Abstract
In Saccharomyces cerevisiae, extracellular amino acids are sensed at the plasma membrane by the SPS sensor, consisting of the transporter homologue Ssy1p, Ptr3p, and the endoprotease Ssy5p. Amino acid sensing results in proteolytic truncation of the transcription factors Stp1p and Stp2p, followed by their relocation from the cytoplasm to the nucleus, where they activate transcription of amino acid permease genes. We screened a transposon mutant library for constitutively signaling mutants, with the aim of identifying down-regulating components of the SPS-mediated pathway. Three isolated mutants were carrying a transposon in the RTS1 gene, which encodes a regulatory subunit of protein phosphatase 2A. We investigated the basal activity of the AGP1 and BAP2 promoters in rts1Δ cells and found increased transcription from these promoters, as well as increased Stp1p processing, even in the absence of amino acids. Based on our findings we propose that the phosphatase complex containing Rts1p keeps the SPS-mediated pathway down-regulated in the absence of extracellular amino acids by dephosphorylating a component of the pathway.
The yeast Saccharomyces cerevisiae has developed a complex regulatory network enabling it to control the production of nutrient transporters depending on substrate availability in the environment. Hexose transporters and amino acid transporters are examples of nutrient transporters that are transcriptionally induced by their substrates (14, 24, 44). Amino acids are imported into the cells through amino acid permeases (AAPs) (20). The yeast AAPs belong to the amino acid/polyamine/organocation (APC) superfamily of transporters (28). They exhibit a range of affinities and specificities for all 20 common amino acids and a number of other compounds (42). The presence of extracellular amino acids results in increased transcription of about a third of the AAP genes (14). Amino acids induce transcription of the AAPs with different efficiencies, the most potent amino acid being leucine, while signaling by, e.g., arginine and proline is undetectable (16).
Ssy1p is an AAP homologue with 12 membrane-spanning domains, but compared to other members of the S. cerevisiae AAP family its cytoplasmic N-terminal tail is unusually long (30). Ssy1p has been shown to act as an amino acid sensor able to detect extracellular amino acids at the plasma membrane (8, 26, 32). Ssy1p is part of the so-called SPS sensor complex that includes Ssy1p, Ptr3p, and Ssy5p (13, 41). Ssy5p (30) and Ptr3p (27) are peripherally associated proteins, which, like Ssy1p, are essential for amino acid induction (14).
Transcriptional induction is mediated by the homologous transcription factors Stp1p and Stp2p, which bind to the promoters of the AAP genes at the UASaa (upstream activating sequence) first identified in the BAP3 promoter (6) and then in the BAP2 promoter (37), but later also found in the promoter sequences of the AAP genes GNP1, AGP1, MUP1, TAT1, TAT2, and DIP5 (10). In the absence of amino acids, Stp1p and Stp2p are present mainly in the cytoplasm (3, 10). When amino acids are detected in the environment, 10 kDa of the N terminus are endoproteolytically cleaved off, resulting in relocation of the transcription factors to the nucleus (3). Ssy5p is the endoprotease responsible for processing of Stp1p (1, 2). Signaling and processing are moreover dependent on the F-box protein Grr1p (1, 5, 11, 26), which is part of the E3 ubiquitin ligase SCF (Skp1-Cullin-F-box) complex SCFGrr1 (38). Though SCFGrr1 normally targets proteins for degradation by the 26S proteasome, activation of Stp1p is not dependent on the 26S proteasome (1). Other factors involved in amino induction include casein kinase I (1). Furthermore, Ptr3p is found to interact with Yfr021wp in a two-hybrid screen (18). The same work reports that yfr021wΔ and ypl100wΔ strains exhibit ptr3Δ-like phenotypes, i.e., they are unable to grow at a high concentration of histidine.
Mutants that exhibit constitutive expression of the target AAP genes independently of the components of the SPS sensor have been identified (15). Here we report a screen likewise aimed at identifying factors down-regulating SPS-mediated signaling, but allowing for factors that, when mutated, cause (constitutive) signaling only if the sensor is present. Using a strain deficient in histidine synthesis, we selected for growth on minimal ammonium medium supplemented with the dipeptide Gly-His. Dipeptides, including Gly-His, are taken up by the permease Ptr2p (39), which is under transcriptional control of the amino acid-sensing pathway (4, 8). Subsequent cleavage of internalized Gly-His satisfies the growth requirement for histidine (39). Thus, growth on Gly-His reflects activation of the signaling pathway, so that growth in the absence of an inducing amino acid reveals that the amino acid-sensing pathway is turned on constitutively. The constitutive mutants thus found were tested for constitutive induction of the BAP2 gene, encoding a branched-chain amino acid permease, using the β-galactosidase reporter assay. After this secondary screen the mutants included some with transposon insertions in the RTS1 gene. RTS1 encodes a regulatory subunit of protein phosphatase 2A (PP2A). We subsequently studied the role of this gene in SPS-mediated amino acid signaling.
MATERIALS AND METHODS
Strains and media.
