Abstract
In pyrophosphate-dependent glycolysis, the ATP/ADP-dependent enzymes phosphofructokinase (PFK) and pyruvate kinase are replaced by the pyrophosphate-dependent PFK and pyruvate phosphate dikinase (PPDK), respectively. This variant of glycolysis is widespread among bacteria, but it also occurs in a few parasitic anaerobic eukaryotes such as Giardia and Entamoeba spp. We sequenced two genes for PPDK from the amitochondriate oxymonad Streblomastix strix and found evidence for PPDK in Trichomonas vaginalis and other parabasalia, where this enzyme was thought to be absent. The Streblomastix and Giardia genes may be related to one another, but those of Entamoeba and perhaps Trichomonas are distinct and more closely related to bacterial homologues. These findings suggest that pyrophosphate-dependent glycolysis is more widespread in eukaryotes than previously thought, enzymes from the pathway coexists with ATP-dependent more often than previously thought and may be spread by lateral transfer of genes for pyrophosphate-dependent enzymes from bacteria.
Adaptation to anaerobic metabolism is a complex process involving changes to many proteins and pathways of critical function to the cell, but it is found in several distantly related eukaryotic lineages (25, 26), raising questions of how it evolved. Some of these anaerobic protists were previously thought to be ancient lineages that retained anaerobic metabolism as a primitive, premitochondrial trait, but extant eukaryotic anaerobes are now nearly all known to have evolved from mitochondrion-bearing ancestors (8, 10, 15), so the acquisition of anaerobiosis-related traits is most likely secondary.
Embden-Meyerhof-Parnas glycolysis is the starting point of the core carbon metabolism in eukaryotes. In aerobic eukaryotes, glycolysis is a minor source of energy, since ca. 95% of ATP production comes from subsequent tricarboxylic acid cycle reactions and oxidative phosphorylation. In contrast, anaerobic organisms rely almost exclusively on glycolysis and fermentation for ATP production, so optimizing the energy output from glycolysis may be subject to strong selective pressure (21, 22). One significant variation of the standard glycolytic pathway that has been described in some anaerobic protists is pyrophosphate-dependent glycolysis, which uses pyrophosphate (PPi) instead of ATP as a phosphate donor (21). This version of glycolysis has been best studied in Entamoeba histolytica (29, 30). In this parasite, two key glycolytic enzymes, phosphofructokinase (PFK) and pyruvate kinase (PK), have been replaced by the PPi-dependent versions, PPi-phosphofructokinase (PFP) and pyruvate-phosphate dikinase (PPDK), respectively (for a comparison of these reactions, see reference 21). Energy efficiency seems to be the chief reason for adopting a PPi-dependent glycolysis, since PPi-dependent glycolysis can in theory yield five ATP molecules instead of the two yielded by standard glycolysis. This increase of 2.5-fold can be crucial under severe energy-limiting conditions (21, 22).
PPDK catalyzes the interconversion between phosphoenolpyruvate (PEP) and pyruvate. Unlike the reaction catalyzed by PK, this reaction has a small ΔG and is reversible, so it can proceed either in the catabolic or the gluconeogenic directions (34). This highly versatile enzyme has been appointed to many functions in different eukaryotes, although its coexistence in some cells with the ATP-dependent enzyme PK makes it difficult to address its role (21, 26). Involvement of PPDK in glycolysis has been shown or suggested in Giardia (28), Entamoeba (30), and Phytophtora (18). PPKD also works in the opposite direction in chloroplasts of C4 plants, where it catalyzes the regeneration of PEP in the stroma of leaf mesophyll cells (6). A cytoplasmic isoenzyme of unclear function also exists in C3 and C4 plants, and it has been suggested that it may have a glycolytic role, especially in oxygen-starved tissues (22). Trypanosomatids target PPDK to glycosomes where it fulfills a thermodynamic role of pyrophosphate recycling in place of pyrophosphatase (1).
At present, studies on the molecular evolution of PPi-linked glycolysis have focused almost exclusively on PFK and PFP (3, 27). These proteins are homologous, and the gene family has had a complex evolutionary history including multiple events of duplication, loss, and lateral gene transfer (3, 27). In contrast, the phylogeny of PPDK has not been extensively explored. Previous studies using limited taxon sampling addressed the relationships of certain eukaryotic PPDKs, but whether eukaryotic enzymes originated in common or independently has not been clear (it has been shown that a plant cluster is separated from other eukaryotes [17, 28]). In addition, it is not known whether PPDK and PFP are always found together, or if some organisms use one PPi-dependent enzyme in glycolysis but not the other. In Giardia and Entamoeba both PPi-dependent enzymes are used, but in Trichomonas vaginalis, PPi-specific PFK and ATP-specific PK have been found, but no PPDK activity has been detected (24). Interestingly, neither PK nor PPDK activities have been detected in the related Tritrichomonas fetus (14, 21).
