Abstract
Current serologic tests provide the foundation for diagnosis of hepatitis A and hepatitis A virus (HAV) infection. Recent advances in methods to identify and characterize nucleic acid markers of viral infections have provided the foundation for the field of molecular epidemiology and increased our knowledge of the molecular biology and epidemiology of HAV. Although HAV is primarily shed in feces, there is a strong viremic phase during infection which has allowed easy access to virus isolates and the use of molecular markers to determine their genetic relatedness. Molecular epidemiologic studies have provided new information on the types and extent of HAV infection and transmission in the United States. In addition, these new diagnostic methods have provided tools for the rapid detection of food-borne HAV transmission and identification of the potential source of the food contamination.
INTRODUCTION
The disease described as “jaundice” in ancient Greek, Roman, and Chinese literature probably was viral hepatitis. A viral etiology was postulated as the cause of certain forms of jaundice as early as 1912, and the term “infectious hepatitis” was used because the disease often occurred in epidemics (43). Hepatitis A, a term first introduced by Krugman et al. in 1967 (138), is now known to be caused by infection with hepatitis A virus (HAV), one of five viruses, each belonging to a different family, whose primary site of replication is the liver.
Early epidemiological studies further characterized hepatitis into infectious and serum forms, based on patterns of disease transmission (17, 82, 102, 103, 104). Epidemiologic and transmission studies with humans showed that infectious hepatitis, or hepatitis A, was transmitted primarily by the fecal-oral route (136, 137, 138). In 1973, HAV was identified in the stools of infected persons (80), which eventually led to development of diagnostic tests, propagation in cell culture, molecular characterization, and development of a vaccine (80, 193).
HEPATITIS A VIRUS INFECTION
Clinical Features
Infection by HAV is generally self-limited and can produce effects that range from a lack of symptoms to death from fulminant hepatitis. The likelihood of clinically apparent disease associated with HAV infection increases with age. In children <6 years of age, most infections (70%) are asymptomatic (98), and if illness does occur, it is usually anicteric. Among older children and adults, infection is usually symptomatic, with jaundice occurring in >70% of patients (141). After an average incubation period of 28 days (range, 15 to 50 days), most HAV-infected persons developed nonspecific constitutional signs and symptoms followed by gastrointestinal symptoms. Typically, these include fever, malaise, anorexia, nausea, abdominal discomfort, dark urine, and jaundice, all of which usually last <2 months. There is no evidence of chronic liver disease or persistent infection following infection. However, 15 to 20% of the patients may have prolonged or relapsing disease lasting up to 6 months (93, 218), and HAV has been detected in serum for as long as 6 to 12 months after infection (19).
Fulminant hepatitis is a rare complication of hepatitis A. The risk of acute liver failure ranges from 0.015 to 0.5%, and the highest rates occur among young children and older adults with underlying chronic liver disease (2, 31). In a large epidemic in Shanghai, China, involving over 300,000 adolescents and young adults, 47 deaths (0.015%) were reported (100, 263). Other than older age and underlying chronic liver disease, no other host factors have been identified as predisposing patients to fulminant hepatitis A (250). Among persons referred to a tertiary care center with acute hepatic failure in the United States, 20 of 295 (6.8%) had hepatitis A (210). No single viral factor has been associated with fulminant disease. Reported findings among persons with fulminant disease include lower viral load (198) and nucleotide and/or amino acids substitutions in the 5′ untranslated region (5′UTR), P2 region, and P3 region of the HAV genome (86; CDC, unpublished data).
Pathogenesis and Natural History of HAV Infection
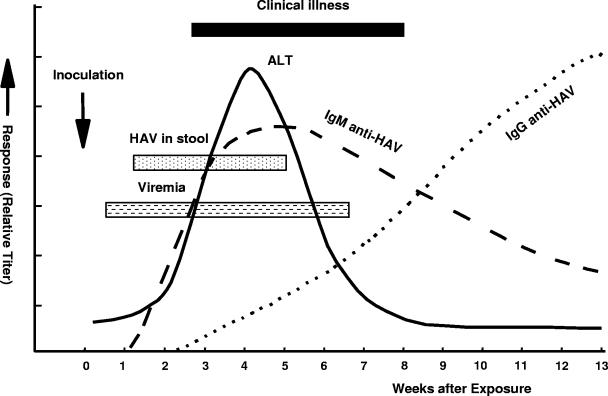
The pathogenetic events that occur during HAV infection have been determined in experimental infection of nonhuman primates and natural infection of humans (Fig. 1). HAV is primarily hepatotropic; it replicates in the liver, produces a viremia, and is excreted in bile and shed in the stools of infected persons. Feces can contain up to 109 infectious virions per gram and is the primary source of HAV infection (221, 241). Peak fecal excretion, and hence infectivity, occurs before the onset of jaundice, symptoms, or elevation of liver enzymes (221, 241) and declines after jaundice appears. Compared to adults, children and infants can shed HAV for longer periods, i.e., up to several months after the onset of clinical illness (35, 241). Fecal shedding of HAV has been shown to occur as late as 6 months after diagnosis of infection in premature infants (205).
FIG. 1.
Virologic, immunologic and biochemical events during the course of experimental hepatitis A virus infection in chimpanzees inoculated intravenously with human HAV, strain HLD2. ALT, alanine aminotransferase. (Adapted from reference 157 with permission of the publisher.)
Viremia occurs within 1 to 2 weeks after HAV exposure and persists through the period of liver enzyme elevation, based on studies in humans and experimentally infected chimpanzees (8, 48, 91, 137, 145, 157). Virus concentrations in serum are 2 to 3 log10 units lower than those in stool (19, 48, 145). An analysis of serum specimens collected prospectively during human and chimpanzee HAV infection showed that HAV RNA was present for ∼3 to 4 weeks before the onset of jaundice and that virus concentrations were highest during the period that precedes onset of liver enzyme elevations (19). Viremia may be present for a much longer period during the convalescent phase of hepatitis A than was previously appreciated (19, 84, 264), although virus concentration is lowest during this period.
The virus is also shed in saliva in most hepatitis A patients. In experimentally infected marmosets (156, 187), the viral load appears to be 1 to 3 log10 units lower than that found in serum (156). However, no epidemiological data suggest that saliva is a significant source of HAV transmission.
A humoral immune response to HAV structural proteins occurs prior to onset of symptoms. Immunoglobulin M (IgM) antibodies to HAV (IgM anti-HAV) are detectable at or prior to onset of clinical illness, decline in about 3 to 6 months, and become undetectable by commercially available diagnostic tests (127). IgG antibodies to HAV (IgG anti-HAV) appear soon after IgM, persist for years after infection, and confer lifelong immunity (220). IgA is also produced during infection for a limited time (4, 152). The role of IgA antibodies in the response against HAV is still unknown. Unlike other Picornaviridae family members, HAV does not seem to elicit an effective intestinal immune response (229).
IgG and IgA anti-HAV are detected in serum, saliva, urine, and feces (39, 60, 115, 123, 151, 152, 177, 178, 183, 214). Saliva tests have been reported as an alternative to conventional serum testing for anti-HAV due to their simplicity of sample collection (115, 177, 178). Several studies have demonstrated the benefits of implementing saliva testing as screening tool in outbreak investigations and epidemiological studies (110, 115, 140). However, the sensitivity of detecting anti-HAV in saliva is 1 to 3 log10 units lower than that with serum (156, 177, 178; CDC, unpublished data).
Antibodies against nonstructural proteins are also produced, although their role in maintenance of immunity is probably less important than that of antibodies to capsid antigens due to their low concentration and lack of neutralization capacity. Antibodies to nonstructural proteins have been detected in humans and experimentally infected chimpanzees but are absent in vaccinated individuals (126, 202). However, because of what appears to be a variable host antibody response during HAV replication, a diagnostic test for these antibodies, which could be used to complement current anti-HAV testing and differentiate previously infected from vaccinated persons, has not been developed (201, 233).
HEPATITIS A VIRUS
HAV is a nonenveloped RNA virus 27 to 32 nm diameter in size, with an icosahedral symmetry, which belongs to the genus Hepatovirus of the Picornaviridae family. Unlike other members of the family, HAV requires a long adaptation period to grow in cell culture, replicates slowly, and rarely produces a cytopathic effect (57, 107, 146). HAV is stable in the environment for at least 1 month (162) and is more resistant to heating and chlorine inactivation than is poliovirus. Inactivation of HAV requires heating foods to >85°C for 1 min, and disinfection of surfaces requires 1 min of contact with a 1:100 dilution of sodium hypochlorite (i.e., household bleach), whereas poliovirus is inactivated at 72°C for 15 s and by treatment with a 1:125 dilution of bleach for 30 s (159, 234, 257).
Genomic Organization
HAV has a positive-polarity, single-stranded 7.5-kb genome (45, 46, 171) that is organized similarly to those of the other picornaviruses (Fig. 2). The 5′UTR is 734 to 740 nucleotides (nt) long and has a covalently linked virus-specific protein (VPg) rather than a cap structure (259). Translation occurs in a cap-independent fashion under control of an internal ribosome entry site located within the 5′UTR (21). The translation terminator sequence, the 3′UTR of 40 to 80 nucleotides, has a poly(A) tract (46) (Table 1). The remainder of the genome is composed of a single open reading frame with three distinct regions (P1, P2, and P3) and is translated as a single polyprotein of 2,225 to 2,227 amino acids (10, 46). This polyprotein subsequently undergoes cleavage mediated by a viral protease (3Cpro) (211), which results in production of four capsid and several nonstructural proteins (Fig. 2; Table 1).
FIG. 2.
Schematic representation of the HAV genome organization, translation products, and regions used for amplification. The area encoding the polyprotein is represented by solid box and the proposed cleavage sites by vertical lines. Regions commonly used for PCR amplifications are as follows: region 1, C terminus of VP3 region (nt 2,020 to nt 2,226); region 2, N terminus of VP1 region (nt 2,172 to nt 2,415); region 3, VP1/P2A junction region (nt 2,984 to nt 3,217); region 4, VP1-P2B region (nt 2,896 to nt 3,289); region 5, entire VP1 region (nt 2,172 to nt 3,125); and region 6, VP3-P2B region (nt 2,133 to nt 3,289). Nucleotide position numbering is according to the HM175 sequence (46).
TABLE 1.
HAV genome organization and open reading framea
| HAV genome segment | Nucleotides | Polyprotein amino acids | Known function(s) |
|---|---|---|---|
| 5′UTR | 1-735 (741)b | Unclear, translation initiation | |
| P1 | 735-3029 | 1-765 | Capsid Proteins |
| 1A | 735-803 | 1-23 | VP4 |
| 1B | 804-1469 | 24-245 | VP2 |
| 1C | 1470-2207 | 246-491 | VP3 |
| 1D | 2208-3029 | 492-765 | VP1 |
| P2 | 3030-5000 | 766-1422 | Nonstructural Proteins |
| 2A | 3030-3242 | 766-836 | Morphogenesis |
| 2B | 3243-3995 | 837-1087 | ? |
| 2C | 3996-5000 | 1088-1422 | RNA synthesis |
| P3 | 5001-7415 | 1423-2227 | Nonstructural proteins and VPg |
| 3A | 5001-5222 | 1423-1496 | Pre-VPg |
| 3B | 5223-5291 | 1497-1519 | VPg |
| 3C | 5292-5948 | 1520-1738 | Protease |
| 3D | 5949-7415 | 1739-2227 | RNA polymerase |
| 3′UTR | 7416-7478 | Unknown, translation terminator, poly(A) |
HAV Genetic Diversity
HAV displays a high degree of antigenic (amino acid) and genetic (nucleotide) conservation throughout the genome (45, 46, 147, 200, 207). However, enough genetic diversity exists to define several HAV genotypes and subgenotypes (200). The entire nucleic acid sequences of several HAV strains have been determined by molecular cloning (10, 45, 46, 96, 171, 182, 243, 244), and a large number of HAV isolates have been characterized by sequencing of short genome segments. The genomic regions most commonly used to define HAV genotypes include (i) the C terminus of the VP3 region (116), (ii) the N terminus of the VP1 region (5, 112, 199, 203), (iii) the 168-bp junction of the VP1/P2A regions (116, 200, 203), (iv) the 390-bp region of the VP1-P2B regions (112, 170), and (v) the entire VP1 region (52, 167) (Fig. 2; Table 2).
TABLE 2.
Oligonucleotide primers used for PCR amplification of HAV RNA from clinical specimens
| Region (reference[s]) | Primer typea | Primerb | Nucleotide sequence (5′→3′) | Positionc |
|---|---|---|---|---|
| C terminus of VP3 (116) | 2020P | ACAGGTATACAAAGTCAG | 2020-2037 | |
| 2226N | CTCCAGAATCATCTCC | 2226-2211 | ||
| N terminus of VP1 (22, 112, 199,202) | External | 2133P | GTGAATGTTTATCTTTCAGCAAT | 2133-2155 |
| 2451N | GATCTGATGTATGTCTGGATTCT | 2451-2429 | ||
| Internal | 2172P | GCTCCTCTTTATCATGCTATGGAT | 2172-2195 | |
| 2415N | CAGGAAATGTCTCAGGTACTTTCT | 2415-2392 | ||
| VP1/P2A junction (22, 116,200) | External | 2950P | TTGTCTGTCACAGAACAATCAG | 2950-2972 |
| 3308N | AGTCACACCTCTCCAGGAAAACTT | 3308-3285 | ||
| Internal | 2984P | TCCCAGAGCTCCATTGAA | 2984-3001 | |
| 3217N | AGGGGGTGGAAGTACTTCATTTGA | 3217-3193 | ||
| VP1-P2B (112,170) | External | 2870P | GACAGATTCTACATTTGGATTGGT | 2870-2893 |
| 3381N | CCATTTCAAGAGTCCACACACT | 3381-3360 | ||
| Internal | 2896P | CTATTCAGATTGCAAATACAAT | 2896-2918 | |
| 3289N | AACTTCATTATTTCATGCTCCT | 3289-3268 | ||
| Entire VP1 (52,167) | External | 2167P | GTTTTGCTCCTCTTTATCATGCTATG | 2167-2192 |
| 3308N | AGTCACACCTCTCCAGGAAAACTT | 3308-3285 | ||
| Internal | 2172P | GCTCCTCTTTATCATGCTATGGAT | 2172-2195 | |
| 3125N | CCTGCATTCTATATGACTCT | 3125-3106 | ||
| VP3-P2B (CDC, unpublished data) | External | 2020P | ACAGGTATACAAAGTCAG | 2020-2037 |
| 3381N | CCATTTCAAGAGTCCACACACT | 3381-3360 | ||
| Internal | 2133P | GTGAATGTTTATCTTTCAGCAAT | 2133-2155 | |
| 3289N | AACTTCATTATTTCATGCTCCT | 3289-3268 |
External primers were used for first-round amplification, and internal primers were used for second-round nested amplification.