The S. cerevisiae strains used in this study are listed in Table 1 and are all derived from strain M4054 (MATa ura3 gap1Δ) (19), which in turn is derived from S288C. The HIS3 and LEU2 genes were disrupted in M4054; the resulting strain was named M4605 and was used as the host for the transposon mutagenesis library, as described below. The histidine-independent strain M5397 (MATa ura3 HIS3 gap1Δ rts1Δ) was constructed by crossing M4599 (MATa ura3 his3 gap1Δ rts1Δ) with the isogenic X2180-1b (MATα). Spores with the desired genotype (ura3 HIS3) were tested for the gap1 phenotype on minimal citrulline medium (20, 43).
TABLE 1.
Strains used in this study
| Strain | Relevant genotype | Reference |
|---|---|---|
| M4054 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ | 19 |
| M4605 | MATaSUC2 mal gal2 CUP1 ura3 his3 leu2 gap1Δ | This work |
| M5397 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ rts1Δ | This work |
| M4871 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ ssy1Δ | P. S. Nielsen |
| M4723 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ ptr3Δ | P. S. Nielsen |
| M4724 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ ssy5Δ | P. S. Nielsen |
| M5470 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ grr1Δ | This work |
| M4600 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ rts1Δ ssy1Δ | This work |
| M5437 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ rts1Δ ptr3Δ | This work |
| M5439 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ rts1Δ ssy5Δ | This work |
| M4608 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ rts1Δ stp1Δ stp2Δ stp3Δ | This work |
| M5471 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ grr1Δ rts1Δ | This work |
| M5447 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ STP1-ZZ | 41 |
| M5445 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ ssy1Δ STP1-ZZ | 41 |
| M5443 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ ptr3Δ STP1-ZZ | 41 |
| M5444 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ ssy5Δ STP1-ZZ | 41 |
| M5559 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ grr1Δ STP1-ZZ | This work |
| M5474 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ rts1Δ STP1-ZZ | This work |
| M5501 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ rts1Δ ptr3Δ STP1-ZZ | This work |
| M5524 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ rts1Δ ssy5Δ STP1-ZZ | This work |
| M5629 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ rts1Δ ssy1Δ STP1-ZZ | This work |
| M5525 | MATaSUC2 mal gal2 CUP1 ura3 gap1Δ rts1Δ grr1Δ STP1-ZZ | This work |
The RTS1/rts1 genotype was tested on SD plates supplemented with uracil (1% [wt/vol] succinic acid, 0.6% [wt/vol] NaOH, 0.17% [wt/vol] Bacto yeast nitrogen base without amino acids, 2% Bacto agar, 2% [wt/vol] glucose, 20 mg/liter uracil), at the center of which a filter was placed with 4 mg of the toxic dipeptide l-leucyl-l-ethionine, as described previously (8). Equal amounts of cells were suspended in water, and 10 μl of each strain was streaked towards the center of the plate with the filter disk. rts1Δ strains are sensitive to l-leucyl-l-ethionine in this test.
To check for a functional SPS amino acid-sensing system, strains were plated on yeast-peptone-dextrose medium (YPD) supplemented with metsulfuron methyl (30).
Strains M5431, M5437, M5470, and M5471 were constructed by PCR amplification of the loxP-kanMX-loxP cassette from plasmid pUG6 (21) with primer overhangs complementary to the parts immediately upstream and downstream of the open reading frame (ORF) to be deleted. Transformants were selected on YPD containing 300 mg/liter Geneticin G-418 sulfate and were tested for correct deletion by diagnostic PCR. The kanMX marker was removed when necessary as described previously (21).
Strains M5443, M5444, M5445, M5447, M5474, M5501, M5524, and M5525 were constructed by replacing in situ the STP1 stop codon by 2,159 bp of a construct carrying a doublet of the immunoglobulin G-binding Z domain of the Staphylococcus aureus protein A and the kanMX marker (40, 41). Transformants were selected with Geneticin and tested by PCR.
Transposon-based mutagenesis.
Mutagenesis was based on insertion of the prokaryotic mTn3 transposon. Propagation of the mutagenized, LEU2-based library, transformation, and isolation of recombinants were carried out essentially as described previously (46). Thirty thousand colonies were screened, and 11 mutants were selected according to the following criteria: their ability to grow on SC medium devoid of leucine before being tested for constitutive amino acid uptake on SD medium supplemented with uracil and 0.5 g/liter Gly-His dipeptide; their ability to grow on SD plus Ura plus 30 mg/liter Ile plus 20 mg/liter His plus 60 mg/liter Val plus 100 mg/liter metsulfuron methyl; and finally by their constitutively active BAP2 promoter.
β-Galactosidase reporter assay.
Strains of interest were transformed with plasmid pRB108 or pTD17 (7, 42), centromere-based plasmids in which 1.0 kb of the amino acid-inducible AGP1 promoter and 683 bp of the BAP2 promoter, respectively, are fused to Escherichia coli lacZ. Transformants were grown overnight in SD medium to an optical density at 600 nm of 0.1 to 0.3. The cultures were then divided in aliquots to which different compounds were added: l-citrulline (Sigma) to a final concentration of 5 mM or an equal volume of water, and l-leucine to a final concentration of 100 μM. Each treatment was performed in duplicate on aliquots originating from the same culture. β-Galactosidase assays were performed as described previously (7). Miller unit values were calculated as follows: (optical density at 420 nm × 1,000)/(cell volume [ml] × reaction time [min] × optical density at 600 nm at harvest).
Western blot analysis.