One group of amitochondriates of which we know relatively little are the oxymonads. These anaerobes are found almost exclusively in association with animals and are generally not available in cultivation. Oxymonad molecular and biochemical data are scarce because they are typically part of a complex community of eukaryotes from which it is difficult to determine which genes or proteins come from which species. We have used the recent documentation of a noncanonical genetic code in one oxymonad, Streblomastix strix, to circumvent this problem (16). S. strix is found exclusively in the hindgut of the Pacific damp-wood termite Zootermopsis angusticollis, an environment it shares with at least six species of parabasalia (37). From this environment we isolated PPDK genes and used the noncanonical genetic code to identify S. strix homologues. We also identified other protist PPDK genes and, comparing them to T. vaginalis genomic data identified the first parabasalian PPDK genes, overturning the notion that T. vaginalis lacks this enzyme. A comprehensive phylogenetic analysis of PPDK shows several independent clades of eukaryotes, overall suggesting at least some lateral gene transfer may have occurred.
MATERIALS AND METHODS
cDNA library construction and EST sequencing.
Termites were collected from a rotten log in Point Grey, Vancouver, Canada. The whole hindgut content of about 60 individuals of Z. angusticollis from a single colony was collected and total RNA was extracted using standard TRIZOL procedure (Invitrogen). A cDNA library was constructed (Amplicon Express) and 5,337 clones were sequenced from the 5′ end. Expressed sequence tags (ESTs) were trimmed for vector and quality, and assembled into 2,595 clusters by PEPdb (http://amoebidia.bcm.umontreal.ca/public/pepdb/agrm.php).
Characterization of S. strix PPDK.
S. strix sequences were recovered from EST sequences by scanning BLASTX outputs to identify putative protein coding sequences containing in-frame TAA and TAG stop codons. Two truncated ESTs with high identity scores to PPDK were recovered according to this criterion. The C-terminal portion of both was acquired by 3′RACE (Ambion) using specific primers and total hindgut RNA. Most of the 5′ portion of the gene was obtained by reverse transcription-PCR (RT-PCR) amplification using a degenerate primer designed from a conserved block located at about 90 amino acids from the start of the G. intestinalis PPDK and gene-specific primers designed from the EST sequences. In all cases, PCR products were cloned by using TOPO cloning system (Invitrogen). Sequences were assembled with Sequencher, and the sequence was confirmed to come from a single gene by amplifying from DNA using primers at the ends of the assembled contig. Parabasalian PPDK and PFP sequences from the EST data were identified by comparing them to the T. vaginalis genome project. T. vaginalis preliminary sequence data was obtained from The Institute for Genomic Research (TIGR; http://www.tigr.org. database).
Sequence and phylogenetic analyses.
Deduced amino acid sequences of new and publicly available PPDK sequences were aligned by using CLUSTALX and edited manually using McClade. Several ongoing eukaryotic genome and EST projects were also searched, from which the T. vaginalis (TIGR) and T. pseudonana (genome.jgi-psf.org/thaps1/thaps1.home.html) were retrieved. Sequences were aligned by using CLUSTALX and adjusted with McClade. Gaps and ambiguously aligned residues were excluded. This resulted in a matrix of 54 sequences with 713 positions. Maximum-likelihood trees were built by using PhyML (http://atgc.lirmm.fr/phyml/) with the WAG model of amino acid substitutions and eight categories of rates, with gamma and proportion of invariant sites estimated from the data. Support of the nodes was estimated by using 100 bootstrap replicates.
RESULTS AND DISCUSSION
Identification of PPDK genes from the oxymonad S. strix and parabasalian symbionts.
From an EST sequence survey of the eukaryotic fauna from the hindgut of the termite Z. angusticollis, we identified three ESTs matching the PPDK family and other related PEP-utilizing enzymes. Two sequences contained several in-frame, TAA and TAG stop codons, identifying them as belonging to the flagellate S. strix, which has an alternate genetic code where TAA and TAG encode glutamine (Q) instead of translational stops (16). One of these sequences was extended by using a combination of RT-PCR with degenerate primers and 3′ RACE so that 88% of the gene was ultimately characterized. Attempts to recover the extreme 5′ end of the gene by 5′ RACE were unsuccessful, probably due to the highly complex nature of the sample and the large size of the gene (the G. intestinalis homologue is 2,655 bp). These procedures resulted in a nucleotide sequence encoding an open reading frame of 780 codons. The second S. strix sequence differed only slightly at the nucleotide level, indicating that at least two gene copies or alleles of this gene exist in this S. strix population and so was not characterized further. Since ppdk belongs to a gene family with at least three functionally differentiated clusters, we wanted to identify which group the S. strix sequences belong to. Phylogenetic analyses of the S. strix gene, including members of the PPDK, water dikinase, and glucan dikinase groups of the family, clearly revealed that the S. strix sequences belong to the PPDK family (not shown).
