Abstract
SPO14, encoding the major Saccharomyces cerevisiae phospholipase D (PLD), is essential for sporulation and mediates synthesis of the new membrane that encompasses the haploid nuclei that arise through meiotic divisions. PLD catalyzes the hydrolysis of phosphatidylcholine to phosphatidic acid (PA) and choline. PA stimulates Arf-GTPase-activating proteins (Arf-GAPs), which are involved in membrane trafficking events and actin cytoskeletal function. To determine if Spo14p-generated PA mediates its biological response through Arf-GAPs, we analyzed the sporulation efficiencies of cells deleted for each of the five known and potential yeast Arf-GAPs. Only gcs1Δ mutants display a sporulation defect similar to that of spo14 mutants: cells deleted for GCS1 initiate the sporulation program but are defective in synthesis of the prospore membrane. Endosome-to-vacuole transport is also impaired in gcs1Δ cells during sporulation. Furthermore, Arf-GAP catalytic activity, but not the pleckstrin homology domain, is required for both prospore membrane formation and endosome-to-vacuole trafficking. An examination of Gcs1p-green fluorescent protein revealed that it is a soluble protein. Interestingly, cells deleted for GCS1 have reduced levels of Spo14p-generated PA. Taken together, these results indicate that GCS1 is essential for sporulation and suggest that GCS1 positively regulates SPO14.
Saccharomyces cerevisiae undergoes sporulation upon transfer to conditions where diploid cells are starved for nitrogen in the presence of a nonfermentable carbon source (35). The developmentally regulated pathway of sporulation consists of meiosis and spore morphogenesis. Meiosis involves a single round of DNA replication, followed by two nuclear divisions, taking place within a single nuclear envelope (35). The nuclei that arise from the meiotic divisions are packaged into four spores.
Spore formation requires de novo synthesis of a double-layered membrane known as the prospore membrane (7). Analyses of secretory mutants with defects in sporulation suggest that the vesicles that fuse to form this prospore membrane are derived from the Golgi (47). The prospore membrane originates on the cytoplasmic face of the four spindle pole bodies (SPBs) as an extension of the outer plaque (7). As meiosis II (MII) proceeds, each prospore membrane extends around the segregated nucleus, thus encapsulating the four haploid nuclei (42). The membrane closest to the daughter nucleus now serves as the plasma membrane of the spore. Exactly how this pathway of membrane formation is regulated and how it differs from other membrane trafficking events are not precisely known.
The closure of each prospore membrane triggers the final step of spore morphogenesis, spore wall formation. Spore wall components are laid down within the luminal space between the two membranes. Once the layers of the spore wall are deposited, the resulting spore is resistant to environmental stress (70).
The SPO14 gene encodes the major yeast phospholipase D (PLD) and is essential for sporulation (15, 25, 61, 79). Spo14p catalyzes the hydrolysis of phosphatidylcholine (PC) to generate phosphatidic acid (PA) and choline; PA is the likely mediator of Spo14p function (67). Analyses of spo14Δ diploids during sporulation revealed that they are defective in completion of the meiotic divisions and are unable to form prospore membranes (25, 61, 65). Furthermore, Spo14p relocalizes from a detergent-resistant cytoplasmic pool to prospore membranes during meiosis (65). One probable target of Spo14p-generated PA is Spo20p, a sporulation-specific t-SNARE (t-soluble NEM-sensitive factor receptor) that mediates the fusion of vesicles with the prospore membrane (45, 47). Spo20p contains an amphipathic helix that mediates membrane binding in vivo and binds acidic phospholipids in vitro (45). However, it is likely that there are additional targets of Spo14p-generated PA.
ADP-ribosylation factors (Arfs) are 21-kDa proteins of the Ras superfamily of GTP-binding proteins. Arfs have been shown to have a number of biochemical activities, including recruitment of coat proteins to membranes, activation of PLD in mammalian cells, and stimulation of phosphatidylinositol 4-phosphate [PI(4)P] kinase (44, 49, 58). These activities contribute to the maintenance of Golgi morphology and the formation of vesicles involved in both secretory and endocytic pathways (8, 18, 34, 80). Arf proteins are therefore involved in a plethora of activities required for membrane trafficking within the cell.
There are three ARF genes in yeast, namely, ARF1, ARF2, and ARF3. ARF1 and ARF2 are 96% identical, functional homologues and together are required for viability. ARF1 produces 90% of the Arf protein in the cell, and thus the deletion of ARF1 significantly impairs Arf function (71). Consistent with a role in membrane trafficking, arf1Δ mutants display altered glycosylation of secreted proteins (72) and also grow slowly, are cold sensitive, and are supersensitive to fluoride (71). ARF3 is most similar to mammalian ARF6 and is not essential for viability (36). Arf3p has been found to play roles in cell polarity (27) and, more recently, in the endocytic process in yeast (12).
Arf proteins have been shown to directly activate mammalian phosphatidylinositol-(4,5)-bisphosphate (PIP2)-dependent PLD enzymes (21, 22). Changes in the lipid composition of membranes mediated through PLD are thought to promote the assembly as well as the release of clathrin-coated vesicles at the trans-Golgi network and the plasma membrane (8, 80). Although Arf1p has been shown to be a potent activator of mammalian PLD1 (21, 22, 50), Arf1p does not influence the catalytic activity of Spo14p in yeast (64). An analysis of a particular allele, arf1-myc, in an arf2Δ mutant showed that this allele supported vegetative growth but that homozygous diploids failed to sporulate (64). Furthermore, visualization of green fluorescent protein (GFP)-Spo14p in arf1-myc arf2Δ cells revealed an improper formation of spore compartments (64). Although Arf1p activity does not appear to directly act through Spo14p, it does play an important role in sporulation. In contrast, deletion of ARF3 has no effect on sporulation efficiency (17).
Arf's activity is dependent on cycling between its GTP- and GDP-bound forms. Arf proteins contain little intrinsic GTPase activity and therefore rely on GTPase-activating proteins (31). Arf-GTPase-activating proteins (Arf-GAPs) are lipid-activated enzymes and are considered key regulators of membrane trafficking events (73). For some Arf-GAPs, the lipids most potent in activation include PA, a direct product of PLD catalytic activity, and diacylglycerol, which can be formed as a result of the dephosphorylation of PA (57, 82). It has been proposed that Arf-GAPs mediate vesicle formation, cargo selection, and uncoating of transport vesicles (37, 74, 82). In addition, Arf-GAPs have Arf-independent roles in cellular functions (58).
Five genes have been identified in yeast which encode polypeptides that share significant sequence similarity to Arf-GAPs: these are AGE2, GCS1, GLO3, SAT1, and SPS18. Age2p, Gcs1p, Glo3p, and Sat1p are known Arf-GAPs (52, 54, 55, 83), while Sps18p, a sporulation-specific protein thought to function during sporulation (11), has not, to our knowledge, been tested for Arf-GAP activity.
Genetic studies have shown that the four known yeast Arf-GAPs have overlapping functions (83). For instance, Glo3p and Gcs1p provide overlapping essential functions for retrograde vesicular transport from the Golgi to the endoplasmic reticulum (53). On the other hand, Gcs1p and Age2p form an essential pair that provides overlapping functions for trans-Golgi network transport (54). Furthermore, these Arf-GAPs have been shown together to be required for both Sec14p-dependent and -independent Golgi secretory function (82). Sec14p is the major yeast phosphatidylinositol transfer protein, which maintains a lipid environment essential for protein transport from the Golgi complex (3, 4, 10). In the absence of Sec14p, Spo14p contributes to the lipid milieu for the maintenance of Golgi function, also through the action of Arf-GAPs (82).
To determine if Arf-GAPs are downstream effectors of Spo14p-generated PA during sporulation, we examined a homozygous mutant of each of the five known or potential Arf-GAPs. Only gcs1Δ diploid cells displayed a similar sporulation defect to that of the spo14Δ mutant. Our findings indicate that Gcs1p plays an important role in prospore membrane formation in budding yeast, in part through the regulation of Spo14p.
MATERIALS AND METHODS
Yeast strains and media.
Routine growth and manipulation of S. cerevisiae strains were performed as described previously (62). The yeast strains used in this study are all derived from the S288C strain background, unless otherwise noted (Table 1). The BY4743 deletions obtained from Research Genetics were confirmed by PCR. DNA-mediated transformations of yeast cells were done by the lithium acetate procedure (29). Gene replacement and disruptions (see below) were performed by the one-step method (63). gcs1::kanMX and ndt80::kanMX, which replace the entire open reading frames with the marker kanMX4 (78), were generated by amplifying a region ∼300 bp upstream and downstream of the disrupted open reading frames from a previously constructed knockout strain obtained from Research Genetics.
TABLE 1.