P, positive-sense primer; N, negative-sense primer.
Positions relative to the genome of HAV strain HM175 (accession no. M14707).
When sequence variation within the VP1/P2A junction is used to define genotypes and subgenotypes, genotypes have >15% nucleotide variation between isolates and subgenotypes have 7 to 7.5% nucleotide variation (200). Seven HAV genotypes have been identified; four genotypes (I, II, III, and VII) are of human origin, and three (IV, V, VI) are of simian origin. Genotypes II and VII were initially defined based on a single isolate for each (34, 200). However, further investigations have reclassified genotype VII as a subgenotype of genotype II (52, 154). Genotypes I and III are the most prevalent genotypes isolated from humans.
The three simian genotypes were each defined by unique nucleotide sequences from the P1 regions of HAV strains recovered from species of Old World monkeys. In addition, all simian HAVs have a distinct signature sequence at the VP3/VP1 junction which distinguishes these strains from human HAVs (20, 167, 248). Genotype IV was recovered from a cynomolgus macaque (Macaca fasicularis) imported from the Philippines (167). The prototype strain of genotype V, AGM27, was isolated from an African green monkey (Cercopithecus aethiops) imported from Kenya (248). Genotype VI was also isolated from a cynomolgus macaque (M. fasicularis) imported from Indonesia (167, 200).
HAV Proteins
Although HAV was successfully adapted to cell culture 25 years ago (193), its protein components have not been completely defined (107, 146). Infected cells contain low titers of virus, and consequently protein chemistry has been limited. The P1 region encodes the three major proteins of the viral capsid, i.e., VP1, VP2, and VP3 (90). A fourth viral capsid protein (VP4), which is essential for virion formation, is not detected in mature viral particles. Capsid proteins are primarily cleaved from the precursor polyprotein by the viral protease 3C (211), which is encoded in the P3 region. However, it remains unclear how the VP2/VP4 cleavage occurs. The native conformation of the VP1 and VP3 capsid proteins forms a single, dominant, antigenic epitope on the viral surface, which elicits a neutralizing antibody response. The predicted functions of nonstructural proteins encoded by the P2 and P3 regions are RNA synthesis and virion formation. VPg is also encoded in the P3 region (259) (Table 1; Fig. 2).
Antigenicity and Serotype
Only a single serotype of HAV exists, despite genetic heterogeneity at the nucleotide level. Individuals infected by HAV in one part of the world are protected from reinfection by HAV from other parts of the world. Immune globulin preparations containing anti-HAV, irrespective of their geographic origin, appear to provide protection from disease, and vaccines prepared from virus isolates originating in Australia or Costa Rica protect from infection worldwide (113, 172).
The antigenic structure of the virus is relatively simple, with a restricted number of overlapping epitopes combining to form a single dominant antigenic site that interacts with virus-neutralizing antibodies. These epitopes are highly conformational and are formed by amino acid residues located on more than one capsid protein (168, 185, 186, 228). Convalescent-phase sera obtained from hepatitis A patents are reactive primarily to VP1 and to a lesser extent to VP0 and VP3 (254). Recombinant VP1 fusion protein expressed in Escherichia coli reacted with rabbit anti-HAV serum (180). However, chimpanzees immunized with this recombinant protein produced antibodies that reacted with VP1 of only denatured and not intact virus, and the animals were not protected when they were challenged with wild-type HAV (CDC, unpublished data). Empty particles appear to be antigenically indistinguishable from infectious, RNA-containing virions, suggesting that antigenicity may depend on assembly of the major capsid proteins or smaller capsid precursors. An accurately processed and assembled recombinant HAV polyprotein has been produced, which was able to elicit neutralizing antibodies detected by commercial assays (139, 262).
Naturally occurring antigenic variants of HAV have been observed only among strains isolated from Old World monkeys (167, 248). These viruses are genetically distinct from human HAV isolates and are not recognized by certain monoclonal antibodies produced against human HAV (128, 167). However, simian HAV binds human polyclonal anti-HAV, and chimpanzees immunized with these viruses had an antibody response that was protective against infection with human HAV challenge (78; CDC, unpublished data). Recently, human HAV isolates with capsid amino acid substitutions and deletions in the immunodominant antigenic site were reported (52, 206). However, it is not clear if capsid antigenicity or virus neutralization was affected by these changes.
DIAGNOSTIC APPROACHES TO HAV DETECTION
Detection of HAV-Specific Antibodies
The humoral immune response plays the pivotal role in the diagnosis of HAV infection and the differentiation of hepatitis A from other types of viral hepatitis. There are a number of commercially available assays for the detection of IgM and total anti-HAV (71, 122, 188). IgM, IgA, and IgG anti-HAV are usually present at the onset of symptoms (Fig. 1). Since hepatitis due to HAV infection is clinically indistinguishable from disease caused by other hepatitis viruses (i.e., HBV, HCV, HDV, and HEV), serologic testing is required to make the diagnosis (230).
Antibodies to structural proteins.
Diagnostically, IgM anti-HAV has been used as the primary marker of acute infection (58); it is comprised mainly of antibodies against capsid proteins. A number of methods have been used to detect this virus-specific antibody class, including radioimmunoassay (194), immunochemical staining (109), enzyme-linked immunosorbent assay (61), immunoblotting (254), and dot blot immunogold filtration (214). IgM anti-HAV enzyme immunoassays are available commercially (188) The commercially available diagnostic assays are configured in such a manner that although IgM antibodies may be present for long periods of time, the lower concentrations found 4 to 6 months after the onset of infection do not produce a positive test result (230). However, the current commercially available IgM assays detect antibody for a short period of time in persons recently administered hepatitis A vaccine (261).
Previous (resolved) HAV infection is diagnosed by detection of IgG anti-HAV. However, commercially available assays detect total anti-HAV (both IgG and IgM antibodies). The presence of total anti-HAV and the absence of IgM anti-HAV can be used to differentiate between past and current infections.
Antibodies to structural proteins are produced following immunization with hepatitis A vaccine. A small proportion (8 to 20%) of vaccinated persons have a transient IgM anti-HAV response (148, 216, 219). IgG anti-HAV is produced by all successfully immunized persons (148, 216, 219). However, unless they are modified, commercially available tests for total anti-HAV are not sensitive enough to detect antibody concentrations in a significant proportion of immunized persons, especially several years after immunization (28, 148).
Antigen Detection
Cell culture propagation.
HAV has been grown in several cell types of human and nonhuman origins, including primary and secondary African green monkey kidney cells (59, 253) and fetal rhesus monkey kidney cells (83). In contrast to most picornaviruses, HAV of human origin requires an extensive adaptation period before it grows in cell culture, and once adapted, HAV produces a persistent infection and becomes attenuated, as shown by not producing disease in experimentally inoculated nonhuman primates (81). In addition, relatively low concentrations of virus and viral antigen are produced compared to other picornaviruses. Mutations in viral nucleic acid may play a major role in the adaptation of HAV in cell culture (47, 76, 77, 88) and attenuation (79, 81). HAV replicates in cell culture without cytopathic signs of infection and without apparent host cell damage. Because of the lack of a cytopathic effect in cell culture, immunological assays are required to detect HAV antigen (217). Methods commonly used to quantitate infectivity include radioimmunofocus assay (144), fluorescent focus assay, in situ radioimmunoassay (217), and in situ hybridization (121).
Cytopathic variants of HAV have been observed in selected cell culture systems. These cytopathic viruses produce an acute rather than persistent infection (55, 173). The replication cycle of these variants is shorter (2 to 3 days) than that observed for noncytopathic HAV, and they produce a much higher viral yield (55).
Detection in clinical and environmental samples.
HAV was first visualized in fecal extracts by electron microscopy using homologous antiserum (80), and similar virus-like particles were observed in the sera and livers of marmosets experimentally infected with human HAV (192). HAV antigen has been detected in stool, cell culture, and environmental samples by using radioimmunoassays and enzyme immunoassays (107, 165). Viremia during HAV infection has been documented both by transmission studies (138) and as a result of outbreaks of posttransfusion hepatitis A (176). However, detection of antigen in blood has been difficult (106) because fibronectin can bind to HAV and mask antigenic determinants required for immunological detection (213). HAV capsid polypeptides and viral RNA have been detected in IgM circulating immune complexes isolated from experimentally infected chimpanzees (158).
Molecular Detection Methods
Nucleic acid detection techniques are more sensitive than immunoassays for viral antigen to detect HAV in samples of different origins (e.g., clinical specimens, environmental samples, or food). HAV has been detected with techniques such as restriction fragment length polymorphism (94), single-strand conformational polymorphism (85, 94), Southern blotting (25, 27, 169, 208), nucleic acid sequencing-based amplification (118, 119, 120), nucleic acid hybridization (266), and reverse transcription-PCR (RT-PCR) and antigen capture RT-PCR (56, 57, 116, 189,199, 200). Amplification of viral RNA by RT-PCR is currently the most sensitive and widely used method for detection of HAV RNA.
Purification of viral RNA from clinical and environmental samples is the first step in RT-PCR, and different extraction platforms are often required, depending on the source of the specimen. Early protocols for RNA extraction from serum or stool included proteinase K digestion followed by phenol-chloroform extraction and ethanol precipitation (167, 199). Subsequently, products which used guanidinium thiocyanate-phenol-chloroform (41) became commercially available for extraction of RNA and total nucleic acids (40) and increased the sensitivity and specificity of HAV detection (65, 112, 170). Antigen capture RT-PCR (116) and magnetic beads coated with anti-HAV (124) have been used to separate virus from potential inhibitors of reverse transcription and PCRs that are often found in environmental and stool samples. Automated RNA extraction protocols have been applied to HAV detection. These have increased specimen throughput, are reproducible and reliable, and have detection sensitivity similar to that of manual extraction methods (142; CDC, unpublished data).
The efficiency of reverse transcription of RNA to cDNA can be reduced by the presence of inhibitors in the source material. Engineered reverse transcriptase with no RNase activity (RNase H) is believed to increase transcription efficiency, although naturally occurring enzymes continue to be used for identification of a variety of targets, including HAV, with high specificity and sensitivity (169, 226, 227). Although thermostable enzymes may improve efficiency of the RT reaction by reducing secondary structure and improving priming, their effect on HAV detection has not been evaluated. Specific or random primers can be used for the reverse transcription reaction (226, 227). Random primers along with specific primers have been routinely used in our laboratory for the detection of HAV because of the increased sensitivity and specificity (CDC, unpublished data).
Primer pairs (Table 2; Fig. 2) spanning the desired genomic region are used to amplify cDNA. Nested PCR, where products obtained from first-round PCR are used as a template for a second round of PCR, has been used to amplify HAV from clinical and environmental samples where the viral load is expected to be low (19, 72, 92, 149, 169). Analysis of the PCR product by probe hybridization also has been shown to increase the sensitivity of detection (62, 95, 116).
The development of single-step RT-PCR methods, in which reverse transcription and PCR are performed together, has considerably reduced the time and handling during cDNA synthesis (72, 149). However, this method appears to reduce detection sensitivity by up to 1 log unit compared to the two-step RT-PCR method (CDC, unpublished data).
Multiplex RT-PCR, where genome sequences of more than one organism are amplified simultaneously, provides the most efficient way to detect multiple agents in clinical and environmental samples compared to conventional RT-PCR amplification for each agent. This method has been developed for simultaneous detection of HAV and HEV (125) and of HAV, rotavirus, and poliovirus (92, 247).
Real-time PCR, which has revolutionized nucleic acid detection by its high speed, sensitivity, and reproducibility and minimization of contamination, has been applied to the detection and quantification of HAV (53). Real-time PCR has been introduced in our laboratory for rapid analysis of specimens in outbreak situations, with <36 h required from amplification to nucleic acid sequence results. The increased speed of real-time PCR is mainly due to reductions in amplification cycles, elimination of post-PCR detection procedures, and availability of devices for sensitive detection of amplified products. Small amplicons are recommended, and this may also play a role in the speed; however, it has been shown that decreasing the size of the PCR fragment does not necessarily improve PCR performance (174). Molecular beacons (1, 155, 240) and TaqMan-based assays (53) are a few of the chemistries that have been used for real-time PCR detection of HAV. Real-time PCR chemistries use primers and probes with separate fluorogenic labels; the generation of a fluorescent signal depends on the interaction between PCR products and/or probes (24, 53, 155, 240). A DNA-binding fluorophore, SYBR green, has been widely used because of its simplicity (129, 131, 166). The advantage of SYBR green over labeled probe-based detection platforms is the detection of highly variable genome regions for which probe design is often difficult (134). SYBR green interference with nucleic acid sequencing can be eliminated by performing a nested PCR without the fluorophore.
Nucleic Acid Sequencing
Nucleic acid sequencing is performed on PCR products to confirm their specificity and provide the ultimate means to identify and characterize the organism. Nucleic acid sequencing of selected genomic regions of HAV has been used to determine the genetic relatedness of isolates (112, 116, 170, 199, 200). The original nucleic acid sequencing methodology described by Sanger et al. (209), which required independent labeling reactions for each nucleotide and conventional gel electrophoresis, has been replaced by high-throughput methods, including fluorescent dyes for label terminators and capillary arrays for electrophoresis, which have improved sequencing speed and accuracy (160, 196).
Molecular Detection from Water and Food
HAV is stable in the environment, especially when associated with organic matter, and is resistant to low pH and heating (107). These characteristics facilitate the likelihood of transmission by contaminated food and water and also improve the likelihood of detection in environmental samples, including water and sewage (15).
Because HAV grows so slowly in cell culture and because of the generally low levels of contamination in environmental samples, virus detection became feasible only with the availability of sensitive nucleic acid detection methods. However, HAV detection in food has not been included as a part of routine analysis of these outbreaks in most parts of the world (9). The primary reason is that because of the long disease incubation period, implicated foods usually have been consumed or discarded by the time the outbreak is recognized (135). The same holds true for detection of HAV in waterborne outbreaks, unless there is ongoing contamination.
Detection in food.
HAV has been successfully isolated from bivalve mollusks (72, 73, 206), oysters (44, 67), mussels (36), and clams (18, 132, 236, 237). Methods for extraction of HAV RNA from food samples have included the use of guanidine isothiocyanate followed by several precipitation steps with polyethylene glycol (PEG) or ultracentrifugation (72, 73, 237). The addition of viral concentration steps (e.g., high pH or PEG precipitation) followed by RNA extraction and purification using poly(dT) magnetic beads has been shown to increase the sensitivity of HAV detection from bivalve mollusk samples (133).
Detection in water.