Cells carrying a fusion (Stp1-ZZ) of an immunoglobulin G-binding domain of the Staphylococcus aureus protein A to the C terminus of Stp1p (40) were cultivated in SD supplemented with 20 mg/liter uracil to the early exponential phase. Cells were harvested and proteins were extracted under denaturing conditions with NaOH and β-mercaptoehanol before being enriched by precipitation with trichloroacetic acid. Protein concentrations were determined using the Bio-Rad protein assay. Proteins were separated on NuPAGE 10% Bis-Tris gels (Invitrogen) and transferred to polyvinylidene difluoride membranes. Chemiluminescent immunodetection was performed as described previously (41). The extent of binding of the antibody against horseradish peroxidase was quantified using the Image Quant software 5.0 (Molecular Dynamics) with a local median-based background correction.
RESULTS
Isolation of mutants constitutive in amino acid signaling.
In order to select mutants with increased activity of the amino acid-sensing pathway, a screen was designed in which a his3 leu2 derivative of the reference strain M4054 was subjected to transposon mutagenesis. The host strain was transformed with a mixture of yeast genomic fragments representing a mutagenized LEU2-based plasmid library (46). Mutants of interest were selected using three different screening criteria. First, we took advantage of the fact that dipeptide uptake (39), as well as branched-chain amino acid uptake (7), is very low in cells grown on minimal ammonium medium in the absence of leucine. Dipeptides, including Gly-His, are taken up by the peptide transporter Ptr2p (39), which is under the transcriptional control of the amino acid-sensing pathway (4). Mutants with constitutive signaling were thus selected on SD medium supplemented with the Gly-His dipeptide, which, upon intracellular cleavage, provides the histidine required for growth (39). A subsequent selection was carried out on SD supplemented with the amino acids Ile, His, and Val, and the sulfonylurea herbicide metsulfuron methyl. This type of compound inhibits synthesis of branched-chain amino acids and therefore prevents growth of cells unable to take up branched-chain amino acids from the medium (53), such as mutants deficient in amino acid signaling (29, 30). Finally, colonies of interest were tested for constitutive acitivity of the BAP2 promoter.
Among the 11 mutants thus isolated, three had transposon insertions in the PP2A subunit-encoding gene RTS1 (34). The remaining transposons were localized in VPS15, VPS34, and VPS16, coding for proteins involved in vacuolar protein sorting, and STH1, coding for the ATPase subunit in the chromatin remodeling complex RSC. Two others had mutations in ORFs with unknown function, YFR044C and YFR045W. Two mutants were not mapped. In the present work, the relationship between the phenotype and the locus affected by transposon insertion was analyzed in more detail for the RTS1 gene.
Deletion of RTS1 results in constitutive activation of BAP2 and AGP1 transcription.
Since transposon insertion very often completely inactivates a gene, we wished to analyze the behavior of a strain deleted for RTS1. One of the selection criteria used in the original screen was the ability to take up Gly-His on SD medium, thereby promoting growth of a histidine-requiring strain because of intracellular cleavage of the dipeptide. To find out if an rts1Δ strain exhibits the same phenotype, we deleted the RTS1 ORF in a his3 derivative of the reference strain M4054. Indeed, the lack of RTS1 enabled growth on the dipeptide as a histidine source (not shown).
In order to be able to analyze the rts1Δ mutation in the absence of extracellular amino acids or dipeptides, we also constructed a HIS3 rts1Δ strain (M5397). First we wished to confirm the activation of target promoters for the amino acid signaling by testing for growth on YPD supplemented with metsulfuron methyl; the strain grew, as expected (not shown). Then we tested whether this activation was constitutive by yet another criterion, using the dipeptide l-leucyl-l-ethionine. If this dipeptide is taken up, it is cleaved to yield the toxic methionine analogue l-ethionine. Also, uptake of the dipeptide l-leucyl-l-ethionine is mediated up by the peptide transporter Ptr2p (39) and is thus under transcriptional control of the amino acid-sensing pathway (4). Insensitivity to l-leucyl-l-ethionine therefore reflects that the pathway is not activated, while growth inhibition in the absence of an inducing amino acid indicates constitutive activation of the amino acid-sensing pathway. The rts1Δ strain M5397 was unable to grow on minimal medium in the presence of l-leucyl-l-ethionine, suggesting constitutive activity of the signaling pathway (not shown).
We also investigated activation of the promoter of the broad-specificity AAP gene AGP1 using a fusion of the promoter to the E. coli lacZ gene. Cells were grown in SD medium and incubated for 40 min with or without 100 μM l-leucine. The experiment was repeated with 5 mM l-citrulline or an equal volume of water. These concentrations have been previously found to fully or almost fully induce the AGP1 promoter (16, 40). The results show that AGP1 transcription in rts1Δ cells is constitutive, i.e., unaffected by leucine or citrulline addition. In the wild-type control experiment, AGP1 promoter activation was low in the absence of amino acids and increased strongly in response to either leucine or citrulline (Table 2).
TABLE 2.
AGP1 promoter activity in wild-type cells and rts1Δ cells in response to leucine and citrulline
| Inducer | Mean β-galactosidase activitya ± SEM
|
|
|---|---|---|
| M4054 (wild type) | M5397 (rts1Δ) | |
| l-Leucine (100 μM) | ||
| − | 1.5 ± 0.1 | 10.5 ± 0.4 |
| + | 8.4 ± 1.0 | 9.9 ± 0.9 |
| l-Citrulline (5 mM) | ||
| − | 0.1 ± 0.1 | 11.4 ± 0.6 |
| + | 8.7 ± 1.4 | 13.5 ± 0.4 |
Activity in Miller units in the absence (−) and 40 minutes after the addition (+) of inducer.