Genotypes of yeast strains
| Strain | Genotype |
|---|---|
| BY4743 | MATa/α his3/his3 leu2/leu2 met15/MET15 LYS2/lys2 ura3/ura3 |
| 32008a | BY4743 but homozygous for sps18::kanMX |
| 33924a | BY4743 but homozygous for gcs1::kanMX |
| 36121a | BY4743 but homozygous for glo3::kanMX |
| 31437a | BY4743 but homozygous for age2::kanMX |
| 34358a | BY4743 but homozygous for sat1::kanMX |
| Y304 | MATaleu2-x his4-x trp1 ura3-1 thr1 lys2 CYH |
| Y305 | MATα leu2-y his4-y trp1 ura3-1 ade2-1 lys2 CYH |
| Y315 | MATa/α leu2-x/leu2-y his4-x/his4-y trp1/trp1 ura3-1/ura3-1 thr1/+ lys2/lys2 CYH/CYH +/ade2-1 |
| Y4403 | Y315 but homozygous for gcs1::kanMX |
| Y4690 | Y315 but homozygous for GCS1-GFP |
| Y4804 | Y315 but homozygous for GCS1PHΔ-GFP |
| Y5408 | Y315 but homozygous for gcs1R54K |
| Y5397 | Y315 but homozygous for gcs1R54K-GFP |
| Y5328 | Y315 but heterozygous for gcs1R54K-GFP |
| Y5609 | Y315 but heterozygous for SNF7-GFP |
| Y5617 | Y4403 but homozygous for SNF7-GFP |
| Y5450 | Y315 but homozygous for SPC42-dsRED |
| Y5466 | Y4403 but homozygous for SPC42-dsRED |
| Y5502 | Y5450 plus pME1096 (GFP-SPO14 2μm LEU2) |
| Y5503 | Y5466 plus pME1096 (GFP-SPO14 2μm LEU2) |
| Y5040 | Y315 but homozygous for spo14::LEU2 |
| Y5492 | Y315 but homozygous for gcs1::kanMX and spo14::LEU2 |
| Y4995 | Y315 but homozygous for ndt80::kanMX |
| Y5664 | Y315 but homozygous for arl1::kanMX |
| Y5204 | Y315 plus pUN105 (CEN LEU2) |
| Y5205 | Y315 plus YEp351 (2μm LEU2) |
| Y5206 | Y315 plus pME2206 (GCS1 CEN LEU2) |
| Y5207 | Y315 plus pME2306 (GCS1 2μm LEU2) |
| Y5209 | Y4403 plus pUN105 (CEN LEU2) |
| Y5210 | Y4403 plus YEp351 (2μm LEU2) |
| Y5211 | Y4403 plus pME2206 (GCS1 CEN LEU2) |
| Y5212 | Y4403 plus pME2306 (GCS1 2μm LEU2) |
| Y5545 | Y4403 plus pME2399 (GCS1-GFP 2μm TRP1) |
| Y4496 | Y4403 plus pME1096 (GFP-SPO14 2μm LEU2) |
| Y5707 | Y5040 plus pME2399 (GCS1-GFP 2μm TRP1) |
| Y5703 | Y315 plus pME2377 (DTR1-GFP 2μm TRP1) |
| Y5697 | Y4403 plus pME2377 (DTR1-GFP 2μm TRP1) |
| Y5700 | Y5040 plus pME2377 (DTR1-GFP 2μm TRP1) |
| Y5701 | Y5492 plus pME2377 (DTR1-GFP 2μm TRP1) |
Record number from Research Genetics homozygous deletion project.
GFP and dsRED (tdimer2 variant of red fluorescent protein) chromosomal fusions were constructed using the reagents described by Longtine et al. (39) and Sheff and Thorn (69), respectively. All integrants were confirmed using PCR and the appropriate synthetic oligonucleotide primers. Gcs1pPHΔ-GFP was created by generating primers that placed the GFP-Kan tag 227 amino acids into the open reading frame. When the corresponding PCR product was transformed into haploid strains, a deletion of 125 amino acid residues at the C terminus of Gcs1p was created.
Plasmids.
Plasmid ME2206 contains the GCS1 complementing region in an ∼2.2-kb SacI-BamHI (PCR or primer generated) fragment at the corresponding sites of pUN105 (16), a low-copy-number (CEN) LEU2 plasmid. The 2.2-kb fragment was cloned into the SacI-BamHI sites of the high-copy-number (2μm) plasmid YEp351 (23) to generate pME2306 (LEU2). Plasmid ME913 (spo14::LEU2, which removes 4,187 bp of the open reading frame) was used in generating the spo14Δ and gcs1Δ spo14Δ chromosomal deletions/disruptions as described by Rudge et al. (65). Plasmid ME913 was targeted for integration in yeast with XhoI-NotI.
Site-directed mutagenesis was performed on pME2207 (GCS1 2μm LEU2) to generate gcs1R54A (pME2221) and gcs1R54K (pME2355), using a QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) as recommended by the manufacturer. A mutagenic upstream primer (5′-CTTGAATGTGCCGGTATCCATGCAGGGCTTGGTGTGCATATATC-3′; the bold residue indicates the introduced mutation) and downstream primer (5′-GATATATGCACACCAAGCCCTGCATGGATACCGGCACATTCAAG-3′) were used to generate the desired R54A point mutation. A different mutagenic upstream primer (5′CTTGAATGTGCCGGTATCCATAAAGGGCTTGGTGTGCATATATC-3′) and down- stream primer (5′GATATATGCACACCAAGCCCTTTATGGATACCGGCACATTCAAG-3′) were used to generate the desired R54K point mutation. The SacI-XbaI N-terminal fragments of pME2221 (containing gcs1R54A) and pME2355 (containing gcs1R54K) and the XbaI-BamHI fragment containing the C-terminal unmutagenized fragment of pME2206 were moved into the SacI and BamHI sites of pUN105 and pUN15 (16) to generate pME2325 (gcs1R54A CEN LEU2) and pME2358 (gcs1R54K CEN TRP1), respectively, which maintained the Spo− phenotype and contained only the R54A or R54K mutation, respectively, as verified by sequencing. The EcoRI-BamHI fragment containing gcs1R54K from pME2358 was moved into the corresponding sites of YIp5, a yeast integrating plasmid, to generate pME2363 and was confirmed by sequencing to only contain the R54K mutation. Plasmid ME2363 was targeted for integration into wild-type haploid strains with XbaI. The MATα gcs1R54K haploid strain was crossed with a strain of the opposite mating type to generate the gcs1R54K homozygous mutant. The original MATα gcs1R54K haploid strain was additionally chromosomally tagged with GFP by the method of Longtine et al. (39). The homozygous GFP-tagged diploid strain was generated as described above, but haploid MATa segregants were isolated by G418 resistance conferred by the kanMX marker.
GFP-SPO14 was expressed from a high-copy-number (2μm) LEU2 plasmid (pME1096) and has been previously described (65). DTR1-GFP (a generous gift from Aaron Neiman, State University of New York at Stony Brook, Stony Brook, NY) was expressed from a high-copy-number (2μm) TRP1 plasmid. HOP1-lacZ, NDT80-lacZ, and DIT1-lacZ (generous gifts from Andrew Vershon, Waksman Institute of Microbiology, Rutgers State University of New Jersey, Piscataway, NJ) were expressed from high-copy-number (2μm) LEU2 plasmids.
To generate GCS1-GFP expressed from a plasmid, the GCS1-GFP-kan cassette was amplified from Y4690 using an upstream primer that binds ∼1,000 bp upstream of the GCS1 open reading frame and a downstream primer containing a KpnI restriction site in its sequence. A SacI-KpnI fragment containing GCS1-GFP-kan was originally moved into the corresponding sites of pUN15, a low-copy-number (CEN) TRP1 plasmid, to generate pME2369, which complemented the gcs1Δ mutant. The ∼4.5-kb SacI-KpnI fragment containing GCS1-GFP-kan was cloned into corresponding sites of pME856, a high-copy-number (2μm) TRP1 plasmid, to generate pME2399, which additionally complements the gcs1Δ mutant.
Analysis of sporulation.
Cells were grown on YPAD (yeast extract-peptone-dextrose plus 10 mM adenine) or a medium lacking appropriate amino acids to maintain the selection of plasmids and were incubated overnight. Cells were replica plated onto sporulation medium and incubated at 30°C for 3 days. Sporulation was monitored by differential interference contrast (DIC) microscopy. For liquid cultures, 2 ml of YPAD or medium lacking appropriate amino acids was inoculated, and cells were grown for 24 h at 30°C. Approximately 1 × 107 to 2 × 107 cells were then transferred to 4 ml YP-acetate medium and incubated at 30°C for 24 h. The cells were collected by centrifugation, washed one time with double-distilled water (ddH2O), and resuspended in 2 ml 2% potassium acetate (KAc) to induce sporulation. For the BY4743 strain background, cells were sporulated in 10 ml SPM medium (0.3% KAc, 0.02% raffinose). In some experiments, the meiotic divisions were analyzed after fixation by staining with 4′,6′-diamidino-2-phenylindole (DAPI) as described previously (40). Final sporulation counts were taken after 3 days.
Analysis of GFP and dsRED fusion proteins.
Strains expressing GFP-SPO14 (65), DTR1-GFP, SPC42-dsRED, GCS1-GFP, gcs1R54K-GFP, and GCS1PHΔ-GFP (Table 1) were grown and sporulated as described above. At various times after induction, live cells were examined by fluorescein isothiocyanate or rhodamine optics on a Zeiss Axioskop 2 fluorescence microscope.
Analysis of FM4-64 uptake.
Cells were induced to sporulate in liquid as described above (sporulating cells), or log-phase cultures grown in YPAD (vegetative cells) were analyzed. FM4-64 [N-(3-thiethylammoniumpropyl)-4-(p-diethylaminophenylhexatrienyl) pyridinium dibromide; Molecular Probes, Inc., Eugene, OR] (77) was added to 1 ml of culture at a final concentration of 2 μg/ml (from a 1-mg/ml stock solution in dimethyl sulfoxide) at 14 h and incubated on ice for 5 min in the dark. The cells were collected by centrifugation, washed once, and inoculated into fresh 2% KAc. The cells were observed 0, 10, 20, 30, and 60 min after incubation with shaking at 30°C.
In vivo BODIPY-PC analysis.
BODIPY-PC (4 μM final concentration; Molecular Probes, Eugene, OR) was added directly to cultures (∼2 × 108 cells) 12 h after induction in sporulation medium. Cells were harvested 3 h later, and lipids were extracted and analyzed by thin-layer chromatography as previously described (66). All assays were performed in triplicate. The percentage of conversion of BODIPY-PC to BODIPY-PA was determined from the pixel intensities obtained from an image of the thin-layer chromatography plate, using AlphaEase FC4.0 imager software (Alpha Innotech).
Transmission electron microscopy (TEM).
Cells were sporulated as described above. Ten milliliters of cells was fixed by the addition of glutaraldehyde to the culture medium at a final concentration of 2.5% and incubation for 1 h at room temperature. Cells were washed two times with ddH2O, resuspended in 1 ml 4% potassium permanganate (KMnO4) (in water), and incubated at room temperature for 30 min. Cells were then washed with ddH2O until the supernatant appeared clear, followed by resuspension in 1 ml saturated uranyl acetate, and were incubated at room temperature for 30 min. Samples were finally dehydrated through serial washes in acetone (five times for 15 min each). The dehydrated samples were embedded in Mollenhauer medium (41), and thin sections were obtained. The images were collected on a Philips CM120 microscope at 80 kV.
Preparation of particulate and cytosolic fractions.