Concentration by filtration is the classic approach for viral detection from water and wastewater, including the use of beef extracts to elute virus from filters and PEG for concentration (223). However, these components can interfere with RT-PCR (212). Ultrafiltration has been used as an alternate method for virus concentration, and vortex flow filtration followed by microcentrifugation has been used to overcome the limitations of conventional concentration methods (246).
Nucleic acid hybridization assays using labeled probes were initially used to detect HAV in contaminated water (121). However, this method had low sensitivity and required several logs of virus for detection. Today, PCR is widely used for HAV detection in environmental samples (62, 95, 124, 212, 246).
MOLECULAR EPIDEMIOLOGY OF HEPATITIS A
Molecular biomarkers have been used to determine the genetic relatedness of organisms, both to identify and track modes and chains of transmission and to characterize the evolution of organisms in host populations. New approaches from disciplines such as bioinformatics and molecular evolution have added to the understanding of both the epidemiology of agents within host populations and the dynamics of genetic changes within the agent itself. Molecular epidemiology has played an important role in improving the understanding of HAV infection by identifying sources of infection and the dynamics of virus evolution.
Overview of Hepatitis A Epidemiology
Worldwide, the endemicity of HAV infection varies according to regional hygienic standards; the highest prevalence of infection occurs in regions with the lowest socioeconomic levels (12, 99). The United States generally has a low endemicity of HAV infection, although high rates of infection occur in certain populations (11, 28, 32). Approximately every decade, epidemics of hepatitis A have occurred in the United States; the last occurred in 1995. Although the incidence of hepatitis A has declined over the past decade, primarily because of hepatitis A immunization (13, 14, 23, 163, 252, 255), it remains a frequently reported disease. In 2003, 7,653 cases of hepatitis A were reported to CDC (30), which, when corrected for underreporting and asymptomatic infections (7, 255), represents an estimated 36,700 cases and 79,600 infections.
Modes of Transmission and Sources of HAV Infection
The most common reported source of infection is household or other close contact with an infected person (11, 28, 32). Other potential sources of infection include men having sex with men (MSM), travel to countries where HAV is endemic, and illicit drug use. Contaminated food and water are an infrequent source of infection, although they have been associated with outbreaks. On rare occasions, HAV infection has been transmitted by transfusion of blood or blood products (145, 224).
Personal contact.
Most transmission occurs among close contacts in households and extended-family settings (225); this mode accounts for at least 25% of infections. No identifiable source for infection is reported for 40 to 50% of cases, and personal contact with an unidentified source shedding HAV is likely to explain most of these cases. A study in Salt Lake City, Utah, showed that 98 of 390 (25%) household contacts of 167 infected cases without an identified infection source had serologic evidence of recent HAV infection (225). Children have the highest incidence of infection, and infected young children with their less scrupulous hygiene probably serve as a major source of transmission (98, 222). Children appear to excrete virus longer than adults (114, 205, 218, 265), which may facilitate transmission, although good epidemiologic studies are lacking in this area.
MSM.
Although early studies did not show a higher prevalence of HAV infection among MSM (238), prospective studies showed a high incidence of infection (49, 54). In addition, hepatitis A outbreaks among MSM have been reported in the United States and Europe (22, 105, 170, 235). HAV isolates with nucleotide sequence identity have been observed within several outbreaks among MSM and between outbreaks which occurred in different geographic locations (22, 105, 170, 235).
Illicit drug use.
Outbreaks of HAV infection among injection drug users (IDUs) have been reported in North America and Scandinavia (97, 101, 143). Several routes of transmission are likely to occur, and these include a combination of person-to-person and percutaneous spread. Injection of drugs is often a group activity, and poor personal hygiene among illicit drug users may be a source of HAV contamination of drugs or drug paraphernalia, as well as direct person-to-person transmission. Fecal contamination of drugs by rectal transportation has been reported but is probably a rare source of infection (143). Percutaneous transmission of HAV by needle sharing can also occur (97).
The complexity of hepatitis A transmission among injection and noninjection drug users is illustrated by an outbreak among methamphetamine users and their contacts in Polk County, Florida, during 2001 and 2002 (252). The significance of IDUs as the source of the outbreak was generally underestimated, since case patients were not willing to admit illicit drug use. However, HAV sequence analysis showed identity or near identity among cases, including cases with no direct links to each other and whose disease onsets varied over time. These findings indicated a limited number of HAV introductions with subsequent person-to-person transmission among IDU networks and their contacts (97, 170). Similar studies in Scandinavia have shown that HAV strains among IDUs involved in outbreaks were significantly different from those of the infected non-drug-using population (97, 143, 232).
International travel.
Persons from regions of low endemicity traveling to regions of high HAV endemicity are at substantial risk for acquiring infection (28, 231). In the United States, about 10% of hepatitis A patients have reported recent travel outside the United States as their only possible source of infection (11, 32, 170). HAV sequences from case patients with history of travel are closely related to the sequence patterns of isolates from countries where the travel occurred (170, 232; CDC, unpublished data).
Food and water.
HAV transmission has been associated with contaminated food and water (63, 112, 175, 263). Outbreaks associated with consumption of mussels (70), clams (18, 100), contaminated lettuce (204), ice slush beverages, raw oysters (67), frozen strawberries (112, 175), blueberries (27), raspberries (195, 197), green onions (3, 63, 260), and other salad items (108, 153, 179) have been reported. The potential for extensive disease transmission is illustrated by the largest recorded hepatitis A outbreak, which occurred from consumption of sewage-contaminated clams and caused illness in 300,000 persons in Shanghai, China (263), and by outbreaks that extended to multiple states in the United States through widespread distribution of HAV-contaminated food (3, 112, 260). Although food- and waterborne outbreaks are often newsworthy, they account for only 2% of the total reported cases in the United States (28, 32).
Waterborne transmission of hepatitis A appears to be less common than transmission by food, and during the past 10 to 15 years contaminated water has not been identified as a source for any cases of hepatitis A in the United States (28, 58). Inadequate sewage management has played an important role in most waterborne outbreaks, which have generally involved contaminated groundwater obtained from wells or rivers (16, 68, 164, 181) and drinking water (125, 190).
Unfortunately, because of the long incubation period of HAV infection, virus detection in food is difficult, unless some of the food was kept or contamination is ongoing. Even more difficult to detect are sporadic cases associated with contaminated food. Only recently have such transmission chains been identified, and then only through the use of molecular epidemiologic investigations (3, 18, 112, 206, 260).
Methods of Molecular Epidemiology
The determination and analysis of molecular biomarkers (e.g., nucleic acid sequencing or phylogenetic analysis) have been used to determine the relatedness of strains from two or more infected persons. These analyses have been greatly facilitated by (i) the ability to easily recover HAV from the serum, rather than stool, of infected persons and (ii) molecular methods (e.g., PCR, DNA sequencing, and sequence analysis) which have been adapted to achieve the rapid throughput of a large number of specimens. These recent advances have resulted in molecular epidemiology becoming an essential tool in the investigation of certain types of hepatitis A outbreaks and in epidemiologic studies of HAV transmission.
The key elements of a molecular epidemiologic analysis include determination of the genotypes and genetic relatedness of HAV isolates obtained from infected persons and a rigorous epidemiologic investigation. To facilitate the molecular analyses of HAV, RNA amplification and nucleic sequencing should be performed on one of the genome regions for which large amounts of comparative data exist (e.g., the VP1-P2B region) (Table 2; Fig. 2).
Distribution of HAV genotypes.
Nucleotide sequence data indicate that HAV genotypes have unique geographic distributions (Table 3; Fig. 3). Genotype I is most prevalent worldwide, and subgenotype IA is more common than IB. Because genotype I is so common, genotyping alone rarely can be used to identify the source of an HAV outbreak or chain of transmission.
TABLE 3.
Geographic distribution of hepatitis A virus genotypes
| Genotype | Host | Distribution of isolates | Reference(s) |
|---|---|---|---|
| IA | Human | United States, Mexico, Canada, Brazil, Argentina, Cuba, Chile, Uruguay, Costa Rica, El Salvador, Spain, former USSR, Russia, Central Asia countries, Estonia, Germany, Greece, The Netherlands, Italy, Denmark, England, Switzerland, Czechoslovakia, Sweden, Norway, France, Albanian, Kosovo, Moldova, China, Korea, Thailand, Israel, Malaysia, India, Japan, North Africa, Tunisia, South Africa | 6, 22, 26, 33, 37, 42,50, 51, 52, 64, 66, 69, 74, 86, 87, 89, 111, 112, 150, 161, 170, 184,191, 199, 200, 203, 206, 232, 239, 242, 245, 249, 251, 256; CDC, unpublished data |
| IB | Human | United States, Brazil, Spain, Germany, Greece, The Netherlands, Italy, Sweden, Norway, France, China, Thailand, Israel, Egypt, Jordan, Iraq, Japan, North Africa, Tunisia, South Africa, Sierra Leone, Nigeria, Morocco, Australia | 37, 38, 46, 51, 52, 64, 170, 200,206, 242, 245, 249, 251, 256, 262; CDC, unpublished data |
| II | Human | The Netherlands, France, Sierra Leone | 34, 154, 200,245 |
| III | Human | United States, Panama, Spain, Central Asia counties, Estonia, The Netherlands, Sweden, Denmark, Norway, Malaysia, India, Sri Lanka, Nepal, Japan | 20, 87, 111, 117, 130, 170, 184, 199, 200,232, 239, 245, 249; CDC, unpublished data |
| IV | Nonhuman primate | Philippines | 167 |
| V | Nonhuman primate | Kenya | 248 |
| VI | Nonhuman primate | Indonesia | 200 |
FIG. 3.
Phylogenetic analysis of 3,582 HAV isolates in the VP1/P2B region (315-bp fragment). The dendrogram (unweighted-pair group method using average linkages) was created by using Kimura's two-parameter model. Isolates were obtained from the United States (n = 2,190) and other parts of the world. The U.S. specimens included specimens from hepatitis A cases from CDC's Sentinel Counties Study of Acute Viral Hepatitis (11, 170), published and unpublished outbreak investigations (30, 63, 112), and others submitted to CDC for investigation. Specimens from the U.S.-Mexico Border Infectious Disease Surveillance (BIDS) project (258) were also included and represented cases primarily from the United States. Specimens from other countries include those from Mexico, Brazil, Israel, Jordan, Egypt, Moldova, and Kazakhstan and were submitted to CDC as part of outbreak or epidemiologic investigations. Sequences from GenBank (Italy, Norway, Spain, Tunisia, and Japan) were also included in the analysis. Only unique sequence patterns are shown in the tree. Genotypes and subgenotypes are indicated on the branches, and the geographical locations of case patients are indicated in different colors.
Countries and their commonly found genotypes are shown in Table 3. Subgenotypes IA and IB are most often found in North and South America, Europe, China, and Japan (200). For subgenotype IIIA, the prototype strain, PA21 (20), was originally isolated from captured Panamanian owl monkeys and thought to be a simian virus. However, once nucleotide sequence analysis was performed many years later, the unique nucleotide and amino acid sequence patterns which differentiate human from simian HAV (i.e., “simian signature”) (167, 200) were not found in this strain. Over the ensuing years, subgenotype IIIA isolates have been recovered only from humans in many parts of the world (Table 3).
Cocirculation of multiple genotypes or subgenotypes has been reported in some regions of the world. Cocirculation of subgenotypes IA and IB is reported in South Africa (242), Brazil (251), and Israel (CDC, unpublished data), and subgenotypes IA and IIIA are reported in India (111) and the Central Asian Republics of the former Soviet Union (CDC, unpublished data). Both subgenotypes IA and IB have been isolated from the United States; however, most IB isolates were found among infected travelers returning from other countries (170; CDC, unpublished data).
Genetic relatedness of HAV.
Hepatitis A virus displays a high degree of antigenic and genetic conservation (146, 147, 200, 207) and does not appear to accumulate the high frequency of genetic changes seen in many RNA viruses. Although precise data on the genome mutation rate are not available, it appears to be lower than those of other RNA viruses (10−4 to 10−5 substitution per base per round of copy) (75). Even in extended common-source outbreaks, viruses isolated from the first cases are genetically the same as those isolated from the last cases (63, 112, 170, 175). However, enough genetic heterogeneity exists in several HAV genome regions to differentiate the relatedness of isolates circulating within and between communities over time, including within subgenotypes (50, 51, 52, 161, 170).
Initially, a 168-nucleotide fragment of the VP1/P2A junction was used for genotype analysis (116, 200). However, this sized fragment was shown not to sufficiently differentiate genotype or relatedness among HAV isolates (52, 206). Currently, the majority of molecular epidemiologic studies conducted at CDC use a 390-nucleotide-long fragment from the VP1-P2B region which includes 2A, one of the most variable regions of the genome (112, 170; CDC, unpublished data) (Table 2; Fig. 2). In a comparative analysis of 240 HAV isolates from a number of communities, using the 350-nucleotide fragment of the VP1-P2B region and a 900-nucleotide fragment from the entire VP1 region, genotype assignment was concordant in all instances and the longer fragment increased the likelihood of identifying a difference between two isolates by only 3% (CDC, unpublished data).
A phylogenetic analysis, restricted to a 315-nucleotide fragment of VP1-P2B, of over 3,000 HAV isolates from the United States and 12 other countries is shown in Fig. 3. The majority of isolates (85%) were obtained from investigations conducted by CDC's Division of Viral Hepatitis; the remaining sequences were obtained from GenBank (accession numbers AB020564 to AB020569, AF050223 to AF050238, AF386846 to AF386888, AF396391 to AF396408, AJ296172, AJ299460 to AJ299467, AJ505561 to AJ505625, AY294047 to AY294049, AY322842 to AY323047, AY875649 to AY875672, and AY753408 to AY753530).Most isolates (80%) belong to subgenotype IA, 17% belong to subgenotype IB, and 3% belong to genotype IIIA. However, these genotype proportions probably do not represent the true worldwide distribution of HAV genotypes, since this is essentially a convenience sample which contains a disproportionate number of isolates from North America. Within a genotype or subgenotype, there was a clustering of HAV isolates by location or geographical regions. For example, isolates from North America, South America, Europe, and Middle East tended to cluster together. Phylogenetic analysis also showed distinct clusters of HAV isolates within subgenotype IA and defined three major “clusters,” i.e., US-IA1, US-IA2, and US-IA3 (Fig. 3). Nucleic acid sequences obtained from HAV isolates from the interior of Mexico and the U.S.-Mexican border were closely related to isolates in US-IA1, while US-IA2 isolates were predominantly from IDUs and US-IA3 isolates were from MSM.
