 relative to the first ATG) with a 4.4-kb plasmid containing the blasticidin resistance gene (1). Transformants were generated by homologous recombination, selected as described previously (29), and verified by Southern blot analysis and by PCR across the homologous recombination junctions. Two independently derived strains were constructed which had identical phenotypes.
Purification of genomic DNA and dot blot analysis.
Genomic DNA was purified using three methods. The CTAB method (47) was used with minor modifications. Nuclei were lysed in 100 mM EDTA and 5% sodium lauryl sarcosyl at 55°C for 20 min. Genomic DNA was incubated in CTAB solution (1% CTAB, 0.7 M NaCl, 10 mM EDTA, 50 mM Tris-HCl [pH 8.0], 0.5% polyvinylpyrrolidone) at 65°C for 5 min and extracted with chloroform and phenol-chloroform and purified by ethanol precipitation. For the sodium dodecyl sulfate (SDS)-proteinase K method, nuclei were lysed in 1% SDS and 0.2 mg/ml proteinase K for 60 min at 60°C. Genomic DNA was extracted with phenol and phenol-chloroform and purified by ethanol precipitation. For extraction with plant DNAzol (Invitrogen), nuclei from vegetative cells or whole cells from developing cultures were treated according to the manufacturer's recommended protocol. Genomic DNA from all three methods was treated with 100 μg/ml RNase A (Sigma) for 1 h at 37°C. The different methods produced essentially identical results, supporting the notion that the signals observed were not an artifact of the purification method. PCR fragments of the thymidine kinase gene thyB (dictyBase identification no. DDB0191436) were used as positive or negative controls for dot blots. To generate a fully methylated positive control fragment, 5-methyl-dCTP (Roche Applied Science) was used instead of dCTP in the PCR. These DNA fragments were used as standards to quantify the amount of 5mC in the genomic DNA samples (see Fig. S1 in the supplemental material).
Detection and quantification of 5mC in genomic DNA were performed by dot blot analysis with an antibody against 5mC. This sensitive immunological method has been used by others to detect 5mC in Entamoeba histolytica (10) and in liver tumors (42) and is especially suitable for detection and quantification of small amounts and small changes in 5mC levels. Genomic DNA (1 to 25 μg) and PCR fragments were denatured with 0.4 N NaOH at 100°C for 10 min, neutralized with ammonium acetate (pH 7.0), and blotted on a nitrocellulose membrane (Schleicher & Schuell). Dot blots were incubated with a 1:750 dilution of anti-5-methylcytosine antibody (Megabase Research), followed by incubation with a 1:5,000 dilution of alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G antibody (Sigma), or with a 1:1,000 dilution of anti-5-methylcytosine antibody (Abcam, Inc.), followed by incubation with a 1:8,000 dilution of alkaline phosphatase-conjugated donkey anti-sheep immunoglobulin G (heavy plus light) antibody (Jackson ImmunoResearch). Signals were detected by chemiluminescence with the Western-Star system (Applied Biosystems) according to the manufacturer's recommended protocol. Antibodies were then removed by incubation with 62.5 mM Tris-HCl (pH 6.8), 2% SDS, and 100 mM 2-mercaptoethanol at 50°C for 30 min, and the blots were hybridized with a radiolabeled DNA probe for DIRS-1 (37) and the control probe thyB.
Analysis of DNA methylation by restriction endonuclease digests and quantitative PCR.
Dictyostelium genomic DNA was digested with methylation-sensitive (HpaII and/or AvaI) or -insensitive endonucleases (MspI and/or BsoBI) and used as templates for PCRs. PCR primer pairs were designed to amplify regions flanking the restriction sites (HpaII/MspI and AvaI/BsoBI) in the guaB gene locus. Quantitative PCR was performed using the SYBR green dye in an MJ Research Opticon 2 instrument according to the manufacturer's recommended protocol. Standard curves were obtained using a control primer set based on serial dilutions of undigested genomic DNA. End products were resolved on a 1% agarose gel to verify the results. For Southern blot analyses, the genomic DNA was also digested with the XbaI restriction endonuclease and the DNA was analyzed as described previously (17) with the DIRS-1 probe or with the guaB probe.
Computational analysis of CpG dinucleotides in the Dictyostelium genome.
We used the “newcpgreport” tool from the EMBOSS package (33) to calculate the observed/expected ratio and to find CpG islands. The definition of CpG islands we used is similar to the one used in mammalian systems with a correction for low G+C content (13). We required the G+C content to be greater than 40% and the observed/expected ratio of CpG to be greater than 0.6. The CpG observed/expected ratios were computed for 100-bp windows, sliding at 1-base increments. The minimal length of a CpG island is 200 bp. We also mapped the codistribution of CpG islands and 124 retrotransposable elements (80 DIRS, 33 SKIPPER, 11 TDD3).
For the computational analysis and corresponding visualizations, we used three different metrics and computed them from the genome sequence of all the genes (13,629) and a subset of 3,745 genes for which expression data are available (43). First, we counted the number of CpG dinucleotides at each position relative to the translational START or STOP sites in the given group of genes, from 10 kb 5′ to 10 kb 3′ of the site and displayed the total number as a histogram, where the width of the bin is 250 bp. Second, the G+C content (%) at a position in a gene was computed by centering a window of 100 bp and then counting the number of G's and C's in the window. This was done for all the genes and then averaged into a single value that was plotted at the corresponding position in a line graph. The values were computed every 30 bp. Third, the observed/expected ratio was computed in each window of 100 bp. We obtained the observed number by counting the number of CpGs in the window. The following formula as implemented in the “newcpgreport” tool was used to compute the expected number of CpGs: (number of G's × number of C's)/window length. The averages of the values were computed every 30 bp and plotted as a line graph.