Extracts were prepared from cells (∼8 × 108) undergoing meiotic divisions as described above (sporulating cells) or from log-phase cultures grown in YPAD (vegetative cells). Cells were harvested and resuspended in 1 to 3 ml lysis buffer (20 mM triethanolamine, 300 mM sorbitol, 5 mM EDTA, 2 mM phenylmethylsulfonyl fluoride, and 1 mM dithiothreitol). One milliliter of the resuspension was added to 100 μl acid-treated glass beads on ice. Cells were vortexed for 40 seconds and placed on ice for 30 seconds, and this was repeated five times. Lysed cells were subjected to a prespin at 1,000 × g for 3 min to remove unlysed cells and cell debris. Whole-cell extracts were taken at this point. The supernatant was then spun at 14,000 × g for 20 min. The supernatant obtained at this speed (14K supernatant) was transferred to a new tube, and the 14K pellet was resuspended in an equivalent volume of lysis buffer. The 14K fractions were isolated at this point. The remaining supernatant was then spun at 100,000 × g for 1 h. The 100K supernatant was transferred to a new tube, and the 100K pellet was resuspended in an equivalent volume of lysis buffer. The above procedure was performed on ice, and centrifugation was done at 4°C. The isolated fractions were combined with an equal volume of 2× sample buffer, boiled at 95°C for 5 min, placed on ice for 5 min, and spun at maximum speed for 1 min. Cell equivalents were used for immunoblot analysis.
Immunoblot analysis.
Cell extracts were prepared as described above and subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis on 8% or 10% SDS-polyacrylamide gels. Proteins were electrophoretically transferred onto nitrocellulose membranes (0.45-μm pore size; Bio-Rad Technologies) for 2 h. Blots were blocked by incubation with 3% nonfat dry milk for 1 h at room temperature. Blots were then probed with an anti-GFP monoclonal antibody (Ab) (Clontech/BD Biosciences, Palo Alto, CA) diluted 1:1,000 in 3% nonfat dry milk in TBS-T (Tris-buffered saline with 0.1% Tween 20 [vol/vol]) or a rabbit polyclonal anti-Gas1p antibody (1:25,000 dilution in 3% nonfat dry milk in TBS-T) to detect the glycosylphosphatidylinositol-anchored plasma membrane 1,3-β-glucanosyl transferase (a gift from H. Riezman) and incubated at room temperature for 1 h. After three washes with TBS-T, the anti-GFP blots were incubated with a horseradish peroxidase-conjugated anti-mouse secondary Ab (Pierce, Rockford, IL) diluted 1:3,000 in 3% nonfat dry milk in TBS-T, the anti-Gas1p blots were incubated with a horseradish peroxidase-conjugated anti-rabbit secondary Ab (Pierce, Rockford, IL) diluted 1:3,000 in 3% nonfat dry milk in TBS-T, and both types of blot were incubated at room temperature for 1 h. After three final washes with TBS-T, proteins were visualized by enhanced chemiluminescence detection (Pierce, Rockford, IL) on preflashed film.
RESULTS
Genetic analysis of Arf-GAPs during sporulation.
To analyze the role of Arf-GAPs in sporulation, we examined a Research Genetics deletion collection strain (BY4743) harboring individual homozygous deletions of all five known or predicted Arf-GAPs. The gene deletions analyzed included age2Δ, gcs1Δ, glo3Δ, sat1Δ, and sps18Δ. A genomic screen previously reported that only a gcs1Δ mutant had a sporulation phenotype; however glo3Δ was not analyzed (17). As shown in Table 2, we have confirmed that gcs1Δ mutants largely fail to form spores when induced in sporulation medium, while age2Δ, sat1Δ, and sps18Δ mutants sporulate similarly to the wild type. In addition, we found that glo3Δ mutants also displayed a sporulation defect (Table 2). The finding that strains deleted for the sporulation-specific SPS18 gene showed no significant sporulation defect led us to investigate this gene further. Since Arf-GAPs have both overlapping and distinct roles in cell function in vegetative cells, we examined the consequence of deleting SPS18 in conjunction with the other Arf-GAP mutants and saw no enhancement of phenotype (data not shown). Thus, the role of SPS18 remains enigmatic.
TABLE 2.
Sporulation of single gene deletion mutants in BY4743 strain background
| Strain | Relevant genotype | % Spore formationa |
|---|---|---|
| BY4743 | 27.7 ± 4.9 | |
| 32008 | sps18Δ | 21.2 ± 13.8 |
| 33924 | gcs1Δ | 1.2 ± 0.9* |
| 36121 | glo3Δ | 6.2 ± 1.6* |
| 31437 | age2Δ | 19.8 ± 4.0 |
| 34358 | sat1Δ | 30.5 ± 9.2 |
A minimum of 500 cells were counted per liquid culture over a minimum of three independent experiments. The final % sporulation includes cells that contained two, three, or four mature spores. Values are means ± standard deviations (SD) from three independent experiments. *, % sporulation was statistically significantly different for gcs1Δ and glo3Δ, with P values of <0.001 and 0.002, respectively.
Cells deleted for GCS1 and GLO3 were examined further to determine where in the sporulation program these mutants stalled. First, we analyzed the ability of the mutants to undergo meiotic divisions. At various times after transfer to sporulation medium, glo3Δ mutants were fixed and stained with DAPI, which revealed that most of the cells failed to undergo either meiotic division (data not shown). To determine if meiosis was initiated in glo3Δ cells, we monitored the expression of HOP1, a sporulation-specific gene that is expressed early in meiosis (24, 76). β-Galactosidase activity was measured for cells harboring a HOP1::lacZ fusion induced in sporulation medium (76). The results of the assay showed that HOP1 was not expressed in glo3Δ cells, indicating that glo3Δ mutants cannot initiate the sporulation program (data not shown). The role of this gene was not investigated further.
In contrast to the case for glo3Δ mutants, cells deleted for GCS1 induced the meiotic program, as HOP1::lacZ was expressed and some cells underwent meiotic divisions, as monitored by DAPI staining (data not shown). This led to increased interest in the gcs1Δ mutant; phenotypic characterization of the gcs1Δ mutant is presented below.
GCS1 plays an essential role in sporulation.
Due to the difficulty in inducing sporulation in BY4743, we moved the gcs1::kanMX deletion cassette into an S288C-like strain background, as in previous studies performed in our lab (13).
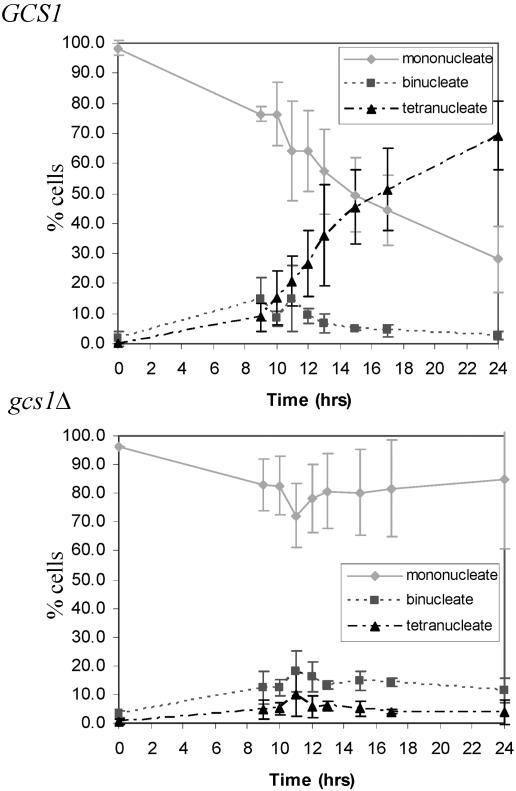
As shown in Table 3, the S288C gcs1Δ mutant sporulated very poorly (<0.1% of gcs1Δ cells versus 69.3% ± 4.0% of GCS1-expressing cells). Analysis of the meiotic divisions by DAPI staining revealed that some gcs1Δ cells underwent meiosis I (MI) but that only about 5% of the cells progressed through MII (Fig. 1). Meiotic progression was also analyzed by examining the expression of sporulation-specific genes, which are regulated temporally and can be grouped into early, middle, mid-late, and late genes (9). We monitored the expression of early (HOP1) (24, 76), middle (NDT80) (9), and mid-late (DIT1) (6) genes by using lacZ fusions. Consistent with the analysis of the meiotic divisions in the gcs1Δ mutant, β-galactosidase assays with strains harboring these fusions revealed that HOP1 and NDT80 were expressed at similar levels to those of the wild type, while DIT1 was expressed at a lower level in cells deleted for GCS1 (data not shown). Therefore, the sporulation program is initiated in gcs1Δ mutants, but the mutants fail to efficiently progress through the meiotic divisions and form mature spores. To ensure that the sporulation defect was due to the deletion of GCS1, complementation analysis was performed, using low-copy-number (CEN) and high-copy-number (2μm) plasmids containing GCS1. Sporulation analyzed on plates was rescued to levels of 34.6% ± 6.8% (CEN) and 28.9% ± 0.8% (2μm), indicating that the deletion of GCS1 is solely responsible for the sporulation phenotype. These same plasmids transformed into wild-type cells had no additional effect on sporulation (Table 4).
TABLE 3.
Sporulation in S288C strain background
| Strain | Relevant genotype | % Spore formationa |
|---|---|---|
| Y315 | GCS1 | 69.3 ± 4.0 |
| Y4403 | gcs1Δ | <0.1 |
| Y5408 | gcs1R54K | <0.1 |
| Y5040 | spo14Δ | <0.1 |
| Y5492 | gcs1Δ spo14Δ | <0.1 |
| Y5664 | arl1Δ | 46.3 ± 9.8 |
| Y4690 | GCS1-GFP | 41.7 ± 3.8 |
| Y4804 | GCS1PHΔ-GFP | 62.7 ± 8.4 |
| Y5396 | gcs1R54K-GFP | <0.1 |
| Y5328 | gcs1R54K-GFP × GCS1 | 70.7 ± 5.4 |
| Y5609 | GCS1 SNF7-GFP | 75.6 ± 8.6 |
| Y5617 | gcs1Δ SNF7-GFP | <0.1 |
A minimum of 300 cells were counted per liquid culture over a minimum of three independent experiments. The final % sporulation includes cells that contained two, three, or four mature spores. Values are means ± SD from three independent experiments.
FIG. 1.
gcs1Δ cells undergo meiotic division with reduced efficiency. Nuclear divisions were analyzed in GCS1-expressing and gcs1Δ cells by staining with the DNA-specific dye DAPI. A minimum of 300 cells were counted over three independent experiments.