The third EST we identified was not highly similar to the S. strix genes and did not contain in-frame stop codons. The other major eukaryotic constituents of this environment are several species of parabasalia, which are thought not to contain PPDK (24). To determine whether this PPDK could come from a parabasalid, we performed extensive BLAST searches against NCBI databases and found 13 ESTs from T. vaginalis with high similarity to PPDK. Twelve partially overlapping ESTs assembled to form an open reading frame corresponding to the last 531 amino acids of the PPDK. The remaining EST encompassed a nonoverlapping stretch from positions 25 to 286 (according to C. symbiosum PPDK numbering). Searching the T. vaginalis genome project at TIGR, we found gene predictions that perfectly matched these ESTs. Interestingly, the T. vaginalis project has at least two PPDK sequences, a finding consistent with previous findings that the T. vaginalis genome contains multiple copies of several genes for metabolic enzymes. Comparing the T. vaginalis PPDKs to our EST data showed that the two were very similar, so we infer that this sequence belongs to one of the six species of parabasalia present in Z. angusticollis (37), and tentatively named it “parabasalian endosymbiont.” The sequence of this EST was extended by RT-PCR to 1,680 bp encoding 653 amino acids.
Conservation of key structural features in S. strix and T. vaginalis PPDKs.
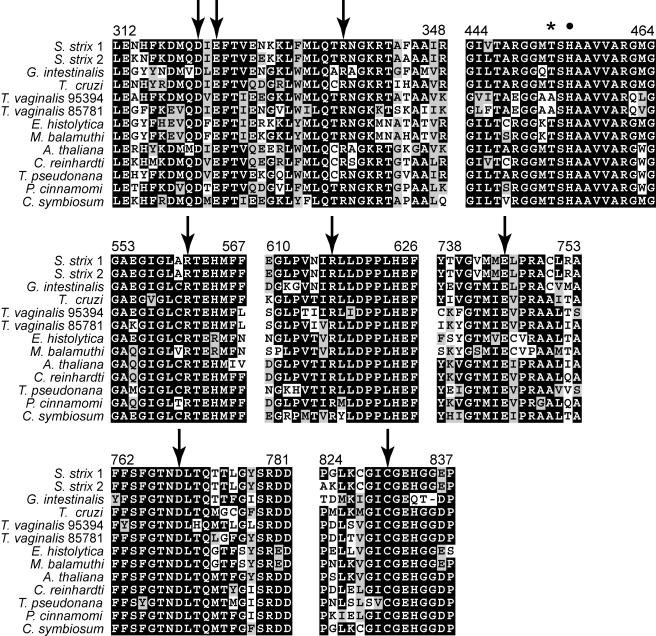
Certain enzymes can change their substrate specificity with a few substitutions at catalytically active sites. A clear example is PFK, which has been shown to have switched from ATP to PPi specificity several times independently (3, 23, 27). This transformation requires only a few substitutions that control the entry of ATP into the active site, so that the position of a sequence in a phylogeny does not necessarily predict its function for these enzymes (3). Although functional assays are needed to ultimately demonstrate the actual substrate specificity, careful mapping of the critical positions are usually helpful to predict the substrate specificity, provided that sufficient information on the structure and physicochemical properties of the enzyme exists. Although no cross-substrate specificity has been reported between the ATP and PPi pyruvate kinases, we examined the S. strix and T. vaginalis sequences and looked for conservation of the residues involved in substrate interaction and enzymatic function. We aligned these with PPDKs from the protists Giardia, Entamoeba, Trypanosoma, along with maize and C. symbiosum. The latter has been subject to detailed mutagenesis and structural analysis, from which the positions essential for catalytic function have been identified (4, 19, 20, 32, 33, 35, 36, 38). All but two positions identified as catalytically important in C. symbiosum are included in the known S. strix sequences, and every one is conserved in S. strix and T. vaginalis sequences, including the reactive center His455 (Fig. 1). One notable difference found between the parabasalian and other PPDK enzymes is two residues upstream of the catalytic His458, where a threonine is conserved in all but the parabasalia (Fig. 1 and Fig. S1 in the supplemental material). In C4 plants, this threonine is directly involved in a particular form of light-dependent regulation by a bifunctional protein kinase/phosphatase (5, 6). Briefly, nocturnal inactivation of the enzyme is achieved by ADP-dependent phosphorylation of Thr453 by the PPDK regulatory protein (RP), whereas the opposite (dephosphorylation) is catalyzed by RP in light conditions. Although this residue is not directly involved in catalysis, and site-directed mutagenesis has shown that some substitutions of Thr453 have little or no negative effect on activity, it is an important potential control point (5, 7). The T. vaginalis and the “parabasalian symbiont” sequences have an alanine at this position (Fig. 1 and Fig. S1 in the supplemental material), and experiments of mutagenesis in maize indicate that activity is lost when bulky or strongly charged residues such as phenylalanine, tyrosine, and the acidic amino acids replace threonine, but not when serine and the aliphatic valine are assayed in place of Thr453 (5, 7). Although functional characterization is ultimately needed, the evidence suggests that the Thr-Ala substitution seen in T. vaginalis would likely not impede PPDK function.