Applications of Molecular Epidemiologic Investigations
The long incubation period of hepatitis A, the epidemic but often sporadic nature of disease transmission, and the often unapparent links between persons involved in outbreaks have made molecular markers important tools for epidemiologic investigations of this infection. The use of nucleotide sequence patterns allows the investigator to determine whether viruses from the same or different locations are related to each other. Case patients with identical sequence patterns and similar epidemiologic characteristics usually suggest a common-source exposure (112, 170). However, there have been instances where sequence analysis suggested that temporally related common-source outbreaks might have been related, when in fact they represented different transmission incidents. This occurred because of the somewhat limited variability of the HAV nucleic acid sequence and the inability of the relatively short sequenced fragments to precisely differentiate the genetic relatedness of HAV isolates. In these instances, outbreak strains had identical or nearly identical sequences which represented similar geographical sources of the infecting virus. However, the epidemiologic investigation showed that the outbreaks were not related (3, 63; CDC, unpublished data).
Molecular epidemiologic investigations have to be planned and conducted with equal attention being paid to the methodological requirements of both disciplines: epidemiology and molecular biology. The use of molecular biomarkers alone will not fix a flawed epidemiologic investigation, and patterns of transmission suggested by patterns of HAV genetic relatedness may not be identified if the epidemiologic information was not collected at the time of the initial investigation.
Population-based molecular epidemiologic studies have shown that HAV is often transmitted within networks of persons with similar risk factors for infection (97, 143, 170, 232, 252). Certain sequence patterns appear to cluster among persons with particular risk factors and may be used as an indicator of the source of infection. For instance, in the United States, outbreaks among MSM have often shared one sequence pattern in subgenotype IA, and most cases associated with injection drug use have shared certain “signature” patterns (170, 249).
Molecular epidemiology has become a particularly powerful tool for the investigation of food-borne outbreaks of disease. The close or identical genetic relatedness of isolates from cases in different locales has provided the link to what previously would have been considered sporadic, yet independent, outbreaks. In addition, the availability of a large and ever-growing database of HAV sequences has allowed the identification of the geographic source of virus contamination (3, 112, 260). Recognition that geographically separated outbreaks may be linked by a food source and identification of the potential geographic origin of the contamination have accelerated the public health response to these outbreaks.
PREVENTION OF HEPATITIS A
The availability of vaccines to provide long-term immunity against HAV infection has the potential to significantly reduce disease incidence and possibly eliminate infection transmission (13, 28, 107, 159). A dramatic effect of widespread childhood hepatitis A vaccination has been observed in the United States. Significant disease reductions have occurred in vaccinated populations that historically had the highest disease incidence, namely, American Indians, Alaskan Natives, and persons living in the western United States (14, 23, 30, 32, 163, 215, 255).
The significant reduction in hepatitis A incidence in the United States due to immunization has also changed the epidemiology of this infection. Prior to childhood hepatitis A vaccination, the highest disease incidence occurred among children (28, 29, 159, 215). Now the highest incidence of disease is among adults, a substantial proportion of whom have risk factors for infection, such as IDUs and MSM (11, 28, 32, 170, 252). In addition, childhood immunization appears to have blunted or possibly even eliminated the cyclical nature of hepatitis A in the United States (13, 29, 163). However, at this time, hepatitis A immunization appears not to have changed the epidemiology of sporadic cases of the disease.
What might become the role of molecular diagnostics in the era of hepatitis A immunization? One role would be the continued use of molecular epidemiology to identify chains of transmission, including the extent of outbreaks among IDUs and MSM. Another would be the expanded use of molecular epidemiology to investigate small outbreaks or sporadic cases in different locales and determine if they are related, such as through the distribution of contaminated food. In addition, most food-borne outbreaks should be investigated to determine the genetic relatedness of HAV isolates to improve the identification of the source of contamination and the extent of the distribution of contaminated food. The use of molecular epidemiology to investigate hepatitis A outbreaks has provided a powerful tool which has improved the public health response to these situations. However, it has also meant that forensic practices (e.g., a documented chain of specimen custody) must be used during the investigation, because these data have eventually been entered into evidence in court cases.
Molecular diagnostics should become more widely used to assess the effectiveness of hepatitis A vaccination as disease incidence declines to very low levels. Because of the high proportion of asymptomatic HAV infections, nucleic acid amplification techniques will be required to determine the extent to which unidentified infection occurs. In addition, it will be very important to characterize the genetic makeup of HAV from immunized persons who may subsequently become infected. Such investigations would identify the possible establishment of antibody-resistant mutants, which may have a selective transmission advantage in immunized populations with continued exposure to HAV.
Acknowledgments
The following people or groups provided specimens or sequences that have not been previously published: Daniel Shouval, Nili Daudi (Liver Unit, Hadassah University Hospital, Jerusalem, Israel), Ala Toukan (University of Jordan, Amman, Jordan), Hend El Sherbini (Cairo University, Cairo, Egypt), Michael O. Favorov (CDC-CAR, Almaty, Kazakhstan), and the following members of the Epidemiology Branch, Division of Viral Hepatitis, CDC: Gregory L. Armstrong, Anthony Fiore, Ian Williams, and Beth P. Bell. We also thank our many other colleagues whose past efforts have made this work possible.
We regretfully report that author Omana V. Nainan passed away on 3 September 2005, and we dedicate this article to her memory and her exceptional contributions on molecular epidemiology of viral hepatitis.
The entire study was funded by the intramural budget of the Division of Viral Hepatitis, Centers for Disease Control and Prevention (CDC). The authors of this paper do not have any commercial association that might pose a conflict of interest.
Use of trade names is for identification only and does not imply endorsement by the Public Health Service or by the U.S. Department of Health and Human Services.
REFERENCES
- 1.Abd El Galil, K. H., M. A. El Sokkary, S. M. Kheira, A. M. Salazar, M. V. Yates, W. Chen, and A. Mulchandani. 2004. Combined immunomagnetic separation-molecular beacon-reverse transcription-PCR assay for detection of hepatitis A virus from environmental samples. Appl. Environ. Microbiol. 70:4371-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akriviadis, E. A., and A. G. Redeker. 1989. Fulminant hepatitis A in intravenous drug users with chronic liver disease. Ann. Intern. Med. 110:838-839. [DOI] [PubMed] [Google Scholar]
- 3.Amon, J. J., R. Devasia, G. Xia, O. V. Nainan, S. Hall, B. Lawson, J. Wolthius, P. D. M. MacDonald, C. Shepard, I. T. Williams, G. Armstrong, J. A. Gabel, P. Erwin, L. Sheeler, W. Kuhnert, P. Patel, G. Vaughan, A. Weltman, A. Craig, B. P. Bell, and A. Fiore. 2005. Molecular epidemiology of foodborne hepatitis A outbreaks in the United States, 2003. J. Infect. Dis. 192:1323-1330. [DOI] [PubMed] [Google Scholar]
- 4.Angarano, G., F. Trotta, L. Monno, T. Santantonio, and G. Pastore. 1985. Serum IgA anti-hepatitis A virus as detected by enzyme-linked immunosorbent assay: diagnostic significance in patients with acute and protracted hepatitis A. Diagn. Microbiol. Infect. Dis. 3:521-523. [DOI] [PubMed] [Google Scholar]
- 5.Apaire-Marchais, V., B. H. Robertson, V. Aubineau-Ferre, M. G. LeRoux, F. Leveque, L. Schwartzbrod, and S. Billaudel. 1995. Direct sequencing of hepatitis A virus strains isolated during an epidemic in France. Appl. Environ. Microbiol. 61:3977-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arauz-Ruiz, P., L. Sundqvist, Z. Garcia, L. Taylor, K. Visona, H. Norder, and L. O. Magnius. 2001. Presumed common source outbreaks of hepatitis A in an endemic area confirmed by limited sequencing within the VP1 region. J. Med. Virol. 65:449-456. [PubMed] [Google Scholar]
- 7.Armstrong, G. L., and B. P. Bell. 2002. Hepatitis A virus infections in the United States: model-based estimates and implications for childhood immunization. Pediatrics 109:839-845. [DOI] [PubMed] [Google Scholar]
- 8.Asher, L. V. S., L. N. Binn, T. L. Mensing, R. H. Marchwicki, R. A. Vassell, and G. D. Young. 1995. Pathogenesis of hepatitis A in orally inoculated owl monkeys (Aotus trivirgatus). J. Med. Virol. 47:260-268. [DOI] [PubMed] [Google Scholar]
- 9.Atmar, R. L., T. G. Metcalf, F. H. Neill, and M. K. Estes. 1993. Detection of enteric viruses in oysters by using the polymerase chain reaction. Appl. Environ. Microbiol. 59:631-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baroudy, B. M., J. R. Ticehurst, T. A. Miele, J. V. Maizel, R. H. Purcell, and S. M. Feinstone. 1985. Sequence analysis of hepatitis A virus cDNA coding for capsid proteins and RNA polymerase. Proc. Natl. Acad. Sci. USA 82:2143-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bell, B. P., C. N. Shapiro, M. J. Alter, L. A. Moyer, F. N. Judson, K. Mottram, M. Fleenor, P. L. Ryder, and H. S. Margolis. 1998. The diverse patterns of hepatitis A epidemiology in the United States—implications for vaccination strategies. J. Infect. Dis. 178:1579-1584. [DOI] [PubMed] [Google Scholar]
- 12.Bell, B. P. 2002. Global epidemiology of hepatitis A: implications for control strategies, p. 9-14. In H. S. Margolis, M. J. Alter, J. T. Liang, and J. L. Dienstag (ed.), Viral hepatitis and liver disease. International Medical Press, London, United Kingdom.
- 13.Bell, B. P., and S. M. Feinstone. 2004. Hepatitis A vaccine, p. 269-297. In S. A. Plotkin, W. A. Orenstein, and P. A. Offit (ed.), Vaccine, 4th ed. Saunders, Philadelphia, Pa.
- 14.Bialek, S. R., D. A. Thoroughman, D. Hu, E. P. Simard, J. Chattin, J. Cheek, and B. P. Bell. 2004. Hepatitis A incidence and hepatitis a vaccination among American Indians and Alaska Natives, 1990-2001. Am. J. Public Health 94:996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biziagos, E., J. Passagot, J. M. Crance, and R. Deloince. 1988. Long-term survival of hepatitis A virus and poliovirus type 1 in mineral water. Appl. Environ. Microbiol. 54:2705-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bloch, A. B., S. L. Stramer, J. D. Smith, H. S. Margolis, H. A. Fields, T. W. McKinley, C. P. Gerba, J. E. Maynard, and R. K. Sikes. 1990. Recovery of hepatitis A virus from a water supply responsible for a common source outbreak of hepatitis A. Am. J. Public Health 80:428-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blumer, G. 1923. Infectious jaundice in the United States. JAMA 81:353-358. [Google Scholar]
- 18.Bosch, A., G. Sanchez, F. Le Guyader, H. Vanaclocha, L. Haugarreau, and R. M. Pinto. 2001. Human enteric viruses in Coquina clams associated with a large hepatitis A outbreak. Water Sci. Technol. 43:61-65. [PubMed] [Google Scholar]
- 19.Bower, W. A., O. V. Nainan, X. Han, and H. S. Margolis. 2000. Duration of viremia in hepatitis A virus infection. J. Infect. Dis. 182:12-17. [DOI] [PubMed] [Google Scholar]
- 20.Brown, E. A., R. W. Jansen, and S. M. Lemon. 1989. Characterization of a simian hepatitis A virus (HAV): antigenic and genetic comparison with human HAV. J. Virol. 63:4932-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown, E. A., S. P. Day, R. W. Jansen, and S. M. Lemon. 1991. The 5′ nontranslated region of hepatitis A virus: secondary atructure and elements required for translation in vitro. J. Virol. 65:5828-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruisten, S. M., J. E. Steenbergen, A. S. Pijl, H. G. Niesters, G. J. van Doornum, and R. A. Coutinho. 2001. Molecular epidemiology of hepatitis A virus in Amsterdam, The Netherlands. J. Med. Virol. 63:88-95. [PubMed] [Google Scholar]
- 23.Bulkow, L. R., R. B. Wainwright, B. J. McMahon, J. P. Middaugh, S. A. Jenkerson, and H. S. Margolis. 1993. Secular trends in hepatitis A virus infection among Alaska Natives. J. Infect. Dis. 168:1017-1020. [DOI] [PubMed] [Google Scholar]
- 24.Bustin, S. A. 2002. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J. Mol. Endocrinol. 29:23-39. [DOI] [PubMed] [Google Scholar]
- 25.Buti, M., R. Jardi, A. Bosch, F. Rodriguez, G. Sanchez, R. Pinto, X. Costa, J. F. Sanchez-Avila, M. Cotrina, R. Esteban, and J. Guardia. 2001. Assessment of the PCR-Southern blot technique for the analysis of viremia in patients with acute hepatitis A. Gastroenterol. Hepatol. 24:1-4. [DOI] [PubMed] [Google Scholar]
- 26.Byun, K. S., J. H. Kim, K. J. Song, L. J. Baek, J. W. Song, S. H. Park, O. S. Kwon, J. E. Yeon, J. S. Kim, Y. T. Bak, and C. H. Lee. 2001. Molecular epidemiology of hepatitis A virus in Korea. J. Gastroenterol. Hepatol. 16:519-524. [DOI] [PubMed] [Google Scholar]
- 27.Calder, L., G. Simmons, C. Thornley, P. Taylor, K. Pritchard, G. Greening, and J. Bishop. 2003. An outbreak of hepatitis A associated with consumption of raw blueberries. Epidemiol. Infect. 131:745-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Centers for Disease Control and Prevention. 1999. Prevention of hepatitis A through active or passive immunization: recommendations of the Advisory Committee on Immunization Practices (ACIP). Morb. Mortal. Wkly. Rep. 48(RR-12):1-37. [PubMed] [Google Scholar]
- 29.Centers for Disease Control and Prevention. 1997. Hepatitis A vaccination programs in communities with high rates of hepatitis A. Morb. Mortal. Wkly. Rep. 46:600-603. [PubMed] [Google Scholar]
- 30.Centers for Disease Control and Prevention. 2004. Final 2003 reports of notifiable diseases. Morb. Mortal. Wkly. Rep. 53:687-705. [Google Scholar]
- 31.Centers for Disease Control and Prevention. 1996. Hepatitis surveillance report no. 56. U.S. Department of Health and Human Services, Public Health Service, CDC, Atlanta, Ga. (Available at http://www.cdc.gov/nciod/diseases/hepatitis/h96surve.htm. Accessed 1 September 1998.)
- 32.Centers for Disease Control and Prevention. 2004. Hepatitis surveillance report no. 59. U.S. Department of Health and Human Services, Public Health Service, CDC., Atlanta, Ga. (Available at http://www.cdc.gov/ncidod/diseases/hepatitis/resource/PDFs/Hep_surveillance_59.pdf. Accessed 25 March 2005.)