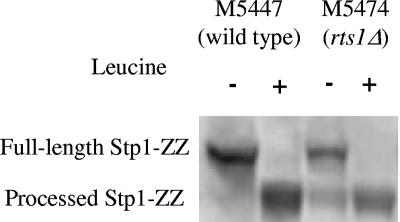
Transcriptional induction of AAP genes was previously shown to involve Stp1p endoproteolysis (3). We used the Stp1-ZZ construct previously described (40), in which the C terminus of Stp1p was fused to a doublet of the IgG-binding Z domain of the S. aureus protein A. This insert was integrated in the wild-type strain and the rts1Δ strain, resulting in strains M5447 (40) and M5474, respectively. The fraction of processed Stp1p in rts1Δ cells in the absence of leucine was greater than in wild-type cells (30% versus 13%), whereas addition of leucine led to almost complete (above 90%) Stp1p processing in both wild-type and rts1Δ cells (Table 3 and Fig. 1). Quantification of antibody binding showed that the relative amount of processed Stp1p in the absence of inducer was more than doubled in rts1Δ cells compared to wild-type cells.
TABLE 3.
Quantification of processed Stp1-ZZ in wild-type and rts1Δ cells in response to leucinea
| Strain | Mean amt of processed Stp1-ZZ ± SEM
|
|
|---|---|---|
| Without inducer | With inducer | |
| M5447 (wild type) | 12.8 ± 1.1 | 90.2 ± 0.4 |
| M5474 (rts1Δ) | 30.4 ± 5 | 92.7 ± 1.6 |
Samples were taken in the absence and 20 minutes after addition of 100 μM l-leucine. Relative amounts of full-length and processed Stp1-ZZ were determined by performing the experiment twice.
FIG. 1.
Quantitative Western analysis of Stp1-ZZ processing in wild-type and rts1Δ cells in response to 100 μM l-leucine.
Remarkably, Stp1p processing to the extent of 30% is sufficient to generate an activity of the AGP1 promoter comparable to that observed in wild-type cells after amino acid addition (compare Table 2 and Table 3). In fact, this is expected from previous comparisons of dose-response relationships using quantification of Stp1p processing versus AGP1 promoter activity as the read-out (40, 41). The sensing of l-leucine exhibits a 50% effective concentration of 12 μM under the growth conditions studied when Stp1p processing is monitored, but only 1 μM when AGP1 promoter-driven lacZ expression is measured (40). The most obvious interpretation is that saturation of the promoter activity occurs already at modest levels of activated Stp1p.
Epistasis relationships.
In order to further investigate the role of RTS1, we constructed strains M4600 (ssy1Δ rts1Δ), M5437 (ptr3Δ rts1Δ), M5439 (ssy5Δ rts1Δ), M5470 (grr1Δ rts1Δ), and M4608 (stp1Δ stp2Δ stp3Δ rts1Δ). STP3 is homologous to STP1 and STP2, and Helge A. Andersen (personal communication) has found that deletion of STP3 further reduces the low, remaining activity of the amino acid signaling pathway in an stp1Δ stp2Δ mutant. All of the resulting strains were sensitive to metsulfuron methyl on YPD, and all were insensitive to l-leucyl-l-ethionine (not shown), suggesting that the amino acid-sensing pathway was inactive.
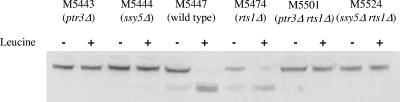
β-Galactosidase assays were performed on extracts from cells in which the E. coli lacZ gene was placed under the control of the AGP1 promoter. β-Galactosidase measurements confirmed that the multiple mutants behaved similarly to the single mutants deficient in signaling, i.e., AGP1 transcription was very low even in the presence of extracellular amino acids (Table 4). Using the Stp1-ZZ construct, we also measured relative amounts of processed and full-length Stp1p in each of the strains, in the absence and presence of amino acids. Stp1p was exclusively present as the full-length protein in all single and multiple mutants (Fig. 2). In other words, ssy1, ptr3, ssy5, and grr1 are epistatic over rts1.
TABLE 4.
AGP1 promoter activity in cells deleted of genes encoding positive factors in amino acid signaling and/or RTS1 in the absence and presence of inducing amino acids
| Inducer | Mean β-galactosidase activitya ± SEM
|
||||||
|---|---|---|---|---|---|---|---|
| M4054 (wild type) | M5397 (rts1Δ) | M4600 (ssy1Δ rts1Δ) | M5437 (ptr3Δ rts1Δ) | M5439 (ssy5Δ rts1Δ) | M5471 (grr1Δ rts1Δ) | M4608 ( rts1Δ stp1Δ stp2Δ stp3Δ) | |
| l-Leucine (100 μM) | |||||||
| − | 1.7 ± 0.1 | 10.1 ± 0.2 | 1.1 ± 0.1 | −0.1 ± 0.0 | −0.8 ± 0.0 | 0.1 ± 0.0 | |
| + | 7.2 ± 0.6 | 9.6 ± 0.2 | 1.1 ± 0.0 | −0.1 ± 0.0 | −0.6 ± 0.0 | −0.1 ± 0.0 | |
| l-Citrulline (5 mM) | |||||||
| − | 0.1 ± 0.0 | 0.95 ± 0.1 | 0.0 ± 0.0 | ||||
| + | 11.3 ± 1.2 | 0.83 ± 0.1 | 0.0 ± 0.0 | ||||
Activity in Miller units in the absence (−) and 40 minutes after the addition (+) of inducer.