FIG. 1.
Sequence alignment of PPDKs from several eukaryotes and C. symbiosum showing conservation of important sites for catalytic function. Numbers follow the residue position in C. symbiosum. Arrows indicate positions that have shown to be involved in nucleotide and substrate binding. The dot shows the active site histidine, and the asterisk indicates the site involved in regulation of PPDK activity in plants (see the text).
Phylogeny and evolution of PPDK.
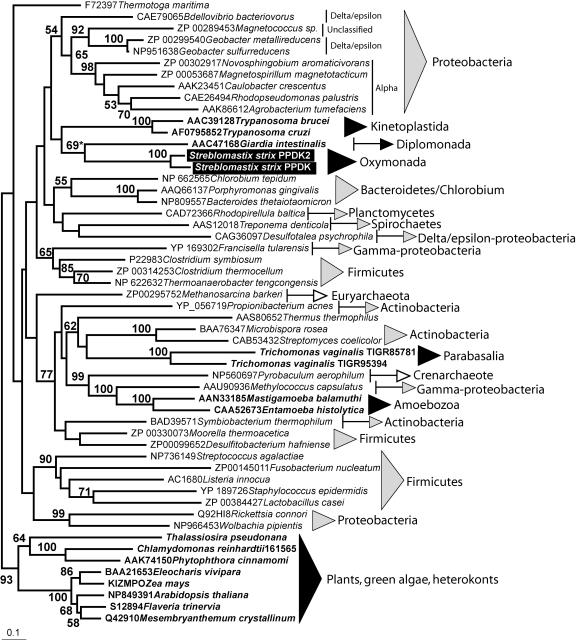
Although they share many metabolic similarities, eukaryotic microbes with PPi-linked glycolysis are not all closely related to one another (26). This was noted some time ago (25, 26), and it was debated whether their metabolism was retained from an ancient ancestor or was a derived adaptations to anaerobic environments. Current data seem to favor the latter interpretation (3). PFK has been extensively sampled (3, 23, 27) and is now known to have a very complex history due to a combination of early duplications and multiple independent losses, as well as some lateral gene transfer events. The phylogeny of PPDK has not been explored in great detail, so we analyzed PPDK sequences from the three domains of life to determine whether the eukaryotic homologues share a common ancestor. A preliminary analysis, including about 100 sequences of purported PPDKs and another member of the extended gene family (water dikinase), confirmed the monophyly of PPDK enzymes (not shown). PPDK genes were subsequently analyzed alone to allow more positions to be included. The overall level of resolution for this phylogeny is not very high, but several nodes are well supported. The maximum-likelihood tree topology (Fig. 2) is generally not consistent with the known relationships within and among different groups of bacteria, archaea, and eukaryotes. For the most part this is probably due to lack of resolution in the phylogeny, but there may also be a case for some ancient duplication events and the incongruent distribution of taxa in some of the well-supported nodes suggests that at least one event of lateral gene transfer probably also occurred (Fig. 2).
FIG. 2.
Maximum-likelihood tree showing the relationships among bacterial, archaeal, and eukaryotic PPDKs. Eukaryotic sequences are in boldface, and the sequences from S. strix are marked with black rectangles. Taxonomic groups are indicated with black, gray, and white triangles for eukaryotes, bacteria, and archaea, respectively. Numbers represent bootstrap values (higher than 50%). The bootstrap support for the node uniting S. strix and G. intestinalis was 69% in the analysis shown but higher (76%) when the unpublished T. pyriformis sequence was included (see text).