- 33.Chen, Y., J. Mao, Y. Hong, L. Yang, Z. Ling, and W. Yu. 2001. Genetic analysis of wild-type hepatitis A virus strains. Chinese Med. J. 114:422-423. [PubMed] [Google Scholar]
- 34.Ching, K. Z., T. Nakano, L. E. Chapman, A. Demby, and B. H. Robertson. 2002. Genetic characterization of wild-type genotype VII hepatitis A virus. J. Gen. Virol. 83:53-60. [DOI] [PubMed] [Google Scholar]
- 35.Chiriaco, P., C. Gaudalupi, M. K. Armigliato, F. Bortolotti, and G. Realdi. 1986. Polyphasic course of hepatitis type A in children. J. Infect. Dis. 154:231. [DOI] [PubMed] [Google Scholar]
- 36.Chironna, M., C. Germinario, D. De Medici, A. Fiore, S. Di Pasquale, M. Quarto, and S. Barbuti. 2002. Detection of hepatitis A virus in mussels from different sources marketed in Puglia region (South Italy). Int. J. Food Microbiol. 75:11-18. [DOI] [PubMed] [Google Scholar]
- 37.Chironna, M., A. Grottola, C. Lanave, E. Villa, S. Barbuti, and M. Quarto. 2003. Genetic analysis of HAV strains recovered from patients with acute hepatitis from Southern Italy. J. Med. Virol. 70:343-349. [DOI] [PubMed] [Google Scholar]
- 38.Chironna, M., P. Lopalco, R. Prato, C. Germinario, S. Barbuti, and M. Quarto. 2004. Outbreak of infection with hepatitis A virus (HAV) associated with a foodhandler and confirmed by sequence analysis reveals a new HAV genotype IB variant. J. Clin. Microbiol. 42:2825-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chitambar, S. D., and M. S. Chadha. 2000. Use of filter paper disks for hepatitis A surveillance. Indian J. Gastroenterol. 19:165-167. [PubMed] [Google Scholar]
- 40.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 41.Chomczynski, P. 1993. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. BioTechniques 15:532-537. [PubMed] [Google Scholar]
- 42.Chudy, M., I. Budek, B. Keller-Stanislawski, K. A. McCaustland, S. Neidhold, B. H. Robertson, C. M. Nubling, R. Seitz, and J. Lower. 1999. A new cluster of hepatitis A infection in hemophiliacs traced to a contaminated plasma pool. J. Med. Virol. 57:91-99. [DOI] [PubMed] [Google Scholar]
- 43.Cockayne, E. A. 1912. Catarrhal jaundice, sporadic and epidemic, and its relation to acute yellow atrophy of the liver. Q. J. Med. 6:1-29. [Google Scholar]
- 44.Coelho, C., A. P. Heinert, C. M. Simoes, and C. R. Barardi. 2003. Hepatitis A virus detection in oysters (Crassostrea gigas) in Santa Catarina State, Brazil, by reverse transcription-polymerase chain reaction. J. Food Prot. 66:507-511. [DOI] [PubMed] [Google Scholar]
- 45.Cohen, J. I., B. Rosenblum, J. R. Ticehurst, R. J. Daemer, S. M. Feinstone, and R. H. Purcell. 1987. complete nucleotide sequence of an attenuated hepatitis A virus: comparison with wild-type virus. Proc. Natl. Acad. Sci. USA 84:2497-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cohen, J. I., J. R. Ticehurst, R. H. Purcell, A. Buckler-White, and B. M. Baroudy. 1987. Complete nucleotide sequence of wild-type hepatitis A virus: Comparison with different strains of hepatitis A virus and other picornaviruses. J. Virol. 61:50-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen, J. I., B. Rosenblum, S. M. Feinstone, J. Ticehurst, and R. H. Purcell. 1989. Attenuation and cell culture adaptation of hepatitis A virus (HAV): a genetic analysis with HAV cDNA. J. Virol. 63:5364-5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cohen, J. I., S. Feinstone, and R. H. Purcell. 1989. Hepatitis A virus infection in a chimpanzee: duration of viremia and detection of virus in saliva and throat swabs. J. Infect. Dis. 160:887-890. [DOI] [PubMed] [Google Scholar]
- 49.Corey, L., and K. K. Holmes. 1980. Sexual transmission of hepatitis A in homosexual men: incidence and mechanism. N. Engl. J. Med. 302:435-438. [DOI] [PubMed] [Google Scholar]
- 50.Costa-Mattioli, M., V. Ferre, S. Monpoeho, L. Garcia, R. Colina, S. Billaudel, I. Vega, R. Perez-Bercoff, and J. Cristina. 2001. Genetic variability of hepatitis A virus in South America reveals heterogeneity and co-circulation during epidemic outbreaks. J. Gen. Virol. 82:2647-2652. [DOI] [PubMed] [Google Scholar]
- 51.Costa-Mattioli, M., S. Monpoeho, C. Schvoerer, B. Besse, M. H. Aleman, S. Billaudel, J. Cristina, and V. Ferre. 2001. Genetic analysis of hepatitis A virus outbreak in France confirms the co-circulation of sub-genotypes Ia, Ib and reveals a new genetic lineage. J. Med. Virol. 65:233-240. [DOI] [PubMed] [Google Scholar]
- 52.Costa-Mattioli, M., J. Cristina, H. Romero, R. Perez-Bercof, D. Casane, R. Colina, L. Garcia, I. Vega, G. Glikman, V. Romanowsky, A. Castello, E. Nicand, M. Gassin, S. Billaudel, and V. Ferre. 2002. Molecular evolution of hepatitis A virus: a new classification based on the complete VP1 protein. J. Virol. 76:9516-9525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Costa-Mattioli, M., S. Monpoeho, E. Nicand, M. H. Aleman, S. Billaudel, and V. Ferre. 2002. Quantification and duration of viraemia during hepatitis A infection as determined by real-time RT-PCR. J. Viral Hepat. 9:101-106. [DOI] [PubMed] [Google Scholar]
- 54.Coutinho, R. A., P. Albrecht-Van lent, T. Rijsdijk, N. Lelie, N. Nagelkerke, and H. Kuipers. 1983. Prevalence and incidence of hepatitis A among male homosexuals. Br. Med. J. (Clin. Res. Ed.) 287:1743-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cromeans, T., M. D. Sobsey, and H. A. Fields. 1987. Development of a plaque assay for a cytopathic, rapidly replicating isolate of hepatitis A virus. J. Med. Virol. 22:45-56. [DOI] [PubMed] [Google Scholar]
- 56.Cromeans, T. L., O. V. Nainan, and H. S. Margolis. 1997. Detection of hepatitis A virus RNA in oyster meat. Appl. Environ. Microbiol. 63:2460-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cromeans, T. L., M. O. Favarov, O. V. Nainan, and H. S. Margolis. 2001. Hepatitis A and E viruses, p. 23-76. In Y. H. Hui, S. A. Sattar, K. D. Murrell, W.-K. Nip, and P. S. Stanfield (ed.), Foodborne disease handbook, 2nd ed., vol. 2. Viruses, parasite, pathogens, and HACCP. Marcel Dekker, New York, N.Y. [Google Scholar]
- 58.Cuthbert, J. A. 2001. Hepatitis A: old and new. Clin. Microbiol. Rev. 14:38-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Daemer, R. J., S. M. Feinstone, I. D. Gust, and R. H. Purcell. 1981. Propagation of human hepatitis A virus in African green monkey kidney cell culture: primary isolation and serial passage. Infect. Immun. 32:388-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Almeida, L. M., R. S. Azevedo, A. A. Guimaraes, S. Coutinho Eda, C. J. Struchiner, and E. Massad. 1999. Detection of antibodies against hepatitis A virus in eluates of blood spotted on filter-paper: a pilot study in Rio de Janeiro, Brazil. Trans. R. Soc. Trop. Med. Hyg. 93:401-404. [DOI] [PubMed] [Google Scholar]
- 61.Delem, A. D. 1992. Comparison of modified HAVAB and ELISA for determination of vaccine-induced anti-HAV response. Biologicals 20:289-291. [DOI] [PubMed] [Google Scholar]
- 62.Deng, M. Y., S. P. Day, and D. O. Cliver. 1994. Detection of hepatitis A virus in environmental samples by antigen-capture PCR. Appl. Environ. Microbiol. 60:1927-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dentiger, C. M., W. A. Bower, O. V. Nainan, S. M. Cotter, G. Myers, L. M. Dubusky, S. Fowler, E. D. Salehi, and B. P. Bell. 2001. An outbreak of hepatitis A associated with green onions. J. Infect. Dis. 183:1273-1276. [DOI] [PubMed] [Google Scholar]
- 64.de Paula, V. S., M. L. Baptista, E. Lampe, C. Niel, and A. M. C. Gaspar. 2002. Characterization of hepatitis A virus isolates from sub-genotypes IA and IB in Rio de Janeiro, Brazil. J. Med. Virol. 66:22-27. [DOI] [PubMed] [Google Scholar]
- 65.de Paula, V. S., L. M. Villar, and A. M. C. Gaspar. 2003. Comparison of four extraction methods to detect hepatitis A virus RNA in serum and stool samples. Braz. J. Infect. Dis. 7:135-141. [DOI] [PubMed] [Google Scholar]
- 66.de Paula, V. S., L. Lu, C. Niel, A. M. C. Gaspar, and B. H. Robertson. 2004. Genetic analysis of hepatitis A virus isolates from Brazil. J. Med. Virol. 73:378-383. [DOI] [PubMed] [Google Scholar]
- 67.Desenclos, J. C., K. C. Klontz, M. H. Wilder, O. V. Nainan, H. S. Margolis, and R. A. Gunn. 1991. A multistate outbreak of hepatitis A caused by the consumption of raw oysters. Am. J. Public Health 81:1268-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.De Serres, G., T. L. Cromeans, B. Levesque, N. Brassard, C. Barthe, M. Dionne, H. Prud'homme, D. Paradis, C. N. Shapiro, O. V. Nainan, and H. S. Margolis. 1999. Molecular confirmation of hepatitis A virus from well water: epidemiology and public health implications. J. Infect. Dis. 179:37-43. [DOI] [PubMed] [Google Scholar]
- 69.Diaz, B. I., C. A. Sariol, A. Normann, L. Rodriguez, and B. Flehmig. 2001. Genetic relatedness of Cuban HAV wild-type isolates. J. Med. Virol. 64:96-103. [DOI] [PubMed] [Google Scholar]
- 70.Dienstag, J. L., I. D. Gust, C. R. Lucas, D. C. Wong, and R. H. Purcell. 1976. Mussel-associated viral hepatitis, type A: serological confirmation. Lancet i:561-564. [DOI] [PubMed] [Google Scholar]
- 71.Dignani, M. C., M. H. Miceli, C. M. Rosa, J. Gatica, J. Martinez-Rolon, and M. Pizzolato. 2003. Loss of hepatitis A virus (HAV) antibodies after peripheral stem cell transplantation (PSCT). Bone Marrow Transplant. 31:809-812. [DOI] [PubMed] [Google Scholar]
- 72.Di Pinto, A., V. T. Forte, G. M. Tantillo, V. Terio, and C. Buonavoglia. 2003. Detection of hepatitis A virus in shellfish (Mytilus galloprovincialis) with RT-PCR. J. Food Prot. 66:1681-1685. [DOI] [PubMed] [Google Scholar]
- 73.Di Pinto, A., M. C. Conversano, V. T. Forte, G. La Salandra, C. Montervino, and G. M. Tantillo. 2004. A comparison of RT-PCR-based assays for the detection of HAV from shellfish. New Microbiol. 27:119-124. [PubMed] [Google Scholar]
- 74.Divizia, M., R. Gabrieli, A. Macaluso, B. Bagnato, L. Palombi, E. Buonomo, F. Cenko, L. Leno, S. Bino, A. Basha, and A. Pana. 2005. Nucleotide correlation between HAV isolates from human patients and environmental samples. J. Med. Virol. 75:8-12. [DOI] [PubMed] [Google Scholar]
- 75.Domingo, E., C. Escarmis, N. Sevilla, A. Moya, S. F. Elena, J. Quer, I. S. Novella, and J. J. Holland. 1996. Basic concepts in RNA virus evolution. FASEB J. 10:859-864. [DOI] [PubMed] [Google Scholar]
- 76.Emerson, S. U., S. A. Tsarev, and R. H. Purcell. 1991. Biological and molecular comparisons of human (HM-175) and simian (AGM-27) hepatitis A viruses. J. Hepatol. 13(Suppl. 4):S144-S145. [DOI] [PubMed] [Google Scholar]
- 77.Emerson, S. U., Y. K. Huang, and R. H. Purcell. 1993. 2B and 2C mutations are essential but mutations throughout the genome of HAV contribute to adaptation to cell culture. Virology 194:475-480. [DOI] [PubMed] [Google Scholar]
- 78.Emerson, S. U., S. A. Tsarev, S. Govindarajan, M. Shapiro, and R. H. Purcell. 1996. A simian strain of hepatitis A virus, AGM-27, functions as an attenuated vaccine for chimpanzees. J. Infect. Dis. 173:592-597. [DOI] [PubMed] [Google Scholar]
- 79.Emerson, S. U., Y. K. Huang, H. Nguyen, A. Brockington, S. Govindarajan, M. S. Claire, M. Shapiro, and R. H. Purcell. 2002. Identification of VP1/2A and 2C as virulence genes of hepatitis A virus and demonstration of genetic instability of 2C. J. Virol. 76:8551-8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Feinstone, S. M., A. Z. Kapikian, and R. H. Purcell. 1973. Hepatitis A: detection by immune electron microscopy of a virus-like antigen associated with acute illness. Science 182:1026-1028. [DOI] [PubMed] [Google Scholar]
- 81.Feinstone, S. M., R. J. Daemer, I. D. Gust, and R. H. Purcell. 1983. Live attenuated vaccine for hepatitis A. Dev. Biol. Stand. 54:429-432. [PubMed] [Google Scholar]
- 82.Findlay, G. M., J. L. Dunlop, and H. C. Brown. 1931. Observations on epidemic catarrhal jaundice. Br. Med. Hyg. 25:7-24. [Google Scholar]
- 83.Flehmig, B. 1980. Hepatitis A-virus in cell culture. I. Propagation of different hepatitis A-virus isolates in a fetal rhesus monkey kidney cell line (FrhK-4). Med. Microbiol. Immunol. 168:239-248. [DOI] [PubMed] [Google Scholar]
- 84.Fujiwara, K., O. Yokosuka, and T. Ehata. 1997. Frequent detection of hepatitis A viral RNA in serum during early convalescent phase of acute hepatitis A. Hepatology 26:1634-1639. [DOI] [PubMed] [Google Scholar]
- 85.Fujiwara, K., O. Yokosuka, T. Ehata, F. Imazeki, and H. Saisho. 2000. PCR-SSCP analysis of 5′-nontranslated region of hepatitis A viral RNA: comparison with clinicopathological features of hepatitis A. Dig. Dis. Sci. 45:2422-2427. [DOI] [PubMed] [Google Scholar]
- 86.Fujiwara, K., O. Yokosuka, K. Fukai, F. Imazeki, H. Saisho, and M. Omata. 2001. Analysis of full-length hepatitis A virus genome in sera from patients with fulminant and self-limited acute type A hepatitis. J. Hepatol. 35:112-119. [DOI] [PubMed] [Google Scholar]
- 87.Fujiwara, K., O. Yokosuka, F. Imazeki, H. Saisho, N. Saotome, K. Suzuki, K. Okita, E. Tanaka, and M. Omata. 2003. Analysis of the genotype-determining region of hepatitis A viral RNA in relation to disease severities. Hepatol. Res. 25:124-134. [DOI] [PubMed] [Google Scholar]
- 88.Funkhouser, A. W., R. H. Purcell, E. D'Hondt, and S. U. Emerson. 1994. Attenuated hepatitis A virus: genetic determinants of adaptation to growth in MRC-5 cells. J. Virol. 68:148-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gabrieli, R., G. Sanchez, A. Macaluso, F. Cenko, S. Bino, L. Palombi, E. Buonomo, R. M. Pinto, A. Bosch, and M. Divizia. 2004. Hepatitis in Albanian children: molecular analysis of hepatitis A virus isolates. J. Med. Virol. 72:533-537. [DOI] [PubMed] [Google Scholar]
- 90.Gauss-Muller, V., F. Lottspeich, and F. Deinhardt. 1986. Characterization of hepatitis A virus structural proteins. Virology 155:732-736. [DOI] [PubMed] [Google Scholar]
- 91.Giles, J. P., H. Liebhaber, S. Krugman, and C. Lattimer. 1964. Early viremia and viruria in infectious hepatitis. Virology 24:107-108. [DOI] [PubMed] [Google Scholar]
- 92.Gilgen, M., D. Germann, J. Luthy, and P. Hubner. 1997. Three-step isolation method for sensitive detection of enterovirus, rotavirus, hepatitis A virus, and small round structured viruses in water samples. Int. J. Food Microbiol. 37:189-199. [DOI] [PubMed] [Google Scholar]
- 93.Glikson, M., E. Galun, R. Oren, R. Tur-Kaspa, and D. Shouval. 1992. Relapsing hepatitis A: review of 14 cases and literature survey. Medicine (Baltimore) 71:14-23. [DOI] [PubMed] [Google Scholar]
- 94.Goswami, B. B., W. Burkhardt III, and T. A. Cebula. 1997. Identification of genetic variants of hepatitis A virus. J. Virol. Methods 65:95-103. [DOI] [PubMed] [Google Scholar]
- 95.Graff, J., J. Ticehurst, and B. Flehmig. 1993. Detection of hepatitis A virus in sewage sludge by antigen capture polymerase chain reaction. Appl. Environ. Microbiol. 59:3165-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Graff, J., A. Normann, S. M. Feinstone, and B. Flehmig. 1994. Nucleotide sequence of wild-type hepatitis A virus GBM in comparison with two cell culture-adapted variants. J. Virol. 68:548-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Grinde, B., K. Stene-Johansen, B. Sharma, T. Hoel, M. Jensenius, and K. Skaug. 1997. Characterisation of an epidemic of hepatitis A virus involving intravenous drug abusers—infection by needle sharing? J. Med. Virol. 53:69-75. [DOI] [PubMed] [Google Scholar]
- 98.Hadler, S. C., H. M. Webster, J. J. Erben, J. E. Swanson, and J. E. Maynard. 1980. Hepatitis A in day-care centers: a community-wide assessment. N. Engl. J. Med. 302:1222-1227. [DOI] [PubMed] [Google Scholar]
- 99.Hadler, S. C. 1991. Global impact of hepatitis A virus infection: changing patterns, p. 14-20. In F. B. Hollinger, S. M. Lemon, and H. S. Margolis (ed.), Viral hepatitis and liver disease, Williams & Wilkins, Baltimore, Md.