RESULTS
Evidence for general methylation of Dictyostelium genomic DNA.
Previous reports suggested that Dictyostelium genomic DNA was not methylated (40), but recent analysis of the Dictyostelium genome suggested otherwise (9). To test whether Dictyostelium DNA contains methylated cytosines, we extracted genomic DNA from cells during growth and during development. The DNA was denatured and blotted onto a nitrocellulose membrane. The presence of 5mC was monitored by incubation with a specific antibody followed by chemiluminescence detection. Figure 1A shows that Dictyostelium genomic DNA does contain 5mC and that the level of methylation was higher in developed cells than in vegetative cells. As controls, we used PCR fragments amplified from the thyB gene with dCTP (negative control) or with 5-methyl-dCTP (positive control). The antibody did not react with the negative control and reacted with the positive control in a dose-dependent manner, indicating high specificity and linearity with the amount of 5mC (see Fig. S1 in the supplemental material). A quantitative analysis comparing the methylation signal of genomic DNA to a dilution series of the positive control suggests that as many as 3 in 10,000 (0.03%) cytosine residues are methylated in vegetative Dictyostelium cells. The methylation level increased to 0.14% of the cytosine residues in developed cells. We treated the genomic DNA with RNase and with proteinase K and confirmed the absence of protein and RNA by electrophoresis and by spectrophotometry (data not shown). We conclude that the signal we observed was indeed due to methylation of cytosine in DNA.
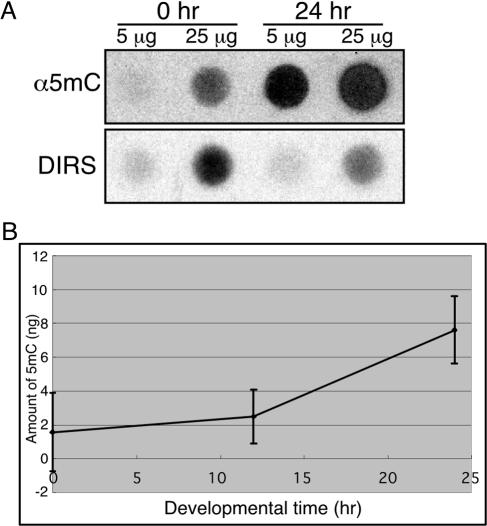
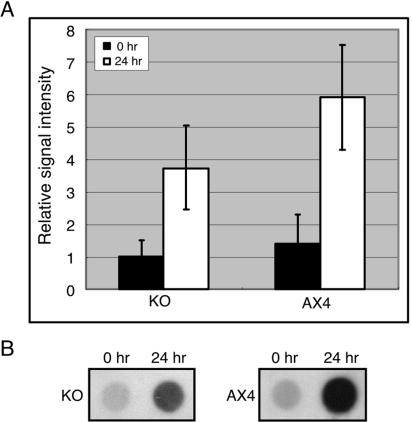
FIG. 1.
DNA methylation is developmentally regulated. Genomic DNA was prepared from cells at different developmental stages and dot blotted on nitrocellulose membranes in the indicated amounts. (A) DNA methylation was detected by reacting the membranes with an antibody directed against 5-methylcytosine (α5mC), and the total amount of DNA was estimated by hybridization with a radioactive probe against DIRS-1 (DIRS). (B) The antibody and hybridization signals were quantified, and methylation levels were normalized to the amounts of DNA and plotted as a function of developmental time. The plot indicates that DNA methylation is increased during Dictyostelium development. Results are the averages and standard deviations from 3 replications.
The dot blots were stripped and hybridized with a probe against the DIRS-1 repetitive element to normalize the signal to the amount of DNA bound to the membrane. Normalized values of the 5mC signal show that basal levels of DNA methylation occur in vegetative cells and are increased about fivefold by the end of development (Fig. 1B). We therefore conclude that Dictyostelium genomic DNA is methylated and that methylation is developmentally regulated.
Unusual distribution of CpG islands in the Dictyostelium genome.
DNA methylation often occurs on cytosines in the dinucleotide CpG. One of the reasons to suspect that Dictyostelium DNA was indeed methylated was the observation that the dinucleotide CpG was underrepresented in the genome (9). In other organisms, methylation of CpG has a profound regulatory role that is associated with the CpG islands (7). CpG islands have been reported around RTEs in other genomes, and the function of methylation at RTEs is a defense mechanism against the expression of these parasitic elements (5, 46). Therefore, we tested whether they were also overrepresented near RTEs in the Dictyostelium genome. Figure 2A shows the distribution of RTEs and CpG islands along the 6 chromosomes and several unassigned contigs in the Dictyostelium genome. We observed that the CpG islands are not distributed evenly throughout the genome and that they are more frequently found around RTEs (Fig. 2A). The most striking correlation is found at the centromeric end of chromosomes 1 and 6 and at the opposite end of chromosome 2 (Dictyostelium chromosomes are telocentric and the map places the centromeres on the left). We also searched for CpG islands using a model that accounts for the bimodal distribution of C+G in coding and noncoding regions, and the results were essentially identical to the ones shown (data not shown).