PPDK is broadly represented in eubacteria, although its representation varies from group to group. For example, PPDK is scarce among proteobacteria in general but ubiquitous in the alpha subdivision. In archaea, PPDK is restricted to a few groups from both crenarchaeotes and euryarchaeotes, even though more than 40 complete genomes are known. This, and the fact that the archaeal sequences do not cluster together are suggestive of lateral acquisition from eubacteria (Fig. 2). Some bacterium-only groupings in the PPDK tree contain unrelated species, but most of these nodes have low bootstrap support (<50), making it impossible to infer anything about their evolution, although it is noteworthy that some groups such as firmicutes, actinobacteria, and proteobacteria show discontinuous distribution (Fig. 2).
Eukaryotic PPDK sequences do not form a monophyletic group. The largest eukaryotic clade comprises plant and green algae, as well as Phytophthora and Thalassiosira sequences. This grouping is well supported by all reconstruction methods used. Phytophthora and Thalassiosira are heterokonts, which diverged from the green lineage deep in eukaryotic history. Two explanations for this observation are possible: plant and heterokont PPDK derive from an ancestral gene in early eukaryotes that was lost elsewhere, or they were independently acquired from related sources. It is tempting to postulate some link to plastid evolution, given its involvement in photosynthesis in some plants and that the diatom is also photosynthetic and plastid-related genes have been found in Phytophthora (2). No cyanobacteria has yet been shown to possess a related PPDK gene, however, and none of these genes encode plastid-targeted proteins, so this link is perhaps coincidence. The sequences from the trypanosomatid parasites T. cruzi and T. brucei branch together, as expected, but show no well-supported association to any other organism in the present sample. The trypanosomatid PPDKs are glycosomal and used to recycle pyrophosphate accumulated from various biosynthetic reactions (1). A controversial hypothesis that trypanosomatids evolved from a plastid-harboring ancestor (13) suggested that PPDK was acquired from the phototrophic endosymbionts (12). However, the trypanosomatid PPDK shows no association with the plant-algal cluster as would have been expected, much like other genes proposed to be endosymbiont-derived (9).
Among amitochondriate eukaryotes, the only relationship that is consistently found by different reconstruction methods is the branch uniting Giardia and Streblomastix. This node is moderately supported by bootstrap (69%); however, adding the PPDK from a sister-lineage to oxymonads, Trimastix pyriformis (A. J. Roger, unpublished data), this value increases to 76% (not shown). Common ancestry between the Giardia and Streblomastix sequences is consistent with the hypothetical eukaryotic supergroup Excavata (11, 31). The PPDK from T. vaginalis, which is also an excavate, does not branch with these sequences but rather with actinobacteria and Thermus spp. with moderate bootstrap support. Finally, PPDK from the amitochondriate amoebae E. hystolytica and M. balamuthi branch within a strongly supported clade containing the gamma-proteobacterium Methylococcus and the crenarchaeon Pyrobaculum. Overall, there is no support for a single origin of amitochondriate PPDK, although the G. intestinalis and S. strix genes likely branch together, and we cannot rule out a common origin with the T. vaginalis gene; the PPDK genes from amoebae have an origin apparently independent from those of other eukaryotes.
Evolution of PPi-dependent glycolysis in amitochondriate eukaryotes.
Three of the main questions surrounding the evolution of PPi-dependent glycolysis are (i) whether the presence of PFP and PPDK in glycolysis are correlated with one another, or if only one or the other may be used; (ii) whether or how analogous PPi- and ATP-dependent enzymes performing the same reaction in one organism are functionally differentiated; and (iii) where did the PPi-dependent enzymes come from and how widespread are they?
Previously nothing was known of oxymonads, and it was thought that parabasalia used PFP but not PPDK. We have now shown that oxymonads and parabasalia both possess PPDK. It is unknown if oxymonads use PFP or PFK to phosphorylate fructose-6-phosphate, or if they also encode PK, but it is possible that their glycolysis is at least partially PPi dependent. In the case of parabasalia, PPDK was undetected in enzyme assays from T. vaginalis, and it was assumed that glycolysis was mixed, such that the phosphorylation of fructose-6-phosphate by PFP was the only PPi-dependent step. However, T. vaginalis and at least one other parabasalian do contain genes for PPDK, further supporting the correlated presence of PFP and PPDK. By analogy to G. intestinalis, E. histolytica, and now also T. vaginalis, we hypothesize that PFP will eventually be found in oxymonads as well. However, G. intestinalis, E. histolytica, and now also T. vaginalis possess both cytosolic PPDK and PK, so these two analogous enzymes clearly can also coexist in the same compartment, raising questions about whether they are functionally distinct. In plants and trypanosomatids PPDK and PK are sequestered in two different compartments, so this is not an issue. It also suggests that other amitochondriates, such as oxymonads, may be found to possess PK as well as PPDK. This growing list of eukaryotes where both kinds of enzyme are found also suggests that a reevaluation of why these enzymes exist in anaerobes and their possible contribution to energy efficiency may be necessary. Biochemical dissection of the role of ATP- versus PPi-dependent enzymes is reemerging as an urgent task for understanding the metabolism of these organisms.