- 100.Halliday, M. L., L. Y. Kang, T. K. Zhou, M. D. Hu, Q. C. Pan, T. Y. Fu, Y. S. Huang, and S. L. Hu. 1991. An epidemic of hepatitis A attributable to the ingestion of raw clams in Shanghai, China. J. Infect. Dis. 164:852-859. [DOI] [PubMed] [Google Scholar]
- 101.Harkess, J., B. Gildon, and G. R. Istre. 1989. Outbreaks of hepatitis A among illicit drug users, Oklahoma, 1984-87. Am. J. Public Health 79:463-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Havens, W. P., Jr., and J. R. Paul. 1945. Prevention of infectious hepatitis with gamma globulin. JAMA 129:270-272. [Google Scholar]
- 103.Havens, W. P., and R. E. Marck, Jr. 1946. The leukocytic response of patients with experimentally induced infectious hepatitis. Am. J. Med. Sci. 212:129-138. [PubMed] [Google Scholar]
- 104.Havens, W. P. 1947. The etiology of infectious hepatitis. JAMA 134:653-655. [DOI] [PubMed] [Google Scholar]
- 105.Henning, K. J., E. Bell, J. Braun, and N. D. Barker. 1995. A communitywide outbreak of hepatitis A: risk factors for infection among homosexual and bisexual men. Am. J. Med. 99:132-136. [DOI] [PubMed] [Google Scholar]
- 106.Hollinger, F. B., D. W. Bradley, J. E. Maynard, G. R. Dreesman, and J. L. Melnick. 1975. Detection of hepatitis A viral antigen by radioimmunoassay. J. Immunol. 115:1464-1466. [PubMed] [Google Scholar]
- 107.Hollinger, F. B., and S. U. Emerson. 2001. Hepatitis A virus, p. 799-840. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, New York, N.Y.
- 108.Hooper, R. R., C. W. Juels, J. A. Routenberg, W. O. Harrison, M. E. Kilpatrick, S. J. Kendra, and J. L. Dienstag. 1977. An outbreak of type A viral hepatitis at the Naval Training Center, San Diego: epidemiologic evaluation. Am. J. Epidemiol. 105:148-155. [DOI] [PubMed] [Google Scholar]
- 109.Huang, S. N., D. Lorenz, and R. J. Gerety. 1979. Electron and immunoelectron microscopic study on liver tissues of marmosets infected with hepatitis A virus. Lab. Investig. 41:63-71. [PubMed] [Google Scholar]
- 110.Hurni, W. M., D. Laufer, W. J. Miller, J. Ryan, and B. Watson. 1993. Anti-hepatitis A in the general population and in hepatitis A vaccinees using saliva and serum as diagnostic media. Ann. N.Y. Acad. Sci. 694:289-292. [DOI] [PubMed] [Google Scholar]
- 111.Hussain, Z., B. C. Dasb, S. A. Husain, M. Asima, S. Chattopadhyay, A. Malik, Y. Poovorawan, A. Theamboonlers, and P. Kar. 2005. Hepatitis A viral genotypes and clinical relevance: clinical and molecular characterization of hepatitis A virus isolates from northern India. Hepatol. Res. 32:16-24. [DOI] [PubMed] [Google Scholar]
- 112.Hutin, Y. J., V. Pool, E. H. Cramer, O. V. Nainan, J. Weth, I. T. Williams, S. T. Goldstein, K. F. Gensheimer, B. P. Bell, C. N. Shapiro, M. J. Alter, H. S. Margolis, et al. 1999. A multistate, foodborne outbreak of hepatitis A. N. Engl. J. Med. 340:595-602. [DOI] [PubMed] [Google Scholar]
- 113.Innis, B. L., R. Snitbhan, P. Kunasol, T. Laorakpongse, W. Poopatanakool, C. A. Kozik, S. Suntayakorn, T. Suknuntapong, A. Safary, D. B. Tang, and J. W. Boslego. 1994. Protection against hepatitis A by an inactivated vaccine. JAMA 271:1328-1334. [PubMed] [Google Scholar]
- 114.Inoue, K., M. Yoshiba, H. Yotsuyanagi, T. Otsuka, K. Sekiyama, and R. Fujita. 1996. Chronic hepatitis A with persistent viral replication. J. Med. Virol. 50:322-324. [DOI] [PubMed] [Google Scholar]
- 115.Jacobson, S. K., R. Buttery, J. V. Parry, K. R. Perry, and T. G. Wreghitt. 1995. Investigation of a hepatitis A outbreak in a primary school by sequential saliva sampling. Clin. Diagn. Virol. 3:173-180. [DOI] [PubMed] [Google Scholar]
- 116.Jansen, R. W., G. Siegl, and S. M. Lemon. 1990. Molecular epidemiology of human hepatitis A virus defined by an antigen-capture polymerase chain reaction method. Proc. Natl. Acad. Sci. USA 87:2867-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jansen, R. W., G. Siegl, and S. M. Lemon. 1991. Molecular epidemiology of human hepatitis A virus (HAV), p. 58-62. In B. N. Hollinger, S. M. Lemon, and H. S. Margolis (ed.), Viral hepatitis and liver disease. Williams & Wilkins, Baltimore, Md.
- 118.Jean, J., B. Blais, A. Darveau, and I. Fliss. 2001. Detection of hepatitis A virus by the nucleic acid sequence-based amplification technique and comparison with reverse transcription-PCR. Appl. Environ. Microbiol. 67:5593-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jean, J., B. Blais, A. Darveau, and I. Fliss. 2002. Simultaneous detection and identification of hepatitis A virus and rotavirus by multiplex nucleic acid sequence-based amplification (NASBA) and microtiter plate hybridization system. J. Virol. Methods 105:123-132. [DOI] [PubMed] [Google Scholar]
- 120.Jean, J., D. H. D'Souza, and L. A. Jaykus. 2004. Multiplex nucleic acid sequence-based amplification for simultaneous detection of several enteric viruses in model ready-to-eat foods. Appl. Environ. Microbiol. 70:6603-6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jiang, X., M. K. Estes, and T. G. Metcalf. 1987. Detection of hepatitis A virus by hybridization with single-stranded RNA probes. Appl. Environ. Microbiol. 53:2487-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jindal, M., S. S. Rana, R. K. Gupta, K. Das, and P. Kar. 2002. Serological study of hepatitis A virus infection amongst the students of a medical college in Delhi and evaluation of the need of vaccination. Indian J. Med. Res. 115:1-4. [PubMed] [Google Scholar]
- 123.Joshi, M. S., S. D. Chitambar, V. A. Arankalle, and M. S. Chadha. 2002. Evaluation of urine as a clinical specimen for diagnosis of hepatitis A. Clin. Diagn. Lab. Immunol. 9:840-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jothikumar, N., D. O. Cliver, and T. W. Mariam. 1998. Immunomagnetic captures PCR for rapid concentration and detection of hepatitis A virus from environmental samples. Appl. Environ. Microbiol. 64:504-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jothikumar, N., R. Paulmurugan, P. Padmanabhan, R. B. Sundari, S. Kamatchiammal, and K. S. Rao. 2000. Duplex RT-PCR for simultaneous detection of hepatitis A and hepatitis E virus isolated from drinking water samples. J. Environ. Monit. 2:587-590. [DOI] [PubMed] [Google Scholar]
- 126.Kabrane-Lazizi, Y., S. U. Emerson, C. Herzog, and R. H. Purcell. 2001. Detection of antibodies to HAV 3C proteinase in experimentally infected chimpanzees and in naturally infected children. Vaccine 19:2878-2883. [DOI] [PubMed] [Google Scholar]
- 127.Kao, H. W., M. Ashcavai, and A. G. Redeker. 1984. The persistence of hepatitis A IgM antibody after acute clinical hepatitis A. Hepatology 4:933-936. [DOI] [PubMed] [Google Scholar]
- 128.Karetnyi, I., A. G. Andzhaparidze, T. M. Orlova, and M. S. Balaian. 1989. Study of human and simian hepatitis A virus isolated by an immunoenzyme method using polyclonal and monoclonal antibodies. Vopr. Virusol. 34:50-53. (In Russian.) [PubMed] [Google Scholar]
- 129.Karlsen, F., H. B. Steen, and J. M. Nesland. 1995. SYBR green I DNA staining increases the detection sensitivity of viruses by polymerase chain reaction. J. Virol. Methods 55:153-156. [DOI] [PubMed] [Google Scholar]
- 130.Khanna, B., J. E. Spelbring, B. L. Innis, and B. H. Robertson. 1992. Characterization of a genetic variant of human hepatitis A virus. J. Med. Virol. 36:118-124. [DOI] [PubMed] [Google Scholar]
- 131.Kiltie, A. E., and A. J. Ryan. 1997. SYBR Green I staining of pulsed field agarose gels is a sensitive and inexpensive way of quantitating DNA double-strand breaks in mammalian cells. Nucleic Acids Res. 25:2945-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kingsley, D. H., G. K. Meade, and G. P. Richards. 2002. Detection of both hepatitis A virus and Norwalk-like virus in imported clams associated with food-borne illness. Appl. Environ. Microbiol. 68:3914-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kingsley, D. H., and G. P. Richards. 2001. Rapid and efficient extraction method for reverse transcription-PCR detection of hepatitis A and Norwalk-like viruses in shellfish. Appl. Environ. Microbiol. 67:4152-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Komurian-Pradel, F., G. Paranhos-Baccala, M. Sodoyer, P. Chevallier, B. Mandrand, V. Lotteau, and P. Andre. 2001. Quantitation of HCV RNA using real-time PCR and fluorimetry. J. Virol. Methods 95:111-119. [DOI] [PubMed] [Google Scholar]
- 135.Koopmans, M., C. H. von Bonsdorff, J. Vinje, D. de Medici, and S. Monroe. 2002. Foodborne viruses. FEMS Microbiol. Rev. 26:187-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Krugman, S., R. Ward, J. P. Giles, D. Bodansky, and A. M. Jacobs. 1959. Infectious hepatitis: detection of virus during the incubation period and in clinically inapparent infection. N. Engl. J. Med. 261:729-734. [DOI] [PubMed] [Google Scholar]
- 137.Krugman, S., R. Ward, and J. P. Giles. 1962. The natural history of infectious hepatitis. Am. J. Med. 32:717-728. [DOI] [PubMed] [Google Scholar]
- 138.Krugman, S., J. P. Giles, and J. Hammond. 1967. Infectious hepatitis. Evidence for two distinctive clinical, epidemiological, and immunological types of infection. JAMA 200:365-373. [DOI] [PubMed] [Google Scholar]
- 139.LaBrecque, F. D., D. R. LaBrecque, D. Klinzman, S. Perlman, J. B. Cederna, P. L. Winokur, J. Q. Han, and J. T. Stapleton. 1998. Recombinant hepatitis A virus antigen: improved production and utility in diagnostic immunoassays. J. Clin. Microbiol. 36:2014-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Laufer, D. S., W. Hurni, B. Watson, W. Miller, J. Ryan, D. Nalin, and L. Brown. 1995. Saliva and serum as diagnostic media for antibody to hepatitis A virus in adults and in individuals who have received an inactivated hepatitis A vaccine. Clin. Infect. Dis. 20:868-871. [DOI] [PubMed] [Google Scholar]
- 141.Lednar, W. M., S. M. Lemon, J. W. Kirkpatrick, R. R. Redfield, M. L. Fields, and P. W. Kelley. 1985. Frequency of illness associated with epidemic hepatitis A virus infections in adults. Am. J. Epidemiol. 122:226-233. [DOI] [PubMed] [Google Scholar]
- 142.Lee, D. H., and A. M. Prince. 2001. Automation of nucleic acid extraction for NAT screening of individual blood units. Transfusion 41:483-487. [DOI] [PubMed] [Google Scholar]
- 143.Leino, T., P. Leinikki, T. Hyypia, M. Ristola, J. Suni, J. Sutinen, A. Holopainen, O. Haikala, M. Valle, and T. Rostila. 1997. Hepatitis A outbreak amongst intravenous amphetamine abusers in Finland. Scand. J. Infect. Dis. 29:213-216. [DOI] [PubMed] [Google Scholar]
- 144.Lemon, S. M., L. N. Binn, and R. H. Marchwicki. 1983. Radioimmunofocus assay for quantitation of hepatitis A virus in cell cultures. J. Clin. Microbiol. 17:834-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lemon, S. M. 1994. The natural history of hepatitis A: the potential for transmission by transfusion of blood or blood products. Vox Sang 67(Suppl 4):19-23. [PubMed] [Google Scholar]
- 146.Lemon, S. M. 1992. HAV: current concepts of the molecular virology, immunobiology and approaches to vaccine development. Rev. Med. Virol. 2:73-87. [Google Scholar]
- 147.Lemon, S. M., R. W. Jansen, and E. A. Brown. 1992. Genetic, antigenic and biological differences between strains of hepatitis A virus. Vaccine 10(Suppl. 1):S40-S44. [DOI] [PubMed] [Google Scholar]