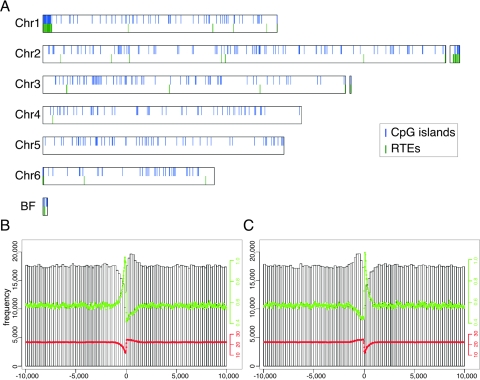
FIG. 2.
Uneven distribution of CpG dinucleotides in the Dictyostelium genome. CpG islands were identified in the Dictyostelium genome. (A) CpG islands (blue vertical lines) and RTEs (green vertical lines) were plotted along the 6 chromosomes, indicating a high degree of overlap. All of the 13,629 predicted genes in the genome were aligned by their ORFs such that the translational start site (B) or the translational termination site (C) was aligned. CpG dinucleotide frequency was computed in a 20-kb region centered on the respective border of the ORF (open bars). The ratio between the observed and expected frequencies of CpG dinucleotides is shown as a green line. The G+C content (%) is indicated by a red line.
Surveys of other genomes have indicated that the dinucleotide CpG has a unique distribution around the translational start site (3, 38). To test whether that was also true in Dictyostelium, we have aligned all the predicted genes in the genome at the translational start and the translational end sites of their respective open reading frames (ORFs). We then calculated the frequency of CpG dinucleotides from all genes and plotted it for a region of 20 kb, centered on the respective borders of the ORF (Fig. 2B and Fig. 2C). We found that the regions 5′ of the ORF start site (Fig. 2B) and 3′ of the ORF end site (Fig. 2C) are significantly deprived of CpGs.
Due to the unique nucleotide composition of the genome, Dictyostelium ORFs have significantly higher G+C contents than their flanking and intergenic regions (9). Plotting the average G+C content around the ORF borders revealed the expected drop 5′ of the start site, followed by a sharp increase 3′ of the start site (Fig. 2B). The end of the ORF exhibited the expected opposite trend with a mild increase 5′, a sharp decline immediately 3′, and a gradual increase back to the average level following the ORF end (Fig. 2C). We therefore tested the ratio between the observed number of CpG dinucleotides and the number expected from the G+C content. We found that the observed/expected ratio increased sharply immediately 5′ of the ORF start site, sharply decreased immediately 3′, and gradually returned to average further 3′ of the start site (Fig. 2B). At the end of the ORF, the CpG ratio gradually decreased 5′ to the end, sharply increased at the end, and then sharply decreased back to the average level 3′ to the end of the ORF (Fig. 2C). We tested the distribution of all other dinucleotides but found none that behaved like CpG (data not shown). Finally, we performed the same analysis on about 3,700 genes that were known to be expressed from microarray data (43). The results were essentially identical to those shown for the entire predicted ORFeome (data not shown). These results support the notion that CpG dinucleotides have a special role around the borders of open reading frames.
Evidence for selective DNA methylation in the Dictyostelium genome.
The correlation between RTEs and CpG islands (Fig. 2A) prompted us to test whether DNA methylation could be observed specifically in RTEs. We digested genomic DNA from growing and developing cells with restriction endonucleases that are sensitive or resistant to the presence of 5mC and separated the restriction fragments by gel electrophoresis. Examination of the banding pattern after ethidium bromide staining revealed that the methylation-sensitive and the methylation-insensitive enzyme reactions were carried out with the same efficiency (Fig. 3A). We therefore conclude that the reactions were carried out to completion. The DNA was then subjected to Southern blot analysis with a probe against one of the most abundant RTEs, DIRS-1 (6, 9). The data in Fig. 3B show the expected multitude of bands but also show that one of the bands was methylated. Comparing the pattern obtained with the 5mC-sensitive endonucleases in lane 3 with the pattern obtained with the 5mC-resistant endonucleases in lane 4, it is clear that at least one type of DIRS RTEs was protected from digestion by the methylation-sensitive endonuclease. This finding was best observed at 16 h of development (Fig. 3B). It is also likely that most of the DIRS RTEs were not methylated on CpG, since their Southern blot patterns were indistinguishable by this method.
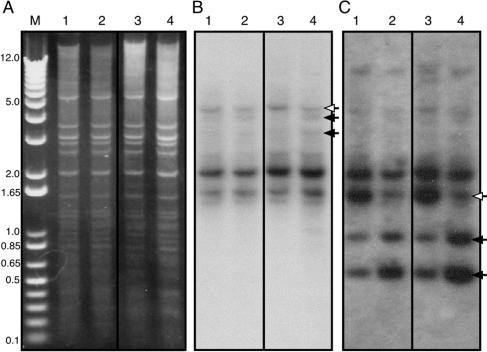
FIG. 3.
Selective DNA methylation in the DIRS retrotransposon and the guaB gene. Genomic DNA was prepared from vegetative (0 h, lanes 1 and 2) and developing (16 h, lanes 3 and 4) cells and digested with the XbaI restriction endonuclease and with restriction endonucleases that are sensitive (both HpaII and AvaI, lanes 1 and 3) or insensitive (both MspI and BsoBI, lanes 3 and 4) to 5mC. Digested DNA was separated by electrophoresis, along with the 1-kb DNA ladder as a size marker, and stained with ethidium bromide to visualize the banding pattern (A). The DNA was analyzed by Southern blotting with the DIRS probe (B) or with the guaB probe (C). Open arrows indicate the full-size fragments, and filled arrows indicate the resulting fragments.