TABLE 4.
Complementation and suppression analysis of gcs1Δ mutant
| Strain | Relevant genotype | % Spore formation |
|---|---|---|
| Y5204 | GCS1 + CEN | 30.3 ± 0.7 |
| Y5205 | GCS1 + 2μm | 30.4 ± 2.9 |
| Y5206 | GCS1 + GCS1 CEN | 35.1 ± 1.7 |
| Y5207 | GCS1 + GCS1 2μm | 28.6 ± 3.3 |
| Y5209 | gcs1Δ + CEN | 0.2 ± 0.4 |
| Y5210 | gcs1Δ + 2μm | 0.1 ± 0.2 |
| Y5211 | gcs1Δ + GCS1 CEN | 34.6 ± 6.8 |
| Y5212 | gcs1Δ + GCS1 2μm | 28.9 ± 0.8 |
| Y5545 | gcs1Δ + GCS1-GFP-2μm | 20.9 ± 6.6 |
| Y4496 | gcs1Δ + GFP-SPO14-2μm | 0.1 ± 0.2 |
| Y5707 | spo14Δ + GCS1-GFP-2μm | <0.1 |
A minimum of 300 cells were counted over three independent experiments performed on plates. The final % sporulation includes cells that contained two, three, or four mature spores. Values are means ± SD from three independent experiments.
gcs1Δ mutant is defective in prospore membrane formation.
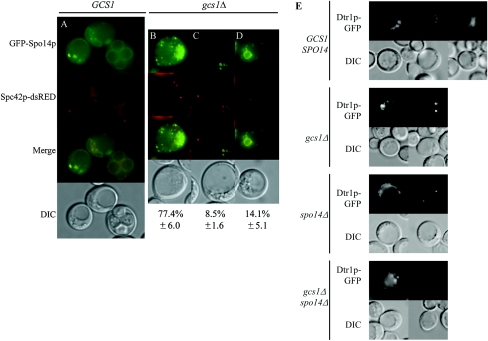
During the second meiotic division, the secretory system is modified to direct the synthesis of a distinct double-layered internal membrane termed the prospore membrane (40). To determine whether gcs1Δ synthesized the prospore membrane, we expressed GFP-SPO14, which is essential for and whose gene product localizes to the prospore membrane (65). Additionally, these strains were marked with SPC42-dsRED. Spc42p is a central plaque component of the SPB (14); thus, the dsRED marker can be used to monitor the meiotic divisions in live cells (48). In the wild type, GFP-Spo14p moves to the SPB and then marks the prospore membrane as it grows to surround the haploid nuclei (Fig. 2A) (30, 43, 64, 65). Visualization of GFP-Spo14p revealed that most cells deleted for GCS1 that formed an MII spindle displayed a punctate pattern of GFP fluorescence (Fig. 2B) and that a small population displayed localization of GFP-Spo14p to the four SPBs (Fig. 2C), similar to what is observed with the spo14Δ mutant (30, 65). In addition, rare cells contained internal membranes; however, these membranes were smaller than those of the wild type, and cells usually contained fewer than four membrane compartments (Fig. 2D). Another prospore membrane marker, Dtr1p-GFP (19), gave a similar pattern of localization in the gcs1Δ mutant (Fig. 2E).
FIG. 2.
gcs1Δ mutants are defective in prospore membrane formation. Fluorescence and DIC images of the same cells are shown. (A) Wild-type cells analyzed at ∼12 h in sporulation medium. (B to D) Localization of GFP-Spo14p to punctate structures (B), the four SPBs (C), and immature prospore membranes (D) in gcs1Δ cells tagged with SPC42-dsRED to indicate the location of the SPBs under sporulation conditions. (E) Dtr1p-GFP localizes similarly to GFP-Spo14p in wild-type and gcs1Δ cells, with the majority of gcs1Δ cells displaying the patterns shown and a small percentage forming immature prospores. Additionally, Dtr1p-GFP localizes to punctate structures or the four SPBs in spo14Δ and gcs1Δ spo14Δ cells. This experiment was repeated a minimum of three times, with similar results.
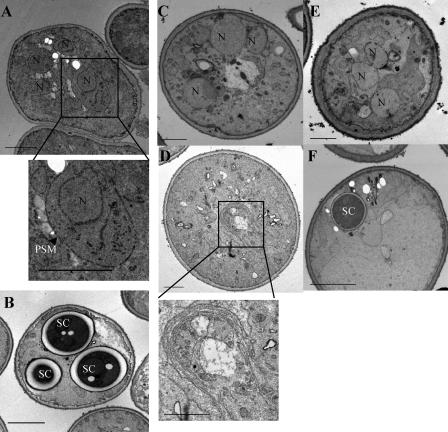
To confirm that gcs1Δ mutants were defective in prospore membrane formation, we examined thin sections of yeast cells induced to sporulate by TEM. In wild-type cells, the presence of prospore membranes encapsulating nuclei after MII, as well as cells containing spore wall compartments, was readily observed (Fig. 3A and B). In contrast, in the majority of cells deleted for GCS1, no prospore membrane or spore structures were seen (Fig. 3C). In addition, many gcs1Δ cells had abnormal morphologies and an abundance of membranous structures. The abnormal membrane structures were reminiscent of what is seen in class E mutants defective in endosome-to-vacuole trafficking (Fig. 3D) (1). Thus, gcs1Δ mutants are defective in prospore membrane formation and display additional defects in cellular membrane compartments.
FIG. 3.
TEM of GCS1-expressing and gcs1 mutant cells during sporulation. (A and B) Wild-type cells show the presence of four nuclei and the corresponding prospore membranes (PSMs), and some cells have reached compartmentalization. (C) gcs1Δ cells largely fail to synthesize the prospore membrane and display abnormal morphology. (D) Some gcs1Δ cells contain aberrant structures reminiscent of “class E compartments.” (E and F) gcs1R54K-expressing cells display less severe morphological defects overall, and one cell with a spore compartment can be observed. N, nuclei; SC, spore compartments; arrowhead, prospore membrane. Bars, 2 μm.
Deletion of GCS1 impairs endosome-to-vacuole trafficking during sporulation.
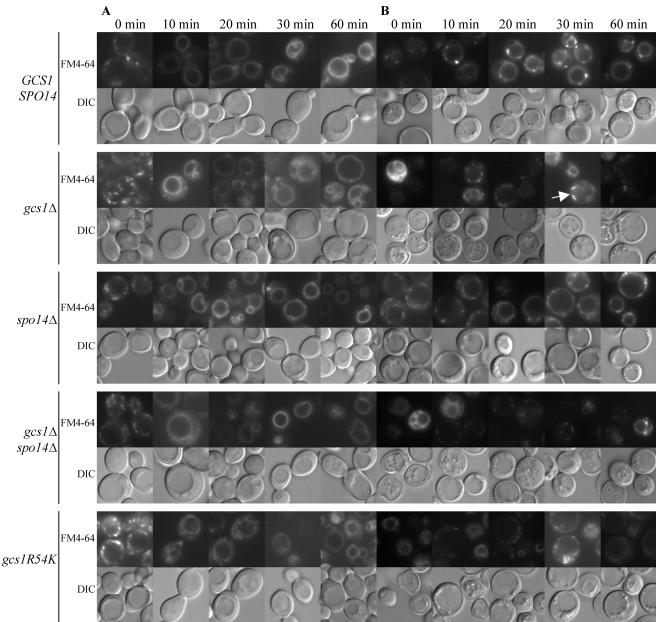
The aberrant membrane structures observed by TEM and the previous report that Gcs1p interacts with the actin cytoskeleton (5) led us to examine FM4-64 uptake by the gcs1Δ mutant. FM4-64 is a lipophilic dye that is internalized by endocytosis at the plasma membrane and is transported through the endosome to the vacuolar membrane (77). In vegetative cells, the kinetics of uptake were similar for wild-type and gcs1Δ mutant cells: FM4-64 was primarily observed in punctate structures immediately after removal from the dye and 10 min later was found at the vacuolar membrane (Fig. 4A).
FIG. 4.
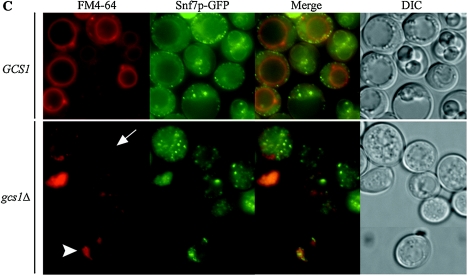
FM4-64 staining in vegetative and sporulating cells. Fluorescence and DIC images of the same cells are shown. (A) FM4-64 uptake by vegetative cells. (B) Cells induced during meiosis for 14 h. An analysis of FM4-64 uptake was performed 0, 10, 20, 30, and 60 min after removal of the cells from the dye. Arrow, bar-like structures. (C) Localization analysis of FM4-64 and the endosomal marker Snf7p-GFP in wild-type (60 min after removal of the dye) and gcs1Δ (30 min [top panel] and 60 min [bottom panel] after removal of the dye) cells. Arrowhead, “class E compartment”; arrow, bar-like structures. These experiments were repeated a minimum of three times, with similar results.
Endocytosis plays an important role in spore morphogenesis (43), and thus we monitored the uptake of FM4-64 during sporulation by the gcs1Δ mutant (Fig. 4B). Cells undergoing meiotic divisions were stained with FM4-64, the dye was removed, and the cells were examined at various times. Wild-type cells rapidly took up FM4-64 into punctate structures and then the vacuolar membrane; >80% of the cells showed vacuolar membrane staining by 60 min after removal from the dye (Table 5). Many cells deleted for GCS1 displayed a very bright signal and the appearance of aberrant vacuoles immediately after removal from the dye. As time progressed, most of the cells showed FM4-64 in punctate or bar-like structures surrounding the vacuole. Some cells showed vacuolar membranes stained with FM4-64 that appeared to be convoluted or fragmented, as previously seen by Zhang et al. (83). Only ∼30% of gcs1Δ cells showed vacuolar membrane staining by 60 min, while ∼33% of the cells showed punctate and bar-like structures (Table 5).
TABLE 5.