- 148.Lemon, S. M. 1993. Immunologic approaches to assessing the response to inactivated hepatitis A vaccine. J. Hepatol. 18(Suppl 2):S15-S19. [DOI] [PubMed] [Google Scholar]
- 149.Li, J. W., T. X. Zhong, W. W. Xin, L. Z. Jin, and H. C. Fu. 2004. Mechanisms of inactivation of hepatitis A virus in water by chlorine dioxide. Water Res. 38:1514-1519. [DOI] [PubMed] [Google Scholar]
- 150.Liu, G., N. Hu, and Y. Hu. 2003. Full-length genome of wild-type hepatitis A virus (DL3) isolated in China. World J. Gastroenterol. 9:499-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Locarnini, S. A., A. G. Coulepis, A. A. Ferris, N. I. Lehmann, and I. D. Gust. 1978. Purification of hepatitis A virus from human feces. Intervirology 10:300-308. [DOI] [PubMed] [Google Scholar]
- 152.Locarnini, S. A., A. G. Coulepis, J. Kaldor, and I. D. Gust. 1980. Coproantibodies in hepatitis A: detection by enzyme-linked immunosorbent assay and immune electron microscopy. J. Clin. Microbiol. 11:710-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lowry, P. W., R. Levine, D. F. Stroup, R. A. Gunn, M. H. Wilder, and C. Konigsberg, Jr. 1989. Hepatitis A outbreak on a floating restaurant in Florida, 1986. Am. J. Epidemiol. 129:155-164. [DOI] [PubMed] [Google Scholar]
- 154.Lu, L., K. Z. Ching, V. S. de Paula, T. Nakano, G. Siegl, M. Weitz, and B. H. Robertson. 2004. Characterization of the complete genomic sequence of genotype II hepatitis A virus (CF53/Berne isolate). J. Gen. Virol. 85:2943-2952. [DOI] [PubMed] [Google Scholar]
- 155.Mackay, I. M., K. E. Arden, and A. Nitsche. 2002. Real-time PCR in virology. Nucleic Acids Res. 30:1292-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mackiewicz, V., E. Dussaix, M. F. Le Petitcorps, and A. M. Roque-Afonso. 2004. Detection of hepatitis A virus RNA in saliva. J. Clin. Microbiol. 42:4329-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Margolis, H. S., O. V. Nainan, K. Krawczynski, D. W. Bradley, J. W. Ebert, J. Spelbring, H. A. Fields, and J. E. Maynard. 1988. Appearance of immune complexes during experimental hepatitis A infection in chimpanzees. J. Med. Virol. 26:315-326. [DOI] [PubMed] [Google Scholar]
- 158.Margolis, H. S., and O. V. Nainan. 1990. Identification of virus components in circulating immune complexes isolated during hepatitis A virus infection. Hepatology 11:31-37. [DOI] [PubMed] [Google Scholar]
- 159.Margolis, H. S. 2000. Viral hepatitis, p. 174-188. In R. B. Wallace and B. N. Doebbeling (ed.), Maxcy-Rosenau-Last Public Health and Preventive Medicine, 14th ed. Appleton & Lange, Stamford, Conn.
- 160.Marziali, A., and M. Akeson. 2001. New DNA sequencing methods. Annu. Rev. Biomed. Eng. 3:195-223. [DOI] [PubMed] [Google Scholar]
- 161.Mbayed, V. A., S. Sookoian, V. Alfonso, and R. H. Campos. 2002. Genetic characterization of hepatitis A virus isolates from Buenos Aires, Argentina. J. Med. Virol. 68:168-174. [DOI] [PubMed] [Google Scholar]
- 162.McCaustland, K. A., W. W. Bond, D. W. Bradley, J. W. Ebert, and J. E. Maynard. 1982. Survival of hepatitis A virus in feces after drying and storage for 1 month. J. Clin. Microbiol. 16:957-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.McMahon, B. J., M. Beller, J. Williams, M. Schloss, H. Tanttila, and L. Bulkow. 1996. A program to control an outbreak of hepatitis A in Alaska by using an inactivated hepatitis A vaccine. Arch. Pediatr. Adolesc. Med. 150:733-739. [DOI] [PubMed] [Google Scholar]
- 164.Morace, G., G. Pisani, M. Divizia, and A. Pana. 1993. Detection of hepatitis A virus in concentrated river water by polymerase chain reaction. Zentralbl. Hyg. Umweltmed. 193:521-527. [PubMed] [Google Scholar]
- 165.Moritsugu, Y., J. L. Dienstag, J. Valdesuso, D. C. Wong, J. Wagner, J. A. Routenberg, and R. H. Purcell. 1976. Purification of hepatitis A antigen from feces and detection of antigen and antibody by immune adherence hemagglutination. Infect. Immun. 13:898-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Morrison, T. B., J. J. Weis, and C. T. Wittwer. 1998. Quantification of low-copy transcripts by continuous SYBR Green I monitoring during amplification. BioTechniques 24:954-958, 960, 962. [PubMed] [Google Scholar]
- 167.Nainan, O. V., H. S. Margolis, B. H. Robertson, M. Balayan, and M. A. Brinton. 1991. Sequence analysis of a new hepatitis A virus naturally infecting cynomolgus macaques (Macacafascicularis). J. Gen. Virol. 72:1685-1689. [DOI] [PubMed] [Google Scholar]
- 168.Nainan, O. V., M. A. Brinton, and H. S. Margolis. 1992. Identification of amino acids located in the antibody binding sites of human hepatitis A virus. Virology 191:984-987. [DOI] [PubMed] [Google Scholar]
- 169.Nainan, O. V., T. L. Cromeans, and H. S. Margolis. 1996. Sequence-specific, single-primer amplification and detection of PCR products for identification of hepatitis viruses. J. Virol. Methods 61:127-134. [DOI] [PubMed] [Google Scholar]
- 170.Nainan, O. V., G. L. Armstrong, X. Han, I. T. Williams, B. P. Bell, and H. S. Margolis. 2005. Hepatitis A molecular epidemiology in the United States, 1996-1997: sources of infection and implications of vaccination policy. J. Infect. Dis. 191:957-963. [DOI] [PubMed] [Google Scholar]
- 171.Najarian, R., D. Caput, W. Gee, S. J. Potter, A. Renard, J. Merryweather, G. Van Nest, and D. Dina. 1985. Primary structure and gene organization of human hepatitis A virus. Proc. Natl. Acad. Sci. USA 82:2627-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Nalin, D. R., B. J. Kuter, L. Brown, C. Patterson, G. B. Calandra, A. Werzberger, D. Shouval, E. Ellerbeck, S. L. Block, and R. Bishop. 1993. Worldwide experience with the CR326F-derived inactivated hepatitis A virus vaccine in pediatric and adult populations: an overview. J. Hepatol. 18(Suppl 2):s51-s55. [DOI] [PubMed] [Google Scholar]
- 173.Nasser, A. M., and T. G. Metcalf. 1987. Production of cytopathology in FRhK-4 cells by BS-C-1-passaged hepatitis A virus. Appl. Environ. Microbiol. 53:2967-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Nitsche, A., N. Steuer, C. A. Schmidt, O. Landt, H. Ellerbrok, G. Pauli, and W. Siegert. 2000. Detection of human cytomegalovirus DNA by real-time quantitative PCR. J. Clin. Microbiol. 38:2734-2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Niu, M. T., L. B. Polish, B. H. Robertson, B. K. Khanna, B. A. Woodruff, C. N. Shapiro, M. A. Miller, J. D. Smith, J. K. Gedrose, M. J. Alter, H. S. Margolis, and the National Hepatitis A Investigation Team. 1992. Multistate outbreak of hepatitis A associated with frozen strawberries. J. Infect. Dis. 166:518-524. [DOI] [PubMed] [Google Scholar]
- 176.Noble, R. C., M. A. Kane, S. A. Reeves, and I. Roeckel. 1984. Posttransfusion hepatitis A in a neonatal intensive care unit. JAMA 252:2711-2715. [PubMed] [Google Scholar]
- 177.Oba, I. T., A. M. Spina, C. P. Saraceni, M. F. Lemos, R. Senhoras, R. C. Moreira, and C. F. Granato. 2000. Detection of hepatitis A antibodies by ELISA using saliva as clinical samples. Rev. Inst. Med. Trop. Sao Paulo 42:197-200. [DOI] [PubMed] [Google Scholar]
- 178.Ochnio, J. J., D. W. Scheifele, M. Ho, and L. A. Mitchell. 1997. New, ultrasensitive enzyme immunoassay for detecting vaccine- and disease-induced hepatitis A virus-specific immunoglobulin G in saliva. J. Clin. Microbiol. 35:98-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Osterholm, M. T., R. J. Kantor, D. W. Bradley, W. N. Hall, D. P. Francis, H. C. Aaron, J. W. Washburn, and D. Velde. 1980. Immunoglobulin M-specific serologic testing in an outbreak of foodborne viral hepatitis, type A. Am. J. Epidemiol. 112:8-16. [DOI] [PubMed] [Google Scholar]
- 180.Ostermayr, R., K. Helm, V. Gauss-Muller, E. L. Winnacker, and F. Deinhardt. 1987. Expression of hepatitis A virus cDNA in Escherichia coli: antigenic VP1 recombinant protein. J. Virol. 61:3645-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Pana, A., M. Divizia, P. de Filippis, and A. di Napoli. 1987. Isolation of hepatitis A virus from polluted river water on FRP/3 cells. Lancet ii:1328. [DOI] [PubMed] [Google Scholar]
- 182.Paul, A. V., H. Tada, K. von der Helm, T. Wissel, R. Kiehn, E. Wimmer, and F. Deinhardt. 1987. The entire nucleotide sequence of the genome of human hepatitis A virus (isolate MBB). Virus Res. 8:153-171. [DOI] [PubMed] [Google Scholar]
- 183.Perry, K. R., J. V. Parry, E. M. Vandervelde, and P. P. Mortimer. 1992. The detection in urine specimens of IgG and IgM antibodies to hepatitis A and hepatitis B core antigens. J. Med. Virol. 38:265-270. [DOI] [PubMed] [Google Scholar]
- 184.Pina, S., M. Buti, R. Jardi, P. Clemente-Casares, J. Jofre, and R. Girones. 2001. Genetic analysis of hepatitis A virus strains recovered from the environment and from patients with acute hepatitis. J. Gen. Virol. 82:2955-2963. [DOI] [PubMed] [Google Scholar]
- 185.Ping, L. H., R. W. Jansen, J. T. Stapleton, J. I. Cohen, and S. M. Lemon. 1988. Identification of an immunodominant antigenic site involving the capsid protein VP3 of hepatitis A virus. Proc. Natl. Acad. Sci. USA 85:8281-8285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ping, L. H., and S. M. Lemon. 1992. Antigenic structure of human hepatitis A virus defined by analysis of escape mutants selected against murine monoclonal antibodies. J. Virol. 66:2208-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Pinto, M. A., R. S. Marchevsky, M. L. Baptista, M. A. de Lima, M. Pelajo-Machado, C. L. Vitral, C. F. Kubelka, J. W. Pissurno, M. S. Franca, H. G. Schatzmayr, and A. M. Gaspar. 2002. Experimental hepatitis A virus (HAV) infection in Callithrix jacchus: early detection of HAV antigen and viral fate. Exp. Toxicol. Pathol. 53:413-420. [DOI] [PubMed] [Google Scholar]
- 188.Poddar, U., B. R. Thapa, A. Prasad, and K. Singh. 2002. Changing spectrum of sporadic acute viral hepatitis in Indian children. J. Trop. Pediatr. 48:210-213. [DOI] [PubMed] [Google Scholar]
- 189.Polish, L. B., B. H. Robertson, B. Khanna, K. Krawczynski, J. Spelbring, F. Olson, and C. N. Shapiro. 1999. Excretion of hepatitis A virus (HAV) in adults: comparison of immunologic and molecular detection methods and relationship between HAV positivity and infectivity in tamarins. J. Clin. Microbiol. 37:3615-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Poonawagul, U., S. Warintrawat, R. Snitbhan, S. Kitisriwarapoj, V. Chaiyakunt, and H. M. Foy. 1995. Outbreak of hepatitis A in a college traced to contaminated water reservoir in cafeteria. Southeast Asian J. Trop. Med. Public Health 26:705-708. [PubMed] [Google Scholar]
- 191.Poovorawan, Y., A. Theamboonlers, V. Chongsrisawat, P. Jantaradsamee, S. Chutsirimongkol, and P. Tangkijvanich. 2005. Clinical features and molecular characterization of hepatitis A virus outbreak in a child care center in Thailand. J. Clin. Virol. 32:24-28. [DOI] [PubMed] [Google Scholar]
- 192.Provost, P. J., B. S. Wolanski, W. J. Miller, O. L. Ittensohn, W. J. McAleer, and M. R. Hilleman. 1975. Physical, chemical and morphologic dimensions of human hepatitis A virus strain CR326 (38578). Proc. Soc. Exp. Biol. Med. 148:532-539. [DOI] [PubMed] [Google Scholar]
- 193.Provost, P. J., and M. R. Hilleman. 1979. Propagation of human hepatitis A virus in cell culture in vitro. Proc. Soc. Exp. Biol. Med. 160:213-221. [DOI] [PubMed] [Google Scholar]