The computational analysis of CpG island distribution (Fig. 2A) revealed several genes that were good candidates for DNA methylation. We selected one of these genes, guaB, for further analysis because it carries convenient restriction sites within CpG islands. The Southern blot described above was stripped and reprobed with the guaB probe (Fig. 3C). We observed a band at 1.6 kb that was digested efficiently into 1.0-kb and 0.6-kb bands by the methylation-insensitive endonucleases (Fig. 3C, lanes 2 and 4) but was partially protected from the methylation-sensitive endonucleases (Fig. 3C, lanes 1 and 3). We speculate that the partial protection was observed because DNA methylation occurred only in some of the cells.
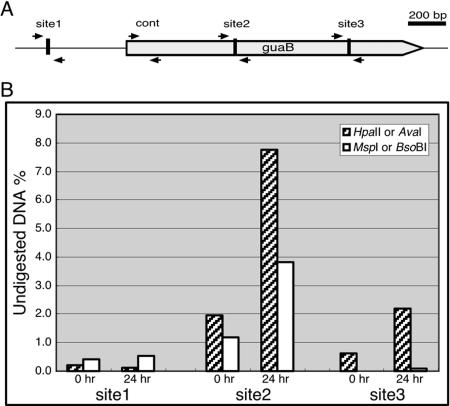
To further test the notion that Dictyostelium DNA methylation is rather sparse, we performed quantitative analysis of DNA methylation at the guaB locus. The guaB locus contains 3 putative methylation sites that can be detected by differential restriction endonuclease digestion (Fig. 4A). We designed PCR primer pairs that flank these sites as well as one pair to amplify a control fragment near the 5′ border of the ORF. Genomic DNA was digested with the appropriate restriction endonucleases and purified, and quantitative PCRs were performed and normalized (Fig. 4B). We found that site 1 was not methylated, since both types of endonuclease digestion eliminated the ability to generate a PCR product. Both site 2 and site 3 were protected from digestion with 5mC-sensitive endonucleases but not protected from the 5mC-insensitive endonuclease. We therefore conclude that the sites within the guaB ORF are methylated, whereas the site 5′ of the ORF is not. The data also support the observation that DNA methylation is developmentally regulated because the phenomenon was more pronounced in DNA from developing cells than in DNA from vegetative cells (Fig. 4B).
FIG. 4.
Quantitative analysis of DNA methylation in the guaB locus. The guaB gene contains several putative methylation sites. (A) A physical map of the guaB locus with the 5′ border on the left. The ORF is indicated by a thick gray arrow. Quantitative PCR was done with four primer sets, indicated by small adjacent black arrows pointing at each other. Sites 1 and 3 are restriction sites for the methylation-sensitive endonuclease HpaII and the methylation-insensitive endonuclease MspI. Site 2 is a restriction site for the methylation-sensitive endonuclease AvaI and the methylation-insensitive endonuclease BsoBI. The control pair of primers (cont) was used for normalization. (B) Genomic DNA from vegetative (0 h) and terminally developed (24 h) cells was digested with methylation-sensitive (hatched bars) or -insensitive (white bars) endonucleases. The fraction (% of total) of DNA that remained undigested was measured by quantitative PCR across the respective sites as indicated in panel A and calculated from the average of two replications. Significant and developmentally regulated DNA methylation can be seen in sites 2 and 3.
The dnmA gene encodes a DNA methyltransferase.
The Dictyostelium genome project predicted the presence of a single copy of dnmA, a DNA methyltransferase gene of the Dnmt2 family (30). To test that prediction, we generated a knockout strain by replacing most of the gene with a blasticidin resistance cassette. We verified the gene disruption by Southern blot analysis (Fig. 5) and by PCR across the relevant junctions (data not shown). Using most of the dnmA gene as a probe (Fig. 5, probe 1), we observed a single band in each of the lanes containing wild-type DNA, verifying the observation that the gene is present as a single copy. These single bands were absent in the lanes containing mutant DNA, indicating that the sequences were deleted from the genome. Probing with the plasmid used to replace the dnmA gene (Fig. 5, probe 2), we found no signal in the lanes containing wild-type DNA and the expected sized single bands in lanes containing the mutant DNA, indicating that a clean replacement has occurred. We then tested the level of DNA methylation in growing (0 h) and in developing (24 h) mutant cells with the anti-5mC antibody (Fig. 6). The data indicate that deletion of dnmA resulted in a decrease in the level of DNA methylation in developing cells. We therefore conclude that dnmA is a bona fide DNA methyltransferase whose activity can account for about 50% of the 5mC in the genome. We also postulate that another activity, encoded by a yet unidentified gene in the genome, must be responsible for the remaining methylation.
FIG. 5.
Physical maps of the dnmA locus. (A) Restriction maps of the dnmA locus in the wild-type strain (AX4) and in the dnmA knockout strain (KO). Probe 1 consists of the dnmA gene, and probe 2 consists of the plasmid DNA used to replace the gene in the knockout mutant. Open boxes and arrowhead indicate the dnmA exons. Restriction sites are indicated as follows: EV, EcoRV; EI, EcoRI; Cl, ClaI. (B) Genomic DNA from the wild-type strain (AX4) and from the dnmA knockout strain (KO) was digested as indicated and subjected to Southern blot analysis with probes 1 and 2.
FIG. 6.
Reduced global DNA methylation in the dnmA knockout strain. Genomic DNA was extracted from vegetative (0 h) and developed (24 h) cells of the wild-type (AX4) and dnmA knockout (KO) strains. DNA methylation was detected by dot blotting with the anti-5mC antibody as described in the legend to Fig. 1. (A) DNA methylation signal intensity during growth (black bar) and development (white bar) are the averages of results from two independent replications of each of two independent knockout strains (KO) and the averages of results from three independent replications of the wild-type strain, showing a reduction in the developmentally regulated methylation. (B) Representative dot blot data comparing the dnmA knockout strain (KO) and the wild-type strain (AX4) signals in growth and development.