As for the origin of the PPi-dependent enzymes, phylogenetic analyses show that ADP-PFK and PPi-PFP are not mutually exclusive and PFK has been shown to have switched phosphate-donor specificity several times (3, 27); thus, PFP originated several times independently. In contrast, the phylogeny of PPDK is less complex but lacks the resolution needed to conclusively determine whether there is a single origin for the eukaryotic protein. The PPDK of amitochondriates G. intestinalis, S. strix, and T. vaginalis may have originated in common, but this is not clearly supported by the phylogeny, and the support for an independent origin of at least the amoebozoan PPDK is very strong. If several independent lateral gene transfers explain the distribution of PPDK in amitochondriates, then perhaps further sampling of prokaryotes will show this more clearly. Until then, only tentative conclusions can be drawn. Overall, however, analyses of PFP and PPDK suggest that PPi-linked glycolysis is a derived adaptation to specific environments or lifestyles rather than a primitive eukaryotic trait, although the known distribution of PPi-dependent enzymes is growing.
Supplementary Material
Acknowledgments
S. strix EST sequencing was supported by the Protist EST Program of Genome Canada/Genome Atlantic. P.J.K. is a fellow of the CIAR and a New Investigator of the CIHR and MSFHR. Sequencing of T. vaginalis was accomplished with support from NIAID, NIH. Preliminary sequence data was obtained from The Institute for Genomic Research (http://www.tigr.org).
We thank Audrey de Koning for valuable help with RNA preparation, Andrew Roger for providing unpublished data from T. pyriformis, and an anonymous reviewer for helpful comments.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Acosta, H., M. Dubourdieu, W. Quinones, A. Caceres, F. Bringaud, and J. L. Concepcion. 2004. Pyruvate phosphate dikinase and pyrophosphate metabolism in the glycosome of Trypanosoma cruzi epimastigotes. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 138:347-356. [DOI] [PubMed] [Google Scholar]
- 2.Andersson, J. O., and A. J. Roger. 2002. A cyanobacterial gene in nonphotosynthetic protists: an early chloroplast acquisition in eukaryotes? Curr. Biol. 12:115-119. [DOI] [PubMed] [Google Scholar]
- 3.Bapteste, E., D. Moreira, and H. Philippe. 2003. Rampant horizontal gene transfer and phospho-donor change in the evolution of the phosphofructokinase. Gene 318:185-191. [DOI] [PubMed] [Google Scholar]
- 4.Carroll, L. J., Y. Xu, S. H. Thrall, B. M. Martin, and D. Dunaway-Mariano. 1994. Substrate binding domains in pyruvate phosphate dikinase. Biochemistry 33:1134-1142. [DOI] [PubMed] [Google Scholar]
- 5.Chastain, C. J., M. Botschner, G. E. Harrington, B. J. Thompson, S. E. Mills, G. Sarath, and R. Chollet. 2000. Further analysis of maize C(4) pyruvate, orthophosphate dikinase phosphorylation by its bifunctional regulatory protein using selective substitutions of the regulatory Thr-456 and catalytic His-458 residues. Arch. Biochem. Biophys. 375:165-170. [DOI] [PubMed] [Google Scholar]
- 6.Chastain, C. J., and R. Chollet. 2003. Regulation of pyruvate, orthophosphate dikinase by ADP-/Pi-dependent reversible phosphorylation in C3 and C4 plants. Plant Physiol. Biochem. 41:523-532. [Google Scholar]
- 7.Chastain, C. J., M. E. Lee, M. A. Moorman, P. Shameekumar, and R. Chollet. 1997. Site-directed mutagenesis of maize recombinant C4-pyruvate, orthophosphate dikinase at the phosphorylatable target threonine residue. FEBS Lett. 413:169-173. [DOI] [PubMed] [Google Scholar]
- 8.Dyall, S. D., and P. J. Johnson. 2000. Origins of hydrogenosomes and mitochondria: evolution and organelle biogenesis. Curr. Opin. Microbiol. 3:404-411. [DOI] [PubMed] [Google Scholar]
- 9.El-Sayed, N. M., P. J. Myler, G. Blandin, M. Berriman, J. Crabtree, G. Aggarwal, E. Caler, H. Renauld, E. A. Worthey, C. Hertz-Fowler, E. Ghedin, C. Peacock, D. C. Bartholomeu, B. J. Haas, A. N. Tran, J. R. Wortman, U. C. Alsmark, S. Angiuoli, A. Anupama, J. Badger, F. Bringaud, E. Cadag, J. M. Carlton, G. C. Cerqueira, T. Creasy, A. L. Delcher, A. Djikeng, T. M. Embley, C. Hauser, A. C. Ivens, S. K. Kummerfeld, J. B. Pereira-Leal, D. Nilsson, J. Peterson, S. L. Salzberg, J. Shallom, J. C. Silva, J. Sundaram, S. Westenberger, O. White, S. E. Melville, J. E. Donelson, B. Andersson, K. D. Stuart, and N. Hall. 2005. Comparative genomics of trypanosomatid parasitic protozoa. Science 309:404-409. [DOI] [PubMed] [Google Scholar]