FIG. 2.
Quantitative Western analysis of Stp1-ZZ processing in rts1Δ, ssy5Δ, and ptr3Δ single mutants and in ssy5Δ rts1Δ and ptr3Δ rts1Δ double mutants, in the absence (−) and 20 min after the addition (+) of 100 μM l-leucine. Upper band: unprocessed Stp1-ZZ; lower band: processed Stp1-ZZ.
DISCUSSION
In this work we have attempted to identify potential negative regulators of the amino acid-sensing pathway. For this purpose we have designed a screen allowing selection of mutants with constitutive activity of the pathway. The screen described in this report was performed in the absence of inducer in cells lacking GAP1, and we investigated transcription levels of a lacZ reporter gene placed under the control of the amino acid-inducible promoters of the AGP1 and BAP2 genes, which are known targets of the SPS-mediated pathway. The genes that were disrupted by transposon insertion encode proteins that could be involved at any level of the signaling.
The isolated mutants include some in which the transposon insertion disrupted the RTS1 gene. This gene encodes a regulatory subunit of PP2A, which is a major serine/threonine phosphatase involved in several nutrient-induced signaling pathways, in cell growth control and cell division control (9, 35, 45, 56). Protein phosphatase 2A exists in several isoforms and is mostly present in cells as a heterotrimeric complex, consisting of a catalytic (C) subunit, encoded by PPH21, PPH22, or PPH3 (45, 49), a scaffolding subunit (A), encoded by TPD3 (55), and a regulatory subunit (B or B′), encoded by CDC55 (22) and RTS1 (47, 48), respectively. The B and B′ subunits are believed to regulate the activity of PP2A by determining its cellular location and modifying the substrate specificity of the C subunit (31, 54). CDC55 is required for correct cell cycle checkpoint control (35), and cdc55Δ cells display a cold-sensitive phenotype but no phenotype at elevated temperatures (22). RTS1 was found in two independent genetic screens, as a multicopy suppressor of hsp60(Ts) mutant alleles (48), and later as a Rox Three Suppressor (12); rts1Δ cells are thermosensitive and exhibit a typical cdc mutant phenotype (47).
We identified RTS1 as a negative component of the SPS-mediated amino acid-sensing pathway. Deletion of RTS1 results in constitutive transcription of AGP1 and BAP2. Moreover, ssy1, ptr3, ssy5, and grr1 were found to be epistatic over rts1. These results indicate that the PP2A is involved in the SPS-mediated pathway and suggest that a dephosphorylation step is required to down-regulate signaling in the absence of extracellular amino acids. Rts1p associated with Tpd3p and the C subunit might thus dephosphorylate one of the SPS proteins. Alternatively, it could dephosphorylate the Stp transcription factors, resulting in a conformational change that perhaps limits accessibility of the cleavage site to the protease, thereby impairing Stp1p and Stp2p proteolysis and activation in the absence of amino acids.
While PP2A appears to down-regulate amino acid signaling, a corresponding kinase should be involved in the activation of the pathway. Casein kinase I is a candidate for this activity, since the amino acid-sensing pathway is inactive in temperature-sensitive mutants affected in the casein kinase I genes YCK1 and YCK2, and these strains exhibit loss of Stp1p processing (1). Casein kinase I and PP2A are known to act on the same substrate in Xenopus embryos (17) and we propose that they do so in yeast as well. This hypothesis is substantiated by the finding that the yeast Cdc55p subunit forms a complex with casein kinase I (23). Moreover, casein kinase I has also been reported to be involved in the glucose-sensing pathway (50), which mediates transcriptional induction of hexose transporter (HXT) genes in response to extracellular glucose and which shares many similarities with the amino acid-sensing pathway. Casein kinase I is indeed required for phosphorylation of Mth1, which is then targeted for degradation by SCFGrr1 (36, 50). Thus, glucose induction of the HXT1 promoter is deficient in a yck1Δ yck2(Ts) mutant; in addition, this mutant lacks glucose-induced degradation of Mth1p and Std1p (36).
Mth1p and Std1p interact with the transcriptional repressor Rgt1p in the absence of glucose: the resulting complex binds promoter DNA, thereby repressing transcription from the HXT genes (33, 52). Likewise, Stp1p is phosphorylated in a casein kinase I-dependent way prior to its endoproteolytic activation (1). SCFGrr1 recognizes phosphorylated substrates; this is the case for Mth1p (50) and for the G1 cyclin Cln2p (25). Although Stp1p is phosphorylated by casein kinase I (1), there is no reason to invoke it as a target for SCFGrr1. A possible target for SCFGrr1 is Ptr3p, which in fact has been found to be subject to posttranslational modification, exhibiting a slower-migrating band (13) that might be due to ubiquitination. Alternatively, the target for SCFGrr1 could be a protein not yet known to be part of the pathway. Further work is needed to propose which protein is a target for PP2A associated with Rts1p. There may, however, be cross talk between the amino acid-sensing and the target of rapamycin (TOR) pathways, since N. Eckert-Boulet (unpublished data) has observed that treatment of rts1Δ cells with rapamycin enhanced the constitutive signal, also at the level of Stp1-ZZ processing.