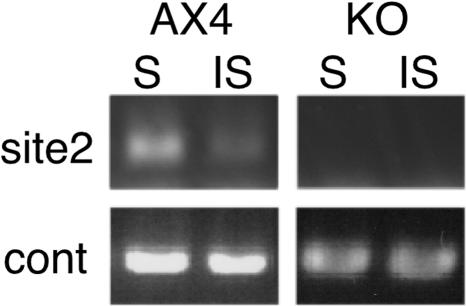
To further test the target specificity of DnmA, we compared the guaB methylation patterns in the mutant and in the wild type. Using the quantitative PCR method described in the legend to Fig. 4, we found that site 2 was completely unmethylated in the mutant but protected from the 5mC-insensitive endonuclease in the wild type (Fig. 7). The guaB expression levels were not affected by the dnmA mutation (data not shown).
FIG. 7.
dnmA is responsible for selective DNA methylation at the guaB locus. Genomic DNA was prepared from developed wild-type (AX4) and dnmA knockout (KO) cells. The DNA was digested with a methylation sensitive (S, AvaI) or a methylation-insensitive (IS, BsoBI) endonuclease and analyzed by PCR using the primer set indicated in the legend to Fig. 4. PCR products were resolved on a 1% agarose gel containing ethidium bromide and visualized by fluorescence under UV light. Photographs are shown, and control PCR amplification products are shown below the experimental lanes.
We conclude that dnmA is responsible for nearly 50% of the 5mC in the genome and that its activity is rather selective, but we cannot link it to gene expression.
dnmA activity is essential for proper culmination.
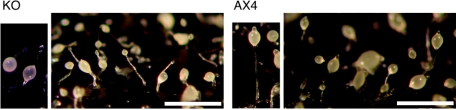
Gross morphological aberrations are a good measure of developmental function in Dictyostelium. We therefore developed the dnmA-knockout mutant side by side with wild-type cells and compared their morphologies. The mutant and wild-type morphological and temporal progression through development were essentially identical for the first 20 h of development (data not shown). During culmination however, the dnmA knockout mutant exhibited subtle but reproducible defects. Many of the mutant sori were fragmented along the stalk, and they were clearer than the wild-type sori (Fig. 8). Clear sori sometimes indicate inefficient sporulation or precocious germination, and we indeed found that the mutant formed fewer spores than the wild type (data not shown). However, the sporulation phenotype was less penetrant and less reproducible than the sorus fragmentation phenotype. Overall, it is clear that dnmA is essential for proper terminal development, consistent with the temporal increase in DNA methylation during development.
FIG. 8.
dnmA is essential for proper development. Wild-type (AX4) and dnmA knockout (KO) cells were starved and developed on black nitrocellulose filters for 24 h. In the mutant cells, sori were found at various points along the stalk, whereas the wild type has one sorus at the top of the stalk. Mutant sori were also more transparent than wild-type sori.
DISCUSSION
DNA methylation is found in both animals and plants (11), indicating that it has evolved before the separation of the two kingdoms. Dictyostelium has evolved from the evolutionary line leading to animals, after the separation from plants (24), so the reported lack of DNA methylation was peculiar (40). Our findings indicate that Dictyostelium is not an exception to the evolution of DNA methylation in that it has at least two mechanisms of DNA methylation. One of the mechanisms depends on the dnmA gene, and the other remains to be identified. DNA methylation was also reported in Candida albicans, which is not known to have any putative Dnmt methyltransferases in its genome (34).
The function of the Dnmt2 family of DNA methyltransferases is somewhat enigmatic. In most organisms, these enzymes have little or no activity, and mutations in the respective genes have almost no consequences (20, 28, 45). DNA methylation in Dictyostelium is fairly rare, in that only 0.03 to 0.14% of the total cytosines are methylated. The fraction of methylated cytosines is higher in other organisms with about 0.1% in Drosophila melanogaster, 2 to 10% in mammals, and more than 30% in some plants (2, 14). In addition, the G+C content in the Dictyostelium genome is very low (22.43%), so there is just one methylated cytosine per 7 to 30 kb in the genome. This low level of DNA methylation is consistent with the weak activity of Dnmt2 enzymes. Nevertheless, mutating dnmA resulted in a subtle yet obvious developmental defect such as fragmentation of the sori midway along the stalk. The positioning of the sorus to the top of the stalk is likely to have a selective advantage in spore dispersal. Therefore, a mutant lacking dnmA would probably have a competitive disadvantage against wild-type cells. These findings provide a demonstration of function for a Dnmt2 DNA methyltransferase. Another recent example is the finding that Drosophila Dnmt2 might be involved in longevity and aging (22).
Dictyostelium DnmA contains all 10 conserved motifs, including the methyl donor (S-adenosyl-l-methionine) binding motif and the active site, and an invariant polypeptide in the target-recognizing domain, TRD, which is the putative DNA recognition site (21). The latter domain is not found in other Dnmt family members, so it is thought that Dnmt2 recognizes a specific kind of target through this TRD domain (8). In this paper, we found that the Dictyostelium DnmA functions as one of the DNA methyltransferase and methylates its targets in a selective manner. In D. melanogaster, most methylation was seen at CpT, and only a small fraction of the 5mC was detected in CpG dinucleotides (25). Overexpression of Dnmt2 in D. melanogaster increased the methylation levels of cytosine in the nonsymmetrical CpT and CpA dinucleotides (20). It is therefore conceivable that the cytosines in CpT and CpA are also recognized by the Dictyostelium DnmA.