Analysis of endocytosis by FM4-64 in sporulating cells
| Strain and time of analysis (min) | Relevant genotype | % FM4-64 signala
|
||
|---|---|---|---|---|
| Punctate | Vacuoleb | No signal | ||
| Y315 | GCS1 SPO14 | |||
| 0 | 59.4 | 3.1 | 37.5 | |
| 10 | 31.0 | 41.0 | 28.0 | |
| 20 | 4.4 | 80.6 | 15.0 | |
| 30 | 1.1 | 76.6 | 22.3 | |
| 60 | 0.0 | 82.9 | 17.1 | |
| Y4403 | gcs1Δ | |||
| 0 | 27.5 | 34.5 | 38.0 | |
| 10 | 32.1 | 10.2 | 57.7 | |
| 20 | 46.6 | 21.9 | 31.5 | |
| 30 | 25.3 | 22.8 | 51.9 | |
| 60 | 32.5 | 29.6 | 37.9 | |
| Y5040 | spo14Δ | |||
| 0 | 83.2 | 3.4 | 13.4 | |
| 10 | 94.5 | 1.6 | 3.9 | |
| 20 | 46.1 | 30.9 | 23.0 | |
| 30 | 1.1 | 95.2 | 3.7 | |
| 60 | 0.0 | 97.7 | 2.3 | |
| Y5492 | gcs1Δ spo14Δ | |||
| 0 | 8.6 | 38.9 | 52.5 | |
| 10 | 23.8 | 41.4 | 34.8 | |
| 20 | 12.7 | 27.6 | 59.7 | |
| 30 | 8.6 | 26.8 | 64.6 | |
| 60 | 9.1 | 38.8 | 52.1 | |
| Y5408 | gcs1R54K | |||
| 0 | 8.4 | 33.7 | 57.9 | |
| 10 | 26.8 | 25.6 | 47.6 | |
| 20 | 32.0 | 23.2 | 44.8 | |
| 30 | 30.7 | 23.2 | 46.1 | |
| 60 | 28.6 | 27.8 | 43.6 | |
Localization of FM4-64 to punctate or vacuolar structures within cells. This experiment was performed multiple times revealing similar results. A minimum of 300 cells was counted in each experiment. These numbers reflect one individual experiment.
Cells showing localization of FM4-64 to normal or fragmented vacuoles.
Colocalization of FM4-64 and Snf7p-GFP, an endosomal marker (2), was analyzed in wild-type and gcs1Δ cells to determine the nature of the punctate and bar-like structures (Fig. 4C). Significant colocalization of FM4-64 and Snf7p-GFP was not observed, although in many cells the signals appeared adjacent to one another. Thus, the punctate and bar-like structures (Fig. 4C, arrow) visualized in gcs1Δ cells were not Snf7p-marked endosomal structures. Additionally, gcs1Δ mutants contained a small population of cells (∼1%) that showed a small ring-like structure next to the vacuole that was labeled with both FM4-64 and Snf7p-GFP (Fig. 4C, arrowhead). This structure most likely represents the “class E compartment,” corresponding to a late endosomal intermediate unable to form intraluminal vesicles (2, 33, 56).
Gcs1p-GFP is a soluble protein that becomes concentrated at spore peripheries during maturation.
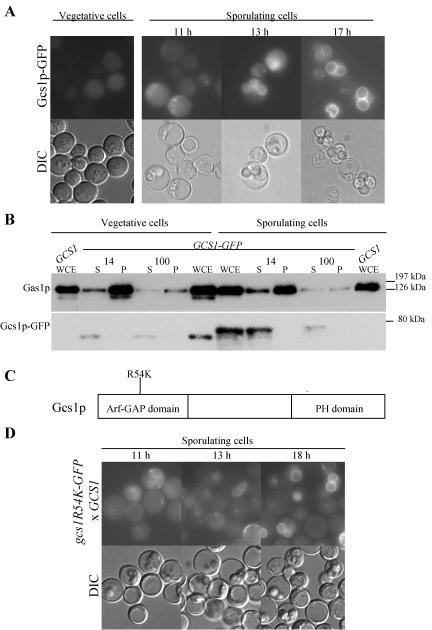
Strains expressing Gcs1p-GFP, either chromosomally or from a high-copy-number plasmid, were constructed, and localization was examined by GFP fluorescence in living cells. The Gcs1p-GFP fusion was largely functional, as assessed by complementation of the sporulation phenotype (Tables 3 and 4). In vegetative cells and early during the sporulation program, Gcs1p-GFP was seen throughout the cytoplasm (Fig. 5A). As sporulation progressed, the GFP signal became concentrated in the developing spore, and during spore maturation, the GFP signal was primarily seen at the spore periphery (Fig. 5A).
FIG. 5.
Gcs1p-GFP is a soluble protein that eventually localizes to mature spore peripheries. (A) Localization of Gcs1p-GFP expressed from a 2μm plasmid in vegetative and sporulating cells. Fluorescence and DIC images of the same cells are shown. (B) Autoradiograph of Western blot analysis of proteins extracted from cells induced during meiosis (14 h) and from vegetative cells probed with anti-GFP to detect Gcs1p-GFP and anti-Gas1p to detect the membrane protein Gas1p. (C) Gcs1p contains 352 amino acids and consists of Arf-GAP and PH domains. A mutation at position 54 (R54K) abolishes Arf-GAP activity. (D) Gcs1pR54K-GFP localizes similarly to the wild-type protein in GCS1-expressing cells. These experiments were repeated a minimum of three times, with similar results.
Cell fractionation was performed to determine whether Gcs1p was membrane associated. The predicted molecular mass of Gcs1p is 39 kDa and that of GFP is 26 kDa. Gcs1p-GFP was specifically detected with anti-GFP antibody as an ∼75-kDa protein that was absent from extracts made from cells lacking the tagged protein (Fig. 5B, GCS1 WCE). Gcs1p-GFP was found exclusively in the soluble fraction after both a 14,000 × g spin and a 100,000 × g spin of extracts from both vegetative and sporulating cells. In contrast, the membrane-anchored Gas1p protein was found predominately in the pellets after spinning at 14,000 × g and 100,000 × g. Therefore, Gcs1p-GFP is a largely soluble protein. In addition, Gcs1p-GFP isolated from sporulating cells migrated slower by SDS-polyacrylamide gel electrophoresis than did Gcs1p-GFP isolated from vegetative cells (Fig. 5B). This shift in molecular mass suggests that the protein undergoes posttranslational modification during sporulation.
Arf-GAP activity, but not the pleckstrin homology (PH) domain, is essential for Gcs1p function in sporulation.
All Arf-GAPs described thus far contain one invariant arginine residue, which corresponds to position 54 in Gcs1p. This residue, when mutated to alanine or lysine in Gcs1p, has been shown to abolish enzymatic activity in vitro (82). Cells expressing gcs1R54K or gcs1R54A as their only source of GCS1 failed to sporulate (Table 3 and data not shown). Thus, GCS1 Arf-GAP activity is crucial for in vivo function during sporulation.
We analyzed strains expressing gcs1R54K for prospore membrane formation and FM4-64 uptake as described above for the null mutant. Like gcs1Δ cells, gcs1R54K-expressing cells largely failed to make prospore membranes, although rarely a cell containing a single spore compartment was observed (Fig. 3E and F; data not shown), and gcs1R54K-expressing cells were additionally defective in FM4-64 trafficking during sporulation (Fig. 4B). Thus, the phenotype of gcs1Δ mutants during sporulation is largely attributable to the absence of GAP activity.
gcs1R54K was chromosomally tagged with GFP to analyze the expression and localization of this mutant. Gcs1pR54K-GFP behaved similarly to Gcs1p-GFP in sporulating GCS1-expressing cells, exhibiting a diffuse staining pattern, that is, concentrated to spore peripheries in maturing wild-type spores, and was expressed at similar levels (Fig. 5D).
In addition to the Arf-GAP region of the protein, Gcs1p contains a PH domain that consists of the C-terminal 125 amino acid residues. PH domains have been implicated in phosphoinositide binding in a variety of proteins (20, 32). Phosphoinositides, such as PIP2, play important roles in meiosis and have been shown to activate Spo14p (67). The PH domain of Gcs1p is dispensable for vegetative growth (82). The same deletion in sporulating cells appears to have no effect on Gcs1p function (Table 3).
Gcs1p does not act through Arl1p during sporulation.
Recently, Gcs1p has been shown to exhibit GAP activity towards Arl1p (Arf-like) as well as Arf1p (38). Thus, it was possible that the unique role Gcs1p plays in sporulation is mediated through Arl1p. To test this hypothesis, we deleted ARL1 in the S288C strain background and examined sporulation in the homozygous mutant. The arl1Δ mutant sporulated with a close-to-wild-type efficiency, indicating that Gcs1p does not act exclusively through Arl1p during sporulation (Table 3).
Relationship between Gcs1p and Spo14p during sporulation.
To determine the relationship between Gcs1p and Spo14p during sporulation, we constructed the gcs1Δ spo14Δ double mutant. Like the case for the spo14Δ mutant, no prospore membranes were formed in the gcs1Δ spo14Δ mutant (Fig. 2E). However, the double mutant behaved similarly to the gcs1Δ mutant with respect to FM4-64 uptake (Fig. 4). Thus, the spo14Δ mutant is epistatic to the gcs1Δ mutant for prospore membrane formation, but Gcs1p plays additional roles independent of Spo14p during sporulation (e.g., endosome-to-vacuole trafficking).
The effect of overexpressing GCS1 in a spo14Δ strain and in a strain with a temperature-sensitive hypomorphic allele of SPO14 was analyzed. High levels of GCS1 were unable to rescue the sporulation defects associated with these spo14 mutants. Likewise, overexpression of SPO14 had no effect on gcs1Δ mutants (Table 4; data not shown).
gcs1Δ has reduced levels of Spo14p-generated PA.