Interestingly, it has recently been found that PP2A is involved in regulating the induction of HXT1 by glucose and that Cdc55p is the regulatory subunit involved. The 14-3-3 proteins Bmh1p and Bmh2p also appear to be involved (51). These observations add to existing similarities between the glucose induction pathway and the amino acid induction pathway. It will be interesting to see whether Bmh1p and Bmh2p also are involved in the SPS-mediated pathway and whether Rts1p interacts with casein kinase I like Cdc55p does.
In summary, we have shown that Rts1p, one of the two known regulatory subunits of PP2A, is a down-regulator of the SPS-mediated amino acid-sensing pathway. This finding illustrates a new role in nutrient-induced signaling for PP2A and further highlights the reuse of components in different signaling pathways.
Acknowledgments
We acknowledge Helge A. Andersen for very fruitful discussions.
Part of this work was financed by the Danish BioInstrument Centre (DABIC). We are grateful for a fellowship from the Knut and Alice Wallenberg Foundation to Katrin Larsson.
REFERENCES
- 1.Abdel-Sater, F., M. El Bakkoury, A. Urrestarazu, S. Vissers, and B. André. 2004. Amino acid signaling in yeast: casein kinase I and the Ssy5 endoprotease are key determinants of endoproteolytic activation of the membrane-bound Stp1 transcription factor. Mol. Cell. Biol. 24:9771-9785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andréasson, C. 2004. Ligand-activated proteolysis in nutrient signaling. Ph.D. thesis. Ludwig Institute for Cancer Research, Stockholm, Sweden.
- 3.Andréasson, C., and P. O. Ljungdahl. 2002. Receptor-mediated endoproteolytic activation of two transcription factors in yeast. Genes Dev. 16:3158-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes, D., W. Lai, M. Breslav, F. Naider, and J. M. Becker. 1998. PTR3, a novel gene mediating amino acid-inducible regulation of peptide transport in Saccharomyces cerevisiae. Mol. Microbiol. 29:297-310. [DOI] [PubMed] [Google Scholar]
- 5.Bernard, F., and B. André. 2001. Ubiquitin and the SCFGrr1 ubiquitin ligase complex are involved in the signalling pathway activated by external amino acids in Saccharomyces cerevisiae. FEBS Lett. 496:81-85. [DOI] [PubMed] [Google Scholar]
- 6.de Boer, M., J.-P. Bebelman, P. M. Gonçalves, J. Maat, H. Van Heerikhuizen, and R. J. Planta. 1998. Regulation of expression of the amino acid transporter gene BAP3 in Saccharomyces cerevisiae. Mol. Microbiol. 30:603-613. [DOI] [PubMed] [Google Scholar]
- 7.Didion, T., M. Grauslund, M. C. Kielland-Brandt, and H. A. Andersen. 1996. Amino acids induce expression of BAP2, a branched-chain amino acid permease gene in Saccharomyces cerevisiae. J. Bacteriol. 178:2025-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Didion, T., B. Regenberg, M. U. Jørgensen, M. C. Kielland-Brandt, and H. A. Andersen. 1998. The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol. Microbiol. 27:643-650. [DOI] [PubMed] [Google Scholar]
- 9.Düvel, K., and J. R. Broach. 2004. The role of phosphatases in TOR signaling in yeast. Curr. Top. Microbiol. Immunol. 279:19-38. [DOI] [PubMed] [Google Scholar]
- 10.Eckert-Boulet, N., P. S. Nielsen, C. Friis, M. M. dos Santos, J. Nielsen, M. C. Kielland-Brandt, and B. Regenberg. 2004. Transcriptional profiling of extracellular amino acid sensing in Saccharomyces cerevisiae and the role of Stp1p and Stp2p. Yeast 21:635-648. [DOI] [PubMed] [Google Scholar]
- 11.Eckert-Boulet, N., B. Regenberg, and J. Nielsen. 2005. Grr1p is required for transcriptional induction of amino acid permease genes and proper transcriptional regulation of genes in carbon metabolism of Saccharomyces cerevisiae. Curr. Genet. 47:139-149. [DOI] [PubMed] [Google Scholar]
- 12.Evangelista, Jr., C. C., A. M. Rodriguez Torres, M. P. Limbach, and R. S. Zitomer. 1996. Rox3 and Rts1 function in the global stress response pathway in baker's yeast. Genetics 142:1083-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsberg, H., and P. O. Ljungdahl. 2001. Genetic and biochemical analysis of the yeast plasma membrane Ssy1p-Ptr3p-Ssy5p sensor of extracellular amino acids. Mol. Cell. Biol. 21:814-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forsberg, H., and P. O. Ljungdahl. 2001. Sensors of extracellular nutrients in Saccharomyces cerevisiae. Curr. Genet. 40:91-109. [DOI] [PubMed] [Google Scholar]