- 10.Embley, T. M., M. van der Giezen, D. S. Horner, P. L. Dyal, S. Bell, and P. G. Foster. 2003. Hydrogenosomes, mitochondria, and early eukaryotic evolution. IUBMB Life 55:387-395. [DOI] [PubMed] [Google Scholar]
- 11.Hampl, V., D. S. Horner, P. Dyal, J. Kulda, J. Flegr, P. Foster, and T. M. Embley. 2005. Inference of the phylogenetic position of oxymonads based on 9 genes: support for Metamonada and Excavata. Mol. Biol. Evol. 12:2508-2518. [DOI] [PubMed] [Google Scholar]
- 12.Hannaert, V., F. Bringaud, F. R. Opperdoes, and P. A. Michels. 2003. Evolution of energy metabolism and its compartmentation in Kinetoplastida. Kinetoplastid Biol. Dis. 2:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hannaert, V., E. Saavedra, F. Duffieux, J. P. Szikora, D. J. Rigden, P. A. Michels, and F. R. Opperdoes. 2003. Plant-like traits associated with metabolism of Trypanosoma parasites. Proc. Natl. Acad. Sci. USA 100:1067-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hrdy, I., E. Mertens, and E. Van Schaftingen. 1993. Identification, purification and separation of different isozymes of NADP-specific malic enzyme from Tritrichomonas fetus. Mol. Biochem. Parasitol. 57:253-260. [DOI] [PubMed] [Google Scholar]
- 15.Keeling, P. J. 1998. A Kingdom's progress: Archezoa and the origin of eukaryotes. Bioessays 20:87-95. [Google Scholar]
- 16.Keeling, P. J., and B. S. Leander. 2003. Characterisation of a non-canonical genetic code in the oxymonad Streblomastix strix. J. Mol. Biol. 326:1337-1349. [DOI] [PubMed] [Google Scholar]
- 17.Maldonado, R. A., and A. H. Fairlamb. 2001. Cloning of a pyruvate phosphate dikinase from Trypanosoma cruzi. Mol. Biochem. Parasitol. 112:183-191. [DOI] [PubMed] [Google Scholar]
- 18.Marshall, J. S., A. R. Ashton, F. Govers, and A. R. Hardham. 2001. Isolation and characterization of four genes encoding pyruvate, phosphate dikinase in the oomycete plant pathogen Phytophthora cinnamomi. Curr. Genet. 40:73-81. [DOI] [PubMed] [Google Scholar]
- 19.McGuire, M., L. J. Carroll, L. Yankie, S. H. Thrall, D. Dunaway-Mariano, O. Herzberg, B. Jayaram, and B. H. Haley. 1996. Determination of the nucleotide binding site within Clostridium symbiosum pyruvate phosphate dikinase by photoaffinity labeling, site-directed mutagenesis, and structural analysis. Biochemistry 35:8544-8552. [DOI] [PubMed] [Google Scholar]
- 20.McGuire, M., K. Huang, G. Kapadia, O. Herzberg, and D. Dunaway-Mariano. 1998. Location of the phosphate binding site within Clostridium symbiosum pyruvate phosphate dikinase. Biochemistry 37:13463-13474. [DOI] [PubMed] [Google Scholar]
- 21.Mertens, E. 1993. ATP versus pyrophosphate: glycolysis revisited in parasitic protists. Parasitol. Today 9:122-126. [DOI] [PubMed] [Google Scholar]
- 22.Mertens, E. 1991. Pyrophosphate-dependent phosphofructokinase, an anaerobic glycolytic enzyme? FEBS Lett. 285:1-5. [DOI] [PubMed] [Google Scholar]
- 23.Mertens, E., U. S. Ladror, J. A. Lee, A. Miretsky, A. Morris, C. Rozario, R. G. Kemp, and M. Muller. 1998. The pyrophosphate-dependent phosphofructokinase of the protist, Trichomonas vaginalis, and the evolutionary relationships of protist phosphofructokinases. J. Mol. Evol. 47:739-750. [DOI] [PubMed] [Google Scholar]
- 24.Mertens, E., E. Van Schaftingen, and M. Muller. 1992. Pyruvate kinase from Trichomonas vaginalis, an allosteric enzyme stimulated by ribose 5-phosphate and glycerate 3-phosphate. Mol. Biochem. Parasitol. 54:13-20. [DOI] [PubMed] [Google Scholar]
- 25.Muller, M. 1988. Energy metabolism of protozoa without mitochondria. Annu. Rev. Microbiol. 42:465-488. [DOI] [PubMed] [Google Scholar]
- 26.Muller, M. 2003. Energy metabolism. I. Anaerobic protozoa, p. 125-139. In J. Marr, T. Nilsen, and R. Komuniecki (ed.), Molecular medical parasitology. Academic Press, London, England.