To determine whether the deletion of GCS1 had any effect on SPO14 function in sporulating cells, we monitored Spo14p PLD activity by measuring the conversion of internalized BODIPY-PC to BODIPY-PA. BODIPY-PC is a fluorescently labeled substrate that is readily internalized by cells. The hydrolysis of BODIPY-PC occurs almost exclusively through the action of Spo14p, as cells deleted for SPO14 do not generate any appreciable levels of BODIPY-PA (43, 66, 67). The conversion rate of BODIPY-PC to BODIPY-PA in gcs1Δ cells was markedly reduced compared to that in the wild type (0.6% ± 0.3% and 3.0% ± 0.8%, respectively), indicating that the deletion of GCS1 had a significant effect on Spo14p catalytic activity (Table 6). Additionally, gcs1R54K-expressing cells showed a similar reduction in the generation of BODIPY-PA, mimicking the effect seen in gcs1Δ cells. The BODIPY-PA generated in the gcs1Δ mutant is due to the activity of Spo14p, as the gcs1Δ spo14Δ double mutant failed to produce any appreciable BODIPY-PA. Furthermore, similar levels of BODIPY-PC were internalized in all strains analyzed in this experiment (data not shown).
TABLE 6.
Spo14p-catalyzed hydrolysis of internalized BODIPY-PC
| Strain | Relevant genotype | % BODIPY-PAa |
|---|---|---|
| Y5040 | spo14Δ | 0.1 ± 0.1 |
| Y315 | GCS1 SPO14 | 3.0 ± 0.8 |
| Y4403 | gcs1Δ | 0.6 ± 0.3* |
| Y5492 | gcs1Δ spo14Δ | 0.1 ± 0.02 |
| Y5408 | gcs1R54K | 0.5 ± 0.2* |
| Y4995 | ndt80Δ | 2.5 ± 0.9 |
The percentage of conversion of intracellular BODIPY-PC to BODIPY-PA was determined in cells sporulated at 30°C as described in Materials and Methods. Values are means ± SD from three independent experiments. *, % BODIPY-PA is statistically significantly different for gcs1Δ and gcs1R54K, with P values of 0.043 and 0.035, respectively.
A reduction in PA might be expected for cells that are not efficiently progressing through meiosis and generating prospore membranes. To ensure that the reduced levels of PA in gcs1Δ and gcs1R54K-expressing cells were not due to a failure to efficiently progress through meiosis and sporulation, Spo14p activity was monitored in ndt80Δ mutants. Cells deleted for NDT80 arrest prior to the MI division (81). In this assay, ndt80Δ cells generated wild-type levels of Spo14p-generated PA (Table 6). Thus, the reduction in Spo14p-generated PA in gcs1 mutants was not due to a failure to internalize the substrate or efficiently progress through sporulation, but rather the data suggest that Gcs1p regulates the activity of Spo14p.
DISCUSSION
Gcs1p has a unique function during sporulation and regulates Spo14p.
Arf-GAPs have been widely studied in cycling cells, where they play roles in many aspects of membrane trafficking. Sporulation not only needs a functional secretory system, as in vegetative cells (67), but it also requires unique components for the generation of the prospore membrane (47). Furthermore, Spo14p-generated PA plays a central role in the formation of the prospore membrane (67), and given the lipid responsiveness of Arf-GAPs, it seemed likely that a subset, either singly or in combination, functions in this process. We found that deletion of either GCS1 or GLO3 alone impaired sporulation to a significant extent, while deletion of the other Arf-GAPs did not have an effect on sporulation. glo3Δ mutants grow slowly (53), and we found that they do not initiate the sporulation program, suggesting a general defect in cell function. On the other hand, gcs1Δ mutants induce the sporulation program but largely fail to synthesize the prospore membrane. Thus, in contrast to the situation in mitotically dividing cells, where different pairs of Arf-GAPs function redundantly, Gcs1p alone is essential for sporulation. However, it is likely that, although not essential, the other Arf-GAPs function during sporulation and may contribute to the phenotype of the gcs1Δ mutant.
In vegetative cells, in addition to its central role in vesicular transport (53, 54, 83), Gcs1p has been implicated in the maintenance of mitochondrial morphology and actin cytoskeletal organization (5, 26). Furthermore, GCS1 was originally identified as a gene conditionally required for the reentry of cells into the cell cycle after stationary-phase growth (28). Similar to the case in cycling cells, it appears that Gcs1p functions in many processes during sporulation, including meiotic progression, prospore membrane formation, and endosome-to-vacuole transport. Thus, the inactivation of GCS1 impinges on a number of different cellular processes in both vegetative and sporulating cells.
With respect to prospore membrane formation, the phenotype of gcs1 mutants is reminiscent of that of the spo14Δ mutant in that many cells display a punctate pattern of prospore membrane markers within the cytoplasm, and in some cells these markers accumulate at the SPB (30, 65). However, unlike spo14Δ cells, a minor proportion of gcs1Δ cells generate small prospore membranes, and, rarely, even a mature spore is produced. gcs1Δ spo14Δ cells do not form these structures. The presence of small prospore membranes in some cells may be explained by the low but detectable level of Spo14p PLD activity in gcs1Δ mutants.
PA has been shown to directly stimulate Gcs1p Arf-GAP activity in vitro and appears to function downstream of Spo14p during Sec14p-independent growth (82). In contrast, we found a reduction in Spo14p-generated PA levels in gcs1Δ mutants, suggesting that Gcs1p positively regulates Spo14p during sporulation. How Gcs1p influences Spo14p activity is currently unknown, but it is likely to be indirect. Gcs1p converts both Arf1pGTP and Arl1pGTP to their GDP-bound forms (38), and an analysis of gcs1R54K-expressing cells showed that they closely mimicked gcs1Δ cells in all assays, indicating that Arf-GAP activity is specifically required for proper functioning of Gcs1p during sporulation. Given that the deletion of ARL1 has no significant effect on sporulation, Arl1p is unlikely to influence Spo14p. Furthermore, there is no direct stimulation or interaction between Arf1p and Spo14p (64). Taken together, these results indicate that the Arf cycle is important for sporulation and suggest that this cycle impinges on Spo14p, but not directly, as observed in mammalian cells (21, 22, 50). Perhaps the Arf cycle influences Spo14p activity by recruitment of other proteins or by modulating the lipid environment.
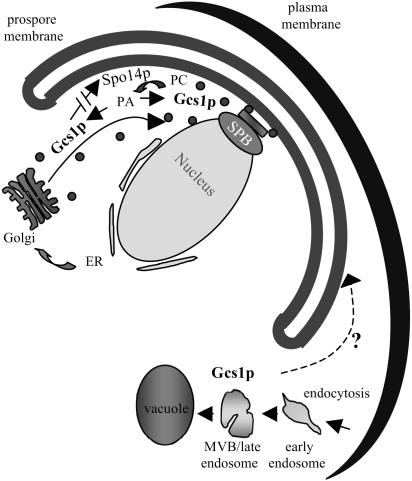
Recent studies indicated that Spo14p is specifically involved in vesicle fusion for prospore membrane formation (46, 60). The results reported here indicate that Gcs1p also plays a role in prospore membrane formation and suggest that there is a feedback loop between these signaling molecules (Fig. 6). Spo14p generates PA, which has been shown to stimulate Gcs1p (82), while in turn Gcs1p converts Arf1pGTP to Arf1pGDP and indirectly influences Spo14p activity. Transport vesicles require coat removal prior to vesicle fusion, which is mediated by hydrolysis of Arf-bound GTP (75). Therefore, the hydrolysis of Arf1pGTP to Arf1pGDP by Gcs1p may be involved in shedding the protein coat and subsequently stimulating the fusion of prospore membrane transport vesicles, which additionally requires Spo14p-generated PA. Therefore, Spo14p, Gcs1p, and the Arf cycle are important and intermingled to facilitate prospore membrane formation.
FIG. 6.
Potential functions of Gcs1p during sporulation. Gcs1p is involved in multiple steps of membrane trafficking during sporulation. Gcs1p may aid in the transport of vesicles from the Golgi and/or in vesicle fusion at the site of prospore membrane synthesis. The interrupted arrow indicates that Gcs1p stimulation of Spo14p is most likely indirect. Additionally, Gcs1p functions in endosome-to-vacuole trafficking. Whether this pathway contributes to the sporulation process is uncertain, as indicated by the dashed arrow and question mark.
Localization and modification of Gcs1p during sporulation.
Previous studies revealed that, depending on growth conditions, the majority of Gcs1p is cytoplasmic (26). Consistent with this, we found that in vegetative cells, Gcs1p-GFP fluorescence was seen throughout the cytoplasm, and in extracts, the protein remained in the soluble fraction after centrifugation. During sporulation, Gcs1p-GFP was present in the cytoplasm of both the mother cell and the developing spore. At a late stage of spore maturation, Gcs1p-GFP became concentrated at the spore peripheries. However, Gcs1p-GFP remained soluble during fractionation. Therefore, the localization of Gcs1p-GFP to the spore peripheries as sporulation progresses perhaps occurs through association with a secondary protein.
Immunoblot analysis of protein extracts from cells expressing GCS1-GFP revealed that Gcs1p-GFP changes its electrophoretic mobility during meiosis, as was seen for Spo14p (65). The change in electrophoretic mobility of Spo14p isolated from sporulating cells was shown to be due to phosphorylation (65). Furthermore, genetic analysis suggested that this modification is specifically important for sporulation (68). Although we cannot rule out the possibility that the change in mobility of Gcs1p-GFP is due to proteolytic breakdown or other modifications, we favor the idea that like Spo14p, Gcs1p is modified by phosphorylation. This suggests that proteins that function during both vegetative growth and sporulation are modified posttranslationally to carry out sporulation-specific functions. The distinct nature of this posttranslational modification and its generality in modifying proteins for sporulation await future investigation.
Role of endosome-to-vacuole transport during sporulation.
In addition to defects in prospore membrane synthesis, we found that Gcs1p facilitates endosome-to-vacuole transport during sporulation. Furthermore, TEM indicated that there are abnormal membrane structures in gcs1Δ sporulating cells. The role of Gcs1p in endosome-to-vacuole transport during vegetative growth has previously been reported (83); however, other studies have not found any defect in this transport step (26). The difference may be due to the strain background or growth conditions. Nonetheless, our studies suggest that sporulation is more sensitive to a loss of Gcs1p with respect to endosome-to-vacuole trafficking. Recently, Morishita and Engebrecht (43) reported that endocytosis is important for spore wall formation and suggested that trafficking between the plasma membrane and prospore membranes is important during the early stages of sporulation. This raises the possibility that Gcs1p may play a role in this trafficking event as well.