- 15.Forsberg, H., M. Hammar, C. Andréasson, A. Molinér, and P. O. Ljungdahl. 2001. Suppressors of ssy1 and ptr3 null mutations define novel amino acid sensor-independent genes in Saccharomyces cerevisiae. Genetics 158:973-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaber, R. F., K. Ottow, H. A. Andersen, and M. C. Kielland-Brandt. 2003. Constitutive and hyperresponsive signaling by mutant forms of Saccharomyces cerevisiae amino acid sensor Ssy1. Eukaryot. Cell. 2:922-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, Z.-H., J. M. Seeling, V. Hill, A. Yochum, and D. M. Virshup. 2002. Casein kinase I phosphorylates and destabilizes the β-catenin degradation complex. Proc. Natl. Acad. USA 99:1182-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Georgakopoulos, T., G. Koutroubas, I. Vakonakis, M. Tzermia, V. Prokova, A. Voutsina, and D. Alexandraki. 2001. Functional analysis of the Saccharomyces cerevisiae YFR021w/YGR223c/YPL100w ORF family suggests relations to mitochondrial/peroxisomal functions and amino acid signalling pathways. Yeast 18:1155-1171. [DOI] [PubMed] [Google Scholar]
- 19.Grauslund, M., T. Didion, M. C. Kielland-Brandt, and H. A. Andersen. 1995. BAP2, a gene encoding a permease for branched-chain amino acids in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1269:275-280. [DOI] [PubMed] [Google Scholar]
- 20.Grenson, M., C. Hou, and M. Crabeel. 1970. Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. IV. Evidence for a general amino acid permease. J. Bacteriol. 103:770-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Güldener, U., S. Heck, T. Fielder, J. Beinhauer, J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 13:2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Healy, A. M., S. Zolnierowicz, A. E. Stapleton, M. Goebl, A. A. DePaoli-Roach, and J. R. Pringle. 1991. CDC55, a Saccharomyces cerevisiae gene involved in cellular morphogenesis: identification, characterization, and homology to the B subunit of mammalian type 2A protein phosphatase. Mol. Cell. Biol. 11:5767-5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S.-L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sørensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. V. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 24.Horák, J. 1997. Yeast nutrient transporters. Biochim. Biophys. Acta 1331:41-79. [DOI] [PubMed] [Google Scholar]
- 25.Hsiung, Y. G., H.-C. Chang, J.-L. Pellequer, R. La Valle, S. Lanker, and C. Wittenberg. 2001. F-box protein Grr1 interacts with phosphorylated targets via the cationic surface of its leucine-rich repeat. Mol. Cell. Biol. 21:2506-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iraqui, I., S. Vissers, F. Bernard, J.-O. De Craene, E. Boles, A. Urrestarazu, and B. André. 1999. Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol. Cell. Biol. 19:989-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Island, M. D., J. R. Perry, F. Naider, and J. M. Becker. 1991. Isolation and characterization of S. cerevisiae mutants deficient in amino acid-inducible peptide transport. Curr. Genet. 20:457-463. [DOI] [PubMed] [Google Scholar]
- 28.Jack, D. L., I. T. Paulsen, and M. H. Saier. 2000. The amino acid/polyamine/ organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology 146:1797-1814. [DOI] [PubMed] [Google Scholar]
- 29.Jørgensen, M. U., C. Gjermansen, H. A. Andersen, and M. C. Kielland-Brandt. 1997. STP1, a gene involved in pre-tRNA processing in yeast, is important for amino-acid uptake and transcription of the permease gene BAP2. Curr. Genet. 31:241-247. [DOI] [PubMed] [Google Scholar]
- 30.Jørgensen, M. U., M. B. Bruun, T. Didion, and M. C. Kielland-Brandt. 1998. Mutations in five loci affecting GAP1-independent uptake of neutral amino acids in yeast. Yeast 14:103-114. [DOI] [PubMed] [Google Scholar]
- 31.Kamibayashi, C., R. Estes, R. L. Lickteig, S. I. Yang, C. Craft, and M. C. Mumby. 1994. Comparison of heterotrimeric protein phosphatase 2A containing different B subunits. J. Biol. Chem. 269:20139-20148. [PubMed] [Google Scholar]
- 32.Klasson, H., G. R. Fink, and P. O. Ljungdahl. 1999. Ssy1p and Ptr3p are plasma membrane components of a yeast system that senses extracellular amino acids. Mol. Cell. Biol. 19:5405-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lakshmanan, J., A. L. Mosley, and S. Özcan. 2003. Repression of transcription by Rgt1 in the absence of glucose requires Std1 and Mth1. Curr. Genet. 44:19-25. [DOI] [PubMed] [Google Scholar]
- 34.Larsson, K., H. A. Andersen, T. Didion, L. Düring-Olsen, M. Jørgensen, P. L. Madsen, P. S. Nielsen, B. Regenberg, and M. C. Kielland-Brandt. 1999.Amino acid sensing and uptake, p. 51-57. E.B.C. Symposium: Yeast physiology, a new era of opportunity. Monograph 28. Fachverlag Hans Carl, Nürnberg, Germany.