- 27.Muller, M., J. A. Lee, P. Gordon, T. Gaasterland, and C. W. Sensen. 2001. Presence of prokaryotic and eukaryotic species in all subgroups of the PPi-dependent group II phosphofructokinase protein family. J. Bacteriol. 183:6714-6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nevalainen, L., I. Hrdy, and M. Muller. 1996. Sequence of a Giardia lamblia gene coding for the glycolytic enzyme, pyruvate, phosphate dikinase. Mol. Biochem. Parasitol. 77:217-223. [DOI] [PubMed] [Google Scholar]
- 29.Reeves, R. E. 1984. Metabolism of Entamoeba histolytica Schaudinn, 1903. Adv. Parasitol. 23:105-142. [DOI] [PubMed] [Google Scholar]
- 30.Saavedra, E., R. Encalada, E. Pineda, R. Jasso-Chavez, and R. Moreno-Sanchez. 2005. Glycolysis in Entamoeba histolytica: biochemical characterization of recombinant glycolytic enzymes and flux control analysis. FEBS J. 272:1767-1783. [DOI] [PubMed] [Google Scholar]
- 31.Simpson, A. G. 2003. Cytoskeletal organization, phylogenetic affinities and systematics in the contentious taxon Excavata (Eukaryota). Int. J. Syst. Evol. Microbiol. 53:1759-1777. [DOI] [PubMed] [Google Scholar]
- 32.Thrall, S. H., and D. Dunaway-Mariano. 1994. Kinetic evidence for separate site catalysis by pyruvate phosphate dikinase. Biochemistry 33:1103-1107. [DOI] [PubMed] [Google Scholar]
- 33.Thrall, S. H., A. F. Mehl, L. J. Carroll, and D. Dunaway-Mariano. 1993. Characterization of the covalent enzyme intermediates formed during pyruvate phosphate dikinase catalysis. Biochemistry 32:1803-1809. [DOI] [PubMed] [Google Scholar]
- 34.Wood, H. G., E. O'Brien, W., and G. Micheales. 1977. Properties of carboxytransphosphorylase; pyruvate, phosphate dikinase; pyrophosphate-phosphofructikinase and pyrophosphate-acetate kinase and their roles in the metabolism of inorganic pyrophosphate. Adv. Enzymol. Relat. Areas Mol. Biol. 45:85-155. [DOI] [PubMed] [Google Scholar]
- 35.Xu, Y., M. McGuire, D. Dunaway-Mariano, and B. M. Martin. 1995. Separate site catalysis by pyruvate phosphate dikinase as revealed by deletion mutants. Biochemistry 34:2195-2202. [DOI] [PubMed] [Google Scholar]
- 36.Xu, Y., L. Yankie, L. Shen, Y. S. Jung, P. S. Mariano, D. Dunaway-Mariano, and B. M. Martin. 1995. Location of the catalytic site for phosphoenolpyruvate formation within the primary structure of Clostridium symbiosum pyruvate phosphate dikinase. 1. Identification of an essential cysteine by chemical modification with [1-14C]bromopyruvate and site-directed mutagenesis. Biochemistry 34:2181-2187. [DOI] [PubMed] [Google Scholar]
- 37.Yamin, M. A. 1979. Flagellates of the orders Trichomonadida Kirby, Oxymonadida Grassè, and Hypermastigida Grassi & Foà reported from lower termites (Isoptera families Mastotermitidae, Kalotermitidae, Hodotermitidae, Termopsidae, Rhinotermitidae, and Serritermitidae) and from the wood-feeding roach Cryptocercus (Dictyoptera: Cryptocercidae). Sociobiology 4:5-119. [Google Scholar]
- 38.Yankie, L., Y. Xu, and D. Dunaway-Mariano. 1995. Location of the catalytic site for phosphoenolpyruvate formation within the primary structure of Clostridium symbiosum pyruvate phosphate dikinase. 2. Site-directed mutagenesis of an essential arginine contained within an apparent P-loop. Biochemistry 34:2188-2194. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.