While this defect in endosome-to-vacuole trafficking was apparent in gcs1Δ cells, we found that the involvement of Gcs1p in prospore membrane formation is likely the major contribution of this protein during sporulation. TEM revealed aberrant structures in gcs1Δ mutants indicative of “class E compartments” usually seen in cells lacking any of the 17 class E VPS genes (51, 56, 59). Additional analyses of these mutants during sporulation revealed that sporulation was not impaired in most of the mutants in this class (17). Thus, this endosome-to-vacuole trafficking step does not appear to be required for efficient sporulation, and therefore is unlikely to be the main function of Gcs1p in sporulation.
In conclusion, our findings that Spo14p-generated PA is reduced in the absence of GCS1 and that Gcs1p is stimulated by PA (82) suggest that Gcs1p and Spo14p function together in the developmentally regulated transport step that mediates prospore membrane formation.
Acknowledgments
We thank Aaron M. Neiman (SUNY, Stony Brook) for the generous gift of yeast strains and plasmids as well as helpful discussions throughout this project; Andrew Vershon (Rutgers State University of New Jersey) for providing the lacZ fusion plasmids for gene expression studies; Howard Riezman (University of Geneva) for the anti-Gas1p antibodies; and Grete Adamson and Patricia Kysar for technical assistance in the Electron Microscopy Laboratory, Department of Pathology and Laboratory Medicine, School of Medicine, University of California at Davis. We give special thanks to Masayo Morishita for extensive advice as well as the members of the Engebrecht lab for helpful discussions. We additionally thank Masayo Morishita, Aimee Jaramillo-Lambert, Rima Mendonsa, and William J. Rohrbach for critically reading the manuscript.
This work was supported by National Institutes of Health (NIH) research grant GM66124 to J.E.
REFERENCES
- 1.Babst, M., D. J. Katzmann, W. B. Snyder, B. Wendland, and S. D. Emr. 2002. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell 3:283-289. [DOI] [PubMed] [Google Scholar]
- 2.Babst, M., B. Wendland, E. J. Estepa, and S. D. Emr. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17:2982-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bankaitis, V. A., J. R. Aitken, A. E. Cleves, and W. Dowhan. 1990. An essential role for a phospholipid transfer protein in yeast Golgi function. Nature 347:561-562. [DOI] [PubMed] [Google Scholar]
- 4.Bankaitis, V. A., D. E. Malehorn, S. D. Emr, and R. Greene. 1989. The Saccharomyces cerevisiae SEC14 gene encodes a cytosolic factor that is required for transport of secretory proteins from the yeast Golgi complex. J. Cell Biol. 108:1271-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blader, I. J., M. J. Cope, T. R. Jackson, A. A. Profit, A. F. Greenwood, D. G. Drubin, G. D. Prestwich, and A. B. Theibert. 1999. GCS1, an Arf guanosine triphosphatase-activating protein in Saccharomyces cerevisiae, is required for normal actin cytoskeletal organization in vivo and stimulates actin polymerization in vitro. Mol. Biol. Cell 10:581-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briza, P., M. Breitenbach, A. Ellinger, and J. Segall. 1990. Isolation of two developmentally regulated genes involved in spore wall maturation in Saccharomyces cerevisiae. Genes Dev. 4:1775-1789. [DOI] [PubMed] [Google Scholar]
- 7.Byers, B. 1981. Cytology of the yeast life cycle, p. 59-96. In E. Strathern, W. Jones, and J. R. Broach (ed.), Molecular and cellular biology of the yeast Saccharomyces: life cycle and inheritance. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 8.Chen, Y. G., A. Siddhanta, C. D. Austin, S. M. Hammond, T. C. Sung, M. A. Frohman, A. J. Morris, and D. Shields. 1997. Phospholipase D stimulates release of nascent secretory vesicles from the trans-Golgi network. J. Cell Biol. 138:495-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699-705. [DOI] [PubMed] [Google Scholar]
- 10.Cleves, A. E., T. P. McGee, E. A. Whitters, K. M. Champion, J. R. Aitken, W. Dowhan, M. Goebl, and V. A. Bankaitis. 1991. Mutations in the CDP-choline pathway for phospholipid biosynthesis bypass the requirement for an essential phospholipid transfer protein. Cell 64:789-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coe, J. G. S., L. E. Murray, and I. W. Dawes. 1994. Identification of a sporulation-specific promoter regulating divergent transcription of two novel sporulation genes in Saccharomyces cerevisiae. Mol. Gen. Genet. 244:661-672. [DOI] [PubMed] [Google Scholar]
- 12.Costa, R., D. T. Warren, and K. R. Ayscough. 2005. Lsb5p interacts with actin regulators Sla1p and Las17p, ubiquitin and Arf3p to couple actin dynamics to membrane trafficking processes. Biochem. J. 387:649-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis, L., M. Barbera, A. McDonnell, K. McIntyre, R. Sternglanz, Q. Jin, J. Loidl, and J. Engebrecht. 2001. The Saccharomyces cerevisiae MUM2 gene interacts with the DNA replication machinery and is required for meiotic levels of double strand breaks. Genetics 157:1179-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donaldson, A. D., and J. V. Kilmartin. 1996. Spc42p: a phosphorylated component of the S. cerevisiae spindle pole body (SPB) with an essential function during SPB duplication. J. Cell Biol. 132:887-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ella, K. M., J. W. Dolan, C. Qi, and K. E. Meier. 1996. Characterization of Saccharomyces cerevisiae deficient in expression of phospholipase D. Biochem. J. 314:15-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elledge, S. J., and R. W. Davis. 1988. A family of versatile centromeric vectors designed for use in the sectoring-shuffle mutagenesis assay in Saccharomyces cerevisiae. Gene 70:303-312. [DOI] [PubMed] [Google Scholar]
- 17.Enyenihi, A. H., and W. S. Saunders. 2003. Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics 163:47-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faundez, V., J. T. Horng, and R. B. Kelly. 1997. ADP ribosylation factor 1 is required for synaptic vesicle budding in PC12 cells. J. Cell Biol. 138:505-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felder, T., E. Bogengruber, S. Tenreiro, A. Ellinger, I. Sa-Correia, and P. Briza. 2002. Dtr1p, a multidrug resistance transporter of the major facilitator superfamily, plays an essential role in spore wall maturation in Saccharomyces cerevisiae. Eukaryot. Cell 1:799-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibson, T. J., M. Hyvonen, A. Musacchio, M. Saraste, and E. Birney. 1994. PH domain: the first anniversary. Trends Biochem. Sci. 19:349-353. [DOI] [PubMed] [Google Scholar]
- 21.Hammond, S. M., Y. M. Altshuller, T. C. Sung, S. A. Rudge, K. Rose, J. Engebrecht, A. J. Morris, and M. A. Frohman. 1995. Human ADP-ribosylation factor-activated phosphatidylcholine-specific phospholipase D defines a new and highly conserved gene family. J. Biol. Chem. 270:29640-29643. [DOI] [PubMed] [Google Scholar]
- 22.Hammond, S. M., J. M. Jenco, S. Nakashima, K. Cadwallader, Q. Gu, S. Cook, Y. Nozawa, G. D. Prestwich, M. A. Frohman, and A. J. Morris. 1997. Characterization of two alternately spliced forms of phospholipase D1. Activation of the purified enzymes by phosphatidylinositol 4,5-bisphosphate, ADP-ribosylation factor, and Rho family monomeric GTP-binding proteins and protein kinase C-alpha. J. Biol. Chem. 272:3860-3868. [DOI] [PubMed] [Google Scholar]
- 23.Hill, J. E., A. M. Myers, T. J. Koerner, and A. Tzagoloff. 1986. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast 2:163-167. [DOI] [PubMed] [Google Scholar]
- 24.Hollingsworth, N. M., L. Goetsch, and B. Byers. 1990. The HOP1 gene encodes a meiosis-specific component of yeast chromosomes. Cell 61:73-84. [DOI] [PubMed] [Google Scholar]
- 25.Honigberg, S. M., C. Conicella, and R. E. Espositio. 1992. Commitment to meiosis in Saccharomyces cerevisiae: involvement of the SPO14 gene. Genetics 130:703-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, C. F., C. C. Chen, L. Tung, L. M. Buu, and F. J. Lee. 2002. The yeast ADP-ribosylation factor GAP, Gcs1p, is involved in maintenance of mitochondrial morphology. J. Cell Sci. 115:275-282. [DOI] [PubMed] [Google Scholar]
- 27.Huang, C. F., Y. W. Liu, L. Tung, C. H. Lin, and F. J. Lee. 2003. Role for Arf3p in development of polarity, but not endocytosis, in Saccharomyces cerevisiae. Mol. Biol. Cell 14:3834-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ireland, L. S., G. C. Johnston, M. A. Drebot, N. Dhillon, A. J. DeMaggio, M. F. Hoekstra, and R. A. Singer. 1994. A member of a novel family of yeast “Zn-finger” proteins mediates the transition from stationary phase to cell proliferation. EMBO J. 13:3812-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwamoto, M. A., S. R. Fairclough, S. A. Rudge, and J. Engebrecht. 2005. Saccharomyces cerevisiae Sps1p regulates trafficking of enzymes required for spore wall synthesis. Eukaryot. Cell 4:536-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kahn, R. A., and A. G. Gilman. 1984. ADP-ribosylation of Gs promotes the dissociation of its alpha and beta subunits. J. Biol. Chem. 259:6235-6240. [PubMed] [Google Scholar]
- 32.Klarlund, J. K., A. Guilherme, J. J. Holik, J. V. Virbasius, A. Chawla, and M. P. Czech. 1997. Signaling by phosphoinositide-3,4,5-trisphosphate through proteins containing pleckstrin and Sec7 homology domains. Science 275:1927-1930. [DOI] [PubMed] [Google Scholar]
- 33.Kranz, A., A. Kinner, and R. Kolling. 2001. A family of small coiled-coil-forming proteins functioning at the late endosome in yeast. Mol. Biol. Cell 12:711-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ktistakis, N. T., H. A. Brown, M. G. Waters, P. C. Sternweis, and M. G. Roth. 1996. Evidence that phospholipase D mediates ADP ribosylation factor-dependent formation of Golgi coated vesicles. J. Cell Biol. 134:295-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kupiec, M., B. Byers, R. E. Esposito, and A. P. Mitchell. 1997. Meiosis and sporulation in Saccharomyces cerevisiae, p. 889-1036. In J. R. Pringle, J. R. Broach, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces cell cycle, vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 36.Lee, F. J., L. A. Stevens, Y. L. Kao, J. Moss, and M. Vaughan. 1994. Characterization of a glucose-repressible ADP-ribosylation factor 3 (ARF3) from Saccharomyces cerevisiae. J. Biol. Chem. 269:20931-20937. [PubMed] [Google Scholar]