The low level of DNA methylation and the developmental regulation explain why previous studies have failed to find it (40). A random choice of probes for Southern blot analysis would have to be very lucky to reveal a methylated gene, and physical methods like HPLC analysis are limited to fairly high proportions of 5mC. We have tried to analyze Dictyostelium DNA by HPLC, followed by mass spectrometry, but found that the proportion of 5mC in the genome was below the limit of detection of the method (data not shown). We were fortunate that the antibody we used is highly sensitive and highly specific to 5mC in DNA. We also had the advantage of knowing the genome sequence as a guide in searching for good probes for Southern blot analysis.
DNA methylation in Dictyostelium is developmentally regulated, and the highest degree of methylation is observed in cells at the end of development. It is tempting to speculate that the timing of highest methylation and the late morphological phenotype of the dnmA mutant are causally correlated. It is possible that the methylation of a small subset of genes is required for regulation of late gene expression and that disruption of the process leads to developmental defects. Another possibility is that the correlation is indirect. Terminal morphology is the result of proper execution of all developmental pathways. Therefore, we cannot rule out the possibility that the dnmA knockout phenotype is the result of subtle defects in processes that are executed early in development and are manifested only later on.
The unusual distribution of CpG islands in the Dictyostelium genome is also quite interesting. The association of CpG islands with RTEs in other organisms is usually attributed to the silencing effects of DNA methylation (5, 46). It is reasonable to hypothesize that silencing RTE transcription by DNA methylation may have a selective advantage in maintaining genome stability and reducing the metabolic burden associated with expressing the RTE genes. This assertion may have to be reexamined in light of our findings. In Dictyostelium, CpG islands are also associated with RTEs, but most of them are probably not methylated because we could not detect methylation around most DIRS1 RTEs. It is therefore possible that the CpG islands have a role that is independent of their methylation in addition to their proven, methylation-dependent role in the regulation of chromatin structure and gene expression. In that context, CpG dinucleotides have an unusual distribution around the borders of ORFs. It is conceivable that most of them are not methylated in Dictyostelium, so we propose that CpGs and CpG islands have a role that is independent of methylation. What that role might be is currently a matter of speculation, but the fact is that the unusual distribution is also found in other organisms (3, 38).
Finally, we did not find evidence for the role of DNA methylation in the regulation of gene expression in Dictyostelium, but we only examined one gene. A more comprehensive study might reveal whether DNA methylation is important for gene regulation and which genes and pathways are regulated by that mechanism.
In an independent study, Nellen and coworkers have detected 5mC DNA methylation of CpA and CpT dinucleotides. In their hands, deletion of dnmA did not result in morphological defects, but their parental strain (AX2) was different from the one we used (AX4). These researchers found that DNA methylation was necessary for transcriptional silencing of the Skipper RTE and showed evidence for the correlation of DNA methylation and RNA interference (19a). Together, our data establish that Dictyostelium cells utilize DNA methylation for regulation of gene expression and of development.
Supplementary Material
Acknowledgments
We thank Cornelius (Neal) Boerkoel and members of his laboratory for attracting our attention to the anti-5mC antibody, for technical advice, and for helpful discussions. We thank William O'Brien and members of his laboratory for performing the HPLC mass spectrometry analysis and for helpful discussions. We thank Angelique Dousis for excellent technical assistance. We also thank Wolfgang Nellen and his colleagues for communicating data prior to publication.
Work at BCM was supported by a grant from the National Institute of Child Health and Human Development (PO1 HD39691). M.K. is a recipient of a research fellowship from the Uehara Memorial Foundation. Work at University of Ljubljana was supported by a Program Grant (P2-0209) and a Scientific Research Collaboration Grant between the United States and the Republic of Slovenia, both from the Slovenian Research Agency.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Adachi, H., T. Hasebe, K. Yoshinaga, T. Ohta, and K. Sutoh. 1994. Isolation of Dictyostelium discoideum cytokinesis mutants by restriction enzyme-mediated integration of the blasticidin S resistance marker. Biochem. Biophys. Res. Commun. 205:1808-1814. [DOI] [PubMed] [Google Scholar]
- 2.Adams, R. L. 1996. DNA methylation, p. 33-66. In E. E. Bitter (ed.), Principles of medical biology, vol. 5. JAI Press, Inc., New York, N.Y. [Google Scholar]