- 35.Minshull, J., A. Straight, A. D. Rudner, A. F. Dernburg, A. Belmont, and A. W. Murray. 1996. Protein phosphatase 2A regulates MPF activity and sister chromatid cohesion in budding yeast. Curr. Biol. 6:1609-1620. [DOI] [PubMed] [Google Scholar]
- 36.Moriya, H., and M. Johnston. 2004. Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc. Natl. Acad. USA 101:1572-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nielsen, P. S., B. van den Hazel, T. Didion, M. de Boer, M. Jørgensen, R. J. Planta, M. C. Kielland-Brandt, and H. A. Andersen. 2001. Transcriptional regulation of the Saccharomyces cerevisiae amino acid permease gene BAP2. Mol. Gen. Genet. 264:613-622. [DOI] [PubMed] [Google Scholar]
- 38.Patton, E. E., A. R. Willems, and M. Tyers. 1998. Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet. 14:236-243. [DOI] [PubMed] [Google Scholar]
- 39.Perry, J. R., M. A. Basrai, H.-Y. Steiner, F. Naider, and J. M. Becker. 1994. Isolation and characterization of a Saccharomyces cerevisiae peptide transport gene. Mol. Cell. Biol. 14:104-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poulsen, P., B. Wu, R. F. Gaber, K. Ottow, H. A. Andersen, and M. C. Kielland-Brandt. 2005. Amino acid sensing by Ssy1. Biochem. Soc. Trans. 33:261-264. [DOI] [PubMed] [Google Scholar]
- 41.Poulsen, P., B. Wu, R. F. Gaber, and M. C. Kielland-Brandt. 2005. Constitutive signal transduction by mutant Ssy5p and Ptr3p components of the SPS amino acid sensor system in Saccharomyces cerevisiae. Eukaryot. Cell 4:1116-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Regenberg, B., L. Düring-Olsen, M. C. Kielland-Brandt, and S. Holmberg. 1999. Substrate specificity and gene expression of the amino-acid permeases in Saccharomyces cerevisiae. Curr. Genet. 36:317-328. [DOI] [PubMed] [Google Scholar]
- 43.Regenberg, B., and J. Hansen. 2000. GAP1, a novel selection and counter-selection marker for multiple gene disruptions in Saccharomyces cerevisiae. Yeast 16:1111-1119. [DOI] [PubMed] [Google Scholar]
- 44.Rolland, F., J. Winderickx, and J. M. Thevelein. 2001. Glucose-sensing mechanisms in eukaryotic cells. Trends Biochem. Sci. 26:310-317. [DOI] [PubMed] [Google Scholar]
- 45.Ronne, H., M. Carlberg, G.-Z. Hu, and J. O. Nehlin. 1991. Protein phosphatase 2A in Saccharomyces cerevisiae: effects on cell growth and bud morphogenesis. Mol. Cell. Biol. 11:4876-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ross-Macdonald, P., A. Sheehan, G. S. Roeder, and M. Snyder. 1997. A multipurpose transposon system for analyzing protein production, localization, and function in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:190-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shu, Y., H. Yang, E. Hallberg, and R. Hallberg. 1997. Molecular genetic analysis of Rts1p, a B' regulatory subunit of Saccharomyces cerevisiae protein phosphatase 2A. Mol. Cell. Biol. 17:3242-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shu, Y., and R. L. Hallberg. 1995. SCS1, a multicopy suppressor of hsp60-ts mutant alleles, does not encode a mitochondrially targeted protein. Mol. Cell. Biol. 15:5618-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sneddon, A. A., P. T. Cohen, and M. J. Stark. 1990. Saccharomyces cerevisiae protein phosphatase 2A performs an essential cellular function and is encoded by two genes. EMBO J. 9:4339-4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spielewoy, N., K. Flick, T. I. Kalashnikova, J. R. Walker, and C. Wittenberg. 2004. Regulation and recognition of SCFGrr1 targets in the glucose and amino acid signaling pathways. Mol. Cell. Biol. 24:8994-9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tomás-Cobos, L., R. Viana, and P. Sanz. 2005. TOR kinase pathway and 14-3-3 proteins regulate glucose-induced expression of HXT1, a yeast low-affinity glucose transporter. Yeast 22:471-479. [DOI] [PubMed] [Google Scholar]
- 52.Tomás-Cobos, L., and P. Sanz. 2002. Active Snf1 protein kinase inhibits expression of the Saccharomyces cerevisiae HXT1 glucose transporter gene. Biochem. J. 368:657-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tullin, S., C. Gjermansen, and M. C. Kielland-Brandt. 1991. A high-affinity uptake system for branched-chain amino acids in Saccharomyces cerevisiae. Yeast 7:933-941. [DOI] [PubMed] [Google Scholar]
- 54.Turowski, P., A. Fernandez, B. Favre, N. J. C. Lamb, and B. A. Hemmings. 1995. Differential methylation and altered conformation of cytoplasmic and nuclear forms of protein phosphatase 2A during cell cycle progression. J. Cell Biol. 129:397-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Zyl, W. H., N. Wills, and J. R. Broach. 1989. A general screen for mutants of Saccharomyces cerevisiae deficient in tRNA biosynthesis. Genetics 123:55-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zabrocki, P., C. Van Hoof, J. Goris, J. M. Thevelein, J. Winderickx, and S. Wera. 2002. Protein phosphatase 2A on track for nutrient-induced signalling in yeast. Mol. Microbiol. 43:835-842. [DOI] [PubMed] [Google Scholar]