- 37.Lewis, S. M., P. P. Poon, R. A. Singer, G. C. Johnston, and A. Spang. 2004. The ArfGAP Glo3 is required for the generation of COPI vesicles. Mol. Biol. Cell 15:4064-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, Y. W., C. F. Huang, K. B. Huang, and F. J. Lee. 2005. Role for Gcs1p in regulation of Arl1p at trans-Golgi compartments. Mol. Biol. Cell 16:4024-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 40.Lynn, R. R., and P. T. Magee. 1970. Development of the spore wall during ascospore formation in Saccharomyces cerevisiae. J. Cell Biol. 44:688-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mollenhauer, H. H. 1964. Plastic embedding mixtures for use in electron microscopy. Stain Technol. 39:111-114. [PubMed] [Google Scholar]
- 42.Moreno-Borchart, A. C., K. Strasser, M. G. Finkbeiner, A. Shevchenko, A. Shevchenko, and M. Knop. 2001. Prospore membrane formation linked to the leading edge protein (LEP) coat assembly. EMBO J. 20:6946-6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morishita, M., and J. Engebrecht. 2005. End3p-mediated endocytosis is required for spore wall formation in Saccharomyces cerevisiae. Genetics 170:1561-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moss, J., and M. Vaughan. 1998. Molecules in the ARF orbit. J. Biol. Chem. 273:21431-21434. [DOI] [PubMed] [Google Scholar]
- 45.Nakanishi, H., P. de los Santos, and A. M. Neiman. 2004. Positive and negative regulation of a SNARE protein by control of intracellular localization. Mol. Biol. Cell 15:1802-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakanishi, H., M. Morishita, C. E. Schwartz, A. Coluccio, J. Engebrecht, and A. M. Neiman. Phospholipase D and the SNARE, Sso1p, are required to promote vesicle fusion during sporulation in yeast. Submitted for publication. [DOI] [PubMed]
- 47.Neiman, A. M. 1998. Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J. Cell Biol. 140:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nickas, M. E., A. E. Diamond, M. J. Yang, and A. M. Neiman. 2004. Regulation of spindle pole function by an intermediary metabolite. Mol. Biol. Cell 15:2606-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nie, Z., D. S. Hirsch, and P. A. Randazzo. 2003. Arf and its many interactors. Curr. Opin. Cell Biol. 15:396-404. [DOI] [PubMed] [Google Scholar]
- 50.Park, S. K., J. J. Provost, C. D. Bae, W. T. Ho, and J. H. Exton. 1997. Cloning and characterization of phospholipase D from rat brain. J. Biol. Chem. 272:29263-29271. [DOI] [PubMed] [Google Scholar]
- 51.Piper, R. C., A. A. Cooper, H. Yang, and T. H. Stevens. 1995. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J. Cell Biol. 131:603-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poon, P. P., D. Cassel, I. Huber, R. A. Singer, and G. C. Johnston. 2001. Expression, analysis, and properties of yeast ADP-ribosylation factor (ARF) GTPase activating proteins (GAPs) Gcs1 and Glo3. Methods Enzymol. 329:317-324. [DOI] [PubMed] [Google Scholar]
- 53.Poon, P. P., D. Cassel, A. Spang, M. Rotman, E. Pick, R. A. Singer, and G. C. Johnston. 1999. Retrograde transport from the yeast Golgi is mediated by two ARF GAP proteins with overlapping function. EMBO J. 18:555-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poon, P. P., S. F. Nothwehr, R. A. Singer, and G. C. Johnston. 2001. The Gcs1 and Age2 ArfGAP proteins provide overlapping essential function for transport from the yeast trans-Golgi network. J. Cell Biol. 155:1239-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poon, P. P., X. Wang, M. Rotman, I. Huber, E. Cukierman, D. Cassel, R. A. Singer, and G. C. Johnston. 1996. Saccharomyces cerevisiae Gcs1 is an ADP-ribosylation factor GTPase-activating protein. Proc. Natl. Acad. Sci. USA 93:10074-10077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raiborg, C., T. E. Rusten, and H. Stenmark. 2003. Protein sorting into multivesicular endosomes. Curr. Opin. Cell Biol. 15:446-455. [DOI] [PubMed] [Google Scholar]
- 57.Randazzo, P. A. 1997. Resolution of two ADP-ribosylation factor 1 GTPase-activating proteins from rat liver. Biochem. J. 324:413-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Randazzo, P. A., and D. S. Hirsch. 2004. Arf GAPs: multifunctional proteins that regulate membrane traffic and actin remodelling. Cell Signal. 16:401-413. [DOI] [PubMed] [Google Scholar]
- 59.Raymond, C. K., I. Howald-Stevenson, C. A. Vater, and T. H. Stevens. 1992. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell 3:1389-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Riedel, C. G., M. Mazza, P. Maier, R. Korner, and M. Knop. 2005. Differential requirement for phospholipase D/SPO14 and its novel interactor SMA1 for regulation of exocytotic vesicle fusion in yeast meiosis. J. Biol. Chem. 280:37846-37852. [DOI] [PubMed] [Google Scholar]
- 61.Rose, K., S. A. Rudge, M. A. Frohman, A. J. Morris, and J. Engebrecht. 1995. Phospholipase D signaling is essential for meiosis. Proc. Natl. Acad. Sci. USA 92:12151-12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 63.Rothstein, R. J. 1983. One-step gene disruption in yeast. Methods Enzymol. 101:202-211. [DOI] [PubMed] [Google Scholar]
- 64.Rudge, S. A., M. M. Cavenagh, R. Kamath, V. A. Sciorra, A. J. Morris, R. A. Kahn, and J. Engebrecht. 1998. ADP-ribosylation factors do not activate yeast phospholipase Ds but are required for sporulation. Mol. Biol. Cell 9:2025-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rudge, S. A., A. J. Morris, and J. Engebrecht. 1998. Relocalization of phospholipase D activity mediates membrane formation during meiosis. J. Cell Biol. 140:81-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rudge, S. A., T. R. Pettitt, C. Zhou, M. J. Wakelam, and J. A. Engebrecht. 2001. SPO14 separation-of-function mutations define unique roles for phospholipase D in secretion and cellular differentiation in Saccharomyces cerevisiae. Genetics 158:1431-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rudge, S. A., V. A. Sciorra, M. Iwamoto, C. Zhou, T. Strahl, A. J. Morris, J. Thorner, and J. Engebrecht. 2004. Roles of phosphoinositides and of Spo14p (phospholipase D)-generated phosphatidic acid during yeast sporulation. Mol. Biol. Cell 15:207-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rudge, S. A., C. Zhou, and J. Engebrecht. 2002. Differential regulation of Saccharomyces cerevisiae phospholipase D in sporulation and Sec14-independent secretion. Genetics 160:1353-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sheff, M. A., and K. S. Thorn. 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21:661-670. [DOI] [PubMed] [Google Scholar]
- 70.Smits, G. J., H. van den Ende, and F. M. Klis. 2001. Differential regulation of cell wall biogenesis during growth and development in yeast. Microbiology 147:781-794. [DOI] [PubMed] [Google Scholar]
- 71.Stearns, T., R. A. Kahn, D. Botstein, and M. A. Hoyt. 1990. ADP ribosylation factor is an essential protein in Saccharomyces cerevisiae and is encoded by two genes. Mol. Cell. Biol. 10:6690-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stearns, T., M. C. Willingham, D. Botstein, and R. A. Kahn. 1990. ADP-ribosylation factor is functionally and physically associated with the Golgi complex. Proc. Natl. Acad. Sci. USA 87:1238-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Szafer, E., E. Pick, M. Rotman, S. Zuck, I. Huber, and D. Cassel. 2000. Role of coatomer and phospholipids in GTPase-activating protein-dependent hydrolysis of GTP by ADP-ribosylation factor-1. J. Biol. Chem. 275:23615-23619. [DOI] [PubMed] [Google Scholar]
- 74.Szafer, E., M. Rotman, and D. Cassel. 2001. Regulation of GTP hydrolysis on ADP-ribosylation factor-1 at the Golgi membrane. J. Biol. Chem. 276:47834-47839. [DOI] [PubMed] [Google Scholar]
- 75.Tanigawa, G., L. Orci, M. Amherdt, M. Ravazzola, J. B. Helms, and J. E. Rothman. 1993. Hydrolysis of bound GTP by ARF protein triggers uncoating of Golgi-derived COP-coated vesicles. J. Cell Biol. 123:1365-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vershon, A. K., N. M. Hollingsworth, and A. D. Johnson. 1992. Meiotic induction of the yeast HOP1 gene is controlled by positive and negative regulatory sites. Mol. Cell. Biol. 12:3706-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vida, T. A., and S. D. Emr. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128:779-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 79.Waksman, M., Y. Eli, M. Liscovitch, and J. E. Gerst. 1996. Identification and characterization of a gene encoding phospholipase D activity in yeast. J. Biol. Chem. 271:2361-2364. [DOI] [PubMed] [Google Scholar]
- 80.West, M. A., N. A. Bright, and M. S. Robinson. 1997. The role of ADP-ribosylation factor and phospholipase D in adaptor recruitment. J. Cell Biol. 138:1239-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu, L., M. Ajimura, R. Padmore, C. Klein, and N. Kleckner. 1995. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:6572-6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yanagisawa, L. L., J. Marchena, Z. Xie, X. Li, P. P. Poon, R. A. Singer, G. C. Johnston, P. A. Randazzo, and V. A. Bankaitis. 2002. Activity of specific lipid-regulated ADP ribosylation factor-GTPase-activating proteins is required for Sec14p-dependent Golgi secretory function in yeast. Mol. Biol. Cell 13:2193-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang, C. J., J. B. Bowzard, A. Anido, and R. A. Kahn. 2003. Four ARF GAPs in Saccharomyces cerevisiae have both overlapping and distinct functions. Yeast 20:315-330. [DOI] [PubMed] [Google Scholar]