- 3.Aerts, S., G. Thijs, M. Dabrowski, Y. Moreau, and B. De Moor. 2004. Comprehensive analysis of the base composition around the transcription start site in Metazoa. BMC Genomics 5:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bestor, T. H. 2000. The DNA methyltransferases of mammals. Hum. Mol. Genet. 9:2395-2402. [DOI] [PubMed] [Google Scholar]
- 5.Bestor, T. H. 1998. The host defence function of genomic methylation patterns. Novartis Found. Symp. 214:187-199, 228-232. [DOI] [PubMed] [Google Scholar]
- 6.Cappello, J., K. Handelsman, and H. F. Lodish. 1985. Sequence of Dictyostelium DIRS-1: an apparent retrotransposon with inverted terminal repeats and an internal circle junction sequence. Cell 43:105-115. [DOI] [PubMed] [Google Scholar]
- 7.Costello, J. F., and C. Plass. 2001. Methylation matters. J. Med. Genet. 38:285-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong, A., J. A. Yoder, X. Zhang, L. Zhou, T. H. Bestor, and X. Cheng. 2001. Structure of human DNMT2, an enigmatic DNA methyltransferase homolog that displays denaturant-resistant binding to DNA. Nucleic Acids Res. 29:439-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eichinger, L., J. A. Pachebat, G. Glockner, M. A. Rajandream, R. Sucgang, M. Berriman, J. Song, R. Olsen, K. Szafranski, Q. Xu, B. Tunggal, S. Kummerfeld, M. Madera, B. A. Konfortov, F. Rivero, A. T. Bankier, R. Lehmann, N. Hamlin, R. Davies, P. Gaudet, P. Fey, K. Pilcher, G. Chen, D. Saunders, E. Sodergren, P. Davis, A. Kerhornou, X. Nie, N. Hall, C. Anjard, L. Hemphill, N. Bason, P. Farbrother, B. Desany, E. Just, T. Morio, R. Rost, C. Churcher, J. Cooper, S. Haydock, N. van Driessche, A. Cronin, I. Goodhead, D. Muzny, T. Mourier, A. Pain, M. Lu, D. Harper, R. Lindsay, H. Hauser, K. James, M. Quiles, M. Madan Babu, T. Saito, C. Buchrieser, A. Wardroper, M. Felder, M. Thangavelu, D. Johnson, A. Knights, H. Loulseged, K. Mungall, K. Oliver, C. Price, M. A. Quail, H. Urushihara, J. Hernandez, E. Rabbinowitsch, D. Steffen, M. Sanders, J. Ma, Y. Kohara, S. Sharp, M. Simmonds, S. Spiegler, A. Tivey, S. Sugano, B. White, D. Walker, J. Woodward, T. Winckler, Y. Tanaka, G. Shaulsky, M. Schleicher, G. Weinstock, A. Rosenthal, E. C. Cox, R. L. Chisholm, R. Gibbs, W. F. Loomis, M. Platzer, R. R. Kay, J. Williams, P. H. Dear, A. A. Noegel, B. Barrell, and A. Kuspa. 2005. The genome of the social amoeba Dictyostelium discoideum. Nature 435:43-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher, O., R. Siman-Tov, and S. Ankri. 2004. Characterization of cytosine methylated regions and 5-cytosine DNA methyltransferase (Ehmeth) in the protozoan parasite Entamoeba histolytica. Nucleic Acids Res. 32:287-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freitag, M., and E. U. Selker. 2005. Controlling DNA methylation: many roads to one modification. Curr. Opin. Genet. Dev. 15:191-199. [DOI] [PubMed] [Google Scholar]
- 12.Fuks, F. 2005. DNA methylation and histone modifications: teaming up to silence genes. Curr. Opin. Genet. Dev. 15:490-495. [DOI] [PubMed] [Google Scholar]
- 13.Gardiner-Garden, M., and M. Frommer. 1987. CpG islands in vertebrate genomes. J. Mol. Biol. 196:261-282. [DOI] [PubMed] [Google Scholar]
- 14.Gowher, H., O. Leismann, and A. Jeltsch. 2000. DNA of Drosophila melanogaster contains 5-methylcytosine. EMBO J. 19:6918-6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hashimshony, T., J. Zhang, I. Keshet, M. Bustin, and H. Cedar. 2003. The role of DNA methylation in setting up chromatin structure during development. Nat. Genet. 34:187-192. [DOI] [PubMed] [Google Scholar]
- 16.Jones, P. A., and D. Takai. 2001. The role of DNA methylation in mammalian epigenetics. Science 293:1068-1070. [DOI] [PubMed] [Google Scholar]
- 17.Katoh, M., M. Hirono, T. Takemasa, M. Kimura, and Y. Watanabe. 1993. A micronucleus-specific sequence exists in the 5′-upstream region of calmodulin gene in Tetrahymena thermophila. Nucleic Acids Res. 21:2409-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kazazian, H. H., Jr., and J. L. Goodier. 2002. LINE drive. Retrotransposition and genome instability. Cell 110:277-280. [DOI] [PubMed] [Google Scholar]
- 19.Knecht, D. A., S. M. Cohen, W. F. Loomis, and H. F. Lodish. 1986. Developmental regulation of Dictyostelium discoideum actin gene fusions carried on low-copy and high-copy transformation vectors. Mol. Cell. Biol. 6:3973-3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Kuhlmann, M., B. E. Borisova, M Kaller, P. Larsson, D. Stach, J. Na, L. Eichinger, F. Lyko, V. Ambros, F. Soderbom, C. Hammann, and W. Nellen. Silencing of retrotransposons in Dictyostelium by DNA methylation and RNAi. Nuclei Acids Res., in press. [DOI] [PMC free article] [PubMed]
- 20.Kunert, N., J. Marhold, J. Stanke, D. Stach, and F. Lyko. 2003. A Dnmt2-like protein mediates DNA methylation in Drosophila. Development 130:5083-5090. [DOI] [PubMed] [Google Scholar]
- 21.Lauster, R., T. A. Trautner, and M. Noyer-Weidner. 1989. Cytosine-specific type II DNA methyltransferases. A conserved enzyme core with variable target-recognizing domains. J. Mol. Biol. 206:305-312. [DOI] [PubMed] [Google Scholar]
- 22.Lin, M. J., L. Y. Tang, M. N. Reddy, and C. K. Shen. 2005. DNA methyltransferase gene dDnmt2 and longevity of Drosophila. J. Biol. Chem. 280:861-864. [DOI] [PubMed] [Google Scholar]
- 23.Lippman, Z., A. V. Gendrel, M. Black, M. W. Vaughn, N. Dedhia, W. R. McCombie, K. Lavine, V. Mittal, B. May, K. D. Kasschau, J. C. Carrington, R. W. Doerge, V. Colot, and R. Martienssen. 2004. Role of transposable elements in heterochromatin and epigenetic control. Nature 430:471-476. [DOI] [PubMed] [Google Scholar]
- 24.Loomis, W. F., and D. W. Smith. 1990. Molecular phylogeny of Dictyostelium discoideum by protein sequence comparison. Proc. Natl. Acad. Sci. USA 87:9093-9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyko, F., B. H. Ramsahoye, and R. Jaenisch. 2000. DNA methylation in Drosophila melanogaster. Nature 408:538-540. [DOI] [PubMed] [Google Scholar]
- 26.Murrell, A., V. K. Rakyan, and S. Beck. 2005. From genome to epigenome. Hum. Mol. Genet. 14(Spec. No. 1):R3-R10. [DOI] [PubMed]
- 27.Ng, H. H., and A. Bird. 1999. DNA methylation and chromatin modification. Curr. Opin. Genet. Dev. 9:158-163. [DOI] [PubMed] [Google Scholar]
- 28.Okano, M., S. Xie, and E. Li. 1998. Dnmt2 is not required for de novo and maintenance methylation of viral DNA in embryonic stem cells. Nucleic Acids Res. 26:2536-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pang, K. M., M. A. Lynes, and D. A. Knecht. 1999. Variables controlling the expression level of exogenous genes in Dictyostelium. Plasmid 41:187-197. [DOI] [PubMed] [Google Scholar]
- 30.Ponger, L., and W. H. Li. 2005. Evolutionary diversification of DNA methyltransferases in eukaryotic genomes. Mol. Biol. Evol. 22:1119-1128. [DOI] [PubMed] [Google Scholar]
- 31.Rakyan, V. K., T. Hildmann, K. L. Novik, J. Lewin, J. Tost, A. V. Cox, T. D. Andrews, K. L. Howe, T. Otto, A. Olek, J. Fischer, I. G. Gut, K. Berlin, and S. Beck. 2004. DNA methylation profiling of the human major histocompatibility complex: a pilot study for the human epigenome project. PLoS Biol. 2:e405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reddy, M. N., L.-Y. Tang, T.-L. Lee, and C.-K. J. Shen. 2003. A candidate gene for Drosophila genome methylation. Oncogene 22:6301-6303. [DOI] [PubMed] [Google Scholar]
- 33.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 34.Russell, P. J., J. A. Welsch, E. M. Rachlin, and J. A. McCloskey. 1987. Different levels of DNA methylation in yeast and mycelial forms of Candida albicans. J. Bacteriol. 169:4393-4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salozhin, S. V., E. B. Prokhorchuk, and G. P. Georgiev. 2005. Methylation of DNA-one of the major epigenetic markers. Biochemistry (Moscow) 70:525-532. [DOI] [PubMed] [Google Scholar]
- 36.Shaulsky, G., and W. F. Loomis. 1993. Cell type regulation in response to expression of ricin A in Dictyostelium. Dev. Biol. 160:85-98. [DOI] [PubMed] [Google Scholar]
- 37.Shaulsky, G., and W. F. Loomis. 1995. Mitochondrial DNA replication but no nuclear DNA replication during development of Dictyostelium. Proc. Natl. Acad. Sci. USA 92:5660-5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimizu, T. S., K. Takahashi, and M. Tomita. 1997. CpG distribution patterns in methylated and non-methylated species. Gene 205:103-107. [DOI] [PubMed] [Google Scholar]
- 39.Singal, R., and G. D. Ginder. 1999. DNA methylation. Blood 93:4059-4070. [PubMed] [Google Scholar]
- 40.Smith, S. S., and D. I. Ratner. 1991. Lack of 5-methylcytosine in Dictyostelium discoideum DNA. Biochem. J. 277:273-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sussman, M. 1987. Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. Methods Cell Biol. 28:9-29. [DOI] [PubMed] [Google Scholar]
- 42.Tao, L., Y. Li, P. M. Kramer, W. Wang, and M. A. Pereira. 2004. Hypomethylation of DNA and the insulin-like growth factor-II gene in dichloroacetic and trichloroacetic acid-promoted mouse liver tumors. Toxicology 196:127-136. [DOI] [PubMed] [Google Scholar]
- 43.Van Driessche, N., C. Shaw, M. Katoh, T. Morio, R. Sucgang, M. Ibarra, H. Kuwayama, T. Saito, H. Urushihara, M. Maeda, I. Takeuchi, H. Ochiai, W. Eaton, J. Tollett, J. Halter, A. Kuspa, Y. Tanaka, and G. Shaulsky. 2002. A transcriptional profile of multicellular development in Dictyostelium discoideum. Development 129:1543-1552. [DOI] [PubMed] [Google Scholar]
- 44.Wilkins, J. F. 2005. Genomic imprinting and methylation: epigenetic canalization and conflict. Trends Genet. 21:356-365. [DOI] [PubMed] [Google Scholar]
- 45.Wilkinson, C. R., R. Bartlett, P. Nurse, and A. P. Bird. 1995. The fission yeast gene pmt1+ encodes a DNA methyltransferase homologue. Nucleic Acids Res. 23:203-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoder, J. A., C. P. Walsh, and T. H. Bestor. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13:335-340. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, Y. P., J. K. Uyemoto, and B. C. Kirkpatrick. 1998. A small-scale procedure for extracting nucleic acids from woody plants infected with various phytopathogens for PCR assay. J. Virol. Methods 71:45-50. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.