- 194.Purcell, R. H., D. C. Wong, Y. Moritsugu, J. L. Dienstag, J. A. Routenberg, and J. D. Boggs. 1976. A microtiter solid phase radioimmunoassay for hepatitis A antigen and antibody. J. Immunol. 116:349-356. [PubMed] [Google Scholar]
- 195.Ramsay, C. N., and P. A. Upton. 1989. Hepatitis A and frozen raspberries. Lancet i:43-44. [DOI] [PubMed] [Google Scholar]
- 196.Randhawa, J. S., and A. J. Easton. 1999. Demystified. DNA nucleotide sequencing. Mol. Pathol. 52:117-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Reid, T. M., and H. G. Robinson. 1987. Frozen raspberries and hepatitis A. Epidemiol. Infect. 98:109-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Rezende, G., A. M. Roque-Afonso, D. Samuel, M. Gigou, E. Nicand, V. Ferre, E. Dussaix, H. Bismuth, and C. Feray. 2003. Viral and clinical factors associated with the fulminant course of hepatitis A infection. Hepatology 38:613-618. [DOI] [PubMed] [Google Scholar]
- 199.Robertson, B. H., B. Khanna, O. V. Nainan, and H. S. Margolis. 1991. Epidemiologic patterns of wild-type hepatitis A virus determined by genetic variation. J. Infect. Dis. 163:286-292. [DOI] [PubMed] [Google Scholar]
- 200.Robertson, B. H., R. W. Jansen, B. Khanna, A. Totsuka, O. V. Nainan, G. Siegl, A. Widell, H. S. Margolis, S. Isomura, K. Ito, T. Ishizu, Y. Moritsugu, and S. M. Lemon. 1992. Genetic relatedness of hepatitis A virus strains recovered from different geographical regions. J. Gen. Virol. 73:1365-1377. [DOI] [PubMed] [Google Scholar]
- 201.Robertson, B. H., X. Y. Jia, H. Tian, H. S. Margolis, D. F. Summers, and E. Ehrenfeld. 1992. Serological approaches to distinguish immune response to hepatitis A vaccine and natural infection. Vaccine 10(Suppl. 1):S106-S109. [DOI] [PubMed] [Google Scholar]
- 202.Robertson, B. H., X. Y. Jia, H. Tian, H. S. Margolis, D. F. Summers, and E. Ehrenfeld. 1993. Antibody response to nonstructural proteins of hepatitis A virus following infection. J. Med. Virol. 40:76-82. [DOI] [PubMed] [Google Scholar]
- 203.Robertson, B. H., F. Averhoff, T. L. Cromeans, X. H. Han, B. Khoprasert, O. V. Nainan, J. Rosenberg, L. Paikoff, E. DeBess, C. N. Shapiro, and H. S. Margolis. 2000. Genetic relatedness of hepatitis A virus isolates during a community-wide outbreak. J. Med. Virol. 62:144-150. [PubMed] [Google Scholar]
- 204.Rosenblum, L. S., I. R. Mirkin, D. T. Allen, S. Safford, and S. C. Hadler. 1990. A multifocal outbreak of hepatitis A traced to commercially distributed lettuce. Am. J. Public Health 80:1075-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Rosenblum, L. S., M. E. Villarino, O. V. Nainan, M. E. Melish, S. C. Hadler, P. P. Pinsky, W. R. Jarvis, C. E. Ott, and H. S. Margolis. 1991. Hepatitis A outbreak in a neonatal intensive care unit: risk factors for transmission and evidence of prolonged viral excretion among preterm infants. J. Infect. Dis. 164:476-482. [DOI] [PubMed] [Google Scholar]
- 206.Sanchez, G., R. M. Pinto, H. Vanaclocha, and A. Bosch. 2002. Molecular characterization of hepatitis A virus isolates from a transcontinental shellfish-borne outbreak. J. Clin. Microbiol. 40:4148-4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Sanchez, G., A. Bosch, and R. M. Pinto. 2003. Genome variability and capsid structural constraints of hepatitis A virus. J. Virol. 77:452-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Sanchez, G., C. Villena, A. Bosch, and R. M. Pinto. 2004. Hepatitis A virus: molecular detection and typing. Methods Mol. Biol. 268:103-114. [DOI] [PubMed] [Google Scholar]
- 209.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Schiodt, F. V., E. Atillasoy, A. Obaid Shakil, E. R. Schiff, C. Caldwell, K. V. Kowdley, R. Stribling, J. S. Crippen, S. Flamm, K. A. Somberg, H. Rosen, T. M. McCashland, J. E. Hay, W. M. Lee, and the Acute Liver Failure Study Group. 1999. Etiology and outcome for 295 patients with acute liver failure in the United States. Liver Transplant. Surg. 5:29-34. [DOI] [PubMed] [Google Scholar]
- 211.Schultheiss, T., Y. Y. Kusov, and V. Gauss-Muller. 1994. Proteinase 3C of hepatitis A virus (HAV) cleaves the HAV polyprotein P2-P3 at all sites including VP1/2A and 2A/2B. Virology 198:275-281. [DOI] [PubMed] [Google Scholar]
- 212.Schwab, K. J., R. De Leon, and M. D. Sobsey. 1995. Concentration and purification of beef extract mock eluates from water samples for the detection of enteroviruses, hepatitis A virus, and Norwalk virus by reverse transcription-PCR. Appl. Environ. Microbiol. 61:531-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Seelig, R., G. Pott, H. P. Seelig, H. Liehr, P. Metzger, and R. Waldherr. 1984. Virus-binding activity of fibronectin: masking of hepatitis A virus. J. Virol. Methods 8:335-347. [DOI] [PubMed] [Google Scholar]
- 214.Shao, Z. J., D. Z. Xu, Y. P. Yan, J. H. Li, J. X. Zhang, Z. Y. Zhang, and B. R. Pan. 2003. Detection of anti-HAV antibody with dot immunogold filtration assay. World J. Gastroenterol. 9:1508-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Shapiro, C. N., P. J. Coleman, G. M. McQuillan, M. J. Alter, and H. S. Margolis. 1992. Epidemiology of hepatitis A: seroepidemiology and risk groups in the USA. Vaccine 10(Suppl. 1):S59-S62. [DOI] [PubMed] [Google Scholar]
- 216.Shouval, D., Y. Ashur, R. Adler, J. A. Lewis, M. E. Armstrong, J. P. Davide, B. McGuire, B. Kuter, L. Brown, and W. Miller. 1993. Single and booster responses to an inactivated hepatitis A vaccine: comparisons with immune globulin prophylaxis. Vaccine 11(Suppl. 1):S9-S14. [DOI] [PubMed] [Google Scholar]
- 217.Siegl, G., J. deChastonay, and G. Kronauer. 1984. Propagation and assay of hepatitis A virus in vitro. J. Virol. Methods 9:53-67. [DOI] [PubMed] [Google Scholar]
- 218.Sjogren, M. H., H. Tanno, O. Fay, S. Sileoni, B. D. Cohen, D. S. Burke, and R. J. Feighny. 1987. Hepatitis A virus in stool during clinical relapse. Ann. Intern. Med. 106:221-226. [DOI] [PubMed] [Google Scholar]
- 219.Sjogren, M. H., C. H. Hoke, L. N. Binn, K. H. Eckels, D. R. Dubois, L. Lyde, A. Tsuchida, S. Oaks, Jr., R. Marchwicki, W. Lednar, R. Chloupek, J. Ticehurst, and W. H. Bancroft. 1991. Immunogenicity of an inactivated hepatitis A vaccine. Ann. Intern. Med. 114:470-471. [DOI] [PubMed] [Google Scholar]
- 220.Skinhoj, P., F. Mikkelsen, and F. B. Hollinger. 1977. Hepatitis A in Greenland: importance of specific antibody testing in epidemiologic surveillance. Am. J. Epidemiol. 105:140-147. [DOI] [PubMed] [Google Scholar]
- 221.Skinhoj, P., L. R. Mathiesen, P. Kiryger, and A. M. Moller. 1981. Faeccel excretion of hepatitis A virus in patients with symptomatic hepatitis A infection. Scand. J. Gastroenterol. 16:1057-1059. [Google Scholar]
- 222.Smith, P. F., J. C. Grabau, A. Werzberger, R. A. Gunn, H. R. Rolka, S. F. Kondracki, R. J. Gallo, and D. L. Morse. 1997. The role of young children in a community-wide outbreak of hepatitis A. Epidemiol. Infect. 118:243-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Sobsey, M. D., S. E. Oglesbee, and D. A. Wait. 1985. Evaluation of methods for concentrating hepatitis A virus from drinking water. Appl. Environ. Microbiol. 50:1457-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Soucie, J. M., B. H. Robertson, B. P. Bell, K. A. McCaustland, and B. L. Evatt. 1998. Hepatitis A virus infections associated with clotting factor concentrate in the United States. Transfusion 38:573-579. [DOI] [PubMed] [Google Scholar]
- 225.Staes, C. J., T. L. Schlenker, I. Risk, K. G. Cannon, H. Harris, A. T. Pavia, C. N. Shapiro, and B. P. Bell. 2000. Sources of infection among persons with acute hepatitis A and no identified risk factors during a sustained community-wide outbreak. Pediatrics 106:E54. [DOI] [PubMed] [Google Scholar]
- 226.Stahlberg, A., J. Hakansson, X. Xian, H. Semb, and M. Kubista. 2004. Properties of the reverse transcription reaction in mRNA quantification. Clin. Chem. 50:509-515. [DOI] [PubMed] [Google Scholar]
- 227.Stahlberg, A., M. Kubista, and M. Pfaffl. 2004. Comparison of reverse transcriptases in gene expression analysis. Clin. Chem. 50:1678-1680. [DOI] [PubMed] [Google Scholar]
- 228.Stapleton, J. T., and S. M. Lemon. 1987. Neutralization escape mutants define a dominant immunogenic neutralization site on hepatitis A virus. J. Virol. 61:491-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Stapleton, J. T., D. K. Lange, J. W. LeDuc, L. N. Binn, R. W. Jansen, and S. M. Lemon. 1991. The role of secretory immunity in hepatitis A virus infection. J. Infect. Dis. 163:7-11. [DOI] [PubMed] [Google Scholar]
- 230.Stapleton, J. T. 1995. Host immune response to hepatitis A virus. J. Infect. Dis. 171:S9-S14. [DOI] [PubMed] [Google Scholar]
- 231.Steffen, R., M. A. Kane, C. N. Shapiro, N. Billo, K. J. Schoellhorn, and P. van Damme. 1994. Epidemiology and prevention of hepatitis A in travelers. JAMA 272:885-889. [PubMed] [Google Scholar]
- 232.Stene-Johansen, K., K. Skaug, H. Blystad, B. Grinde, et al. 1998. A unique hepatitis A virus strain caused an epidemic in Norway associated with intravenous drug abuse. Scand. J. Infect. Dis. 30:35-38. [DOI] [PubMed] [Google Scholar]
- 233.Stewart, D. R., T. S. Morris, R. H. Purcell, and S. U. Emerson. 1997. Detection of antibodies to the nonstructural 3C proteinase of hepatitis A virus. J. Infect. Dis. 176:593-601. [DOI] [PubMed] [Google Scholar]
- 234.Strazynski, M., J. Kramer, and B. Becker. 2002. Thermal inactivation of poliovirus type 1 in water, milk and yoghurt. Int. J. Food Microbiol. 74:73-78. [DOI] [PubMed] [Google Scholar]
- 235.Sundkvist, T., C. Aitken, G. Duckworth, and D. Jeffries. 1997. Outbreak of acute hepatitis A among homosexual men in East London. Scand. J. Infect. Dis. 29:211-212. [DOI] [PubMed] [Google Scholar]
- 236.Sunen, E., and M. D. Sobsey. 1999. Recovery and detection of enterovirus, hepatitis A virus and Norwalk virus in hardshell clams (Mercenaria mercenaria) by RT-PCR methods. J. Virol. Methods 77:179-187. [DOI] [PubMed] [Google Scholar]
- 237.Sunen, E., N. Casas, B. Moreno, and C. Zigorraga. 2004. Comparison of two methods for the detection of hepatitis A virus in clam samples (Tapes spp.) by reverse transcription-nested PCR. Int. J. Food Microbiol. 91:147-154. [DOI] [PubMed] [Google Scholar]
- 238.Szmuness, W., J. L. Dienstag, R. H. Purcell, E. J. Harley, C. E. Stevens, and D. C. Wong. 1976. Distribution of antibody to hepatitis A antigen in urban adult populations. N. Engl. J. Med. 295:755-759. [DOI] [PubMed] [Google Scholar]
- 239.Tallo, T., H. Norder, V. Tefanova, K. Ott, V. Ustina, T. Prukk, O. Solomonova, J. Schmidt, K. Zilmer, L. Priimagi, and T. Krispin. 2003. Sequential changes in hepatitis A virus genotype distribution in Estonia during 1994 to 2001. J. Med. Virol. 70:187-193. [DOI] [PubMed] [Google Scholar]
- 240.Tapp, I., L. Malmberg, E. Rennel, M. Wik, and A. C. Syvanen. 2000. Homogeneous scoring of single-nucleotide polymorphisms: comparison of the 5′-nuclease TaqMan assay and Molecular Beacon probes. BioTechniques 28:732-738. [DOI] [PubMed] [Google Scholar]
- 241.Tassopoulos, N. C., G. J. Papaevangelou, J. R. Ticehurst, and R. H. Purcell. 1986. Fecal excretion of Greek strains of hepatitis A virus in patients with hepatitis A and in experimentally infected chimpanzees. J. Infect. Dis. 154:231-237. [DOI] [PubMed] [Google Scholar]
- 242.Taylor, M. B. 1997. Molecular epidemiology of South African strains of hepatitis A virus: 1982-1996. J. Med. Virol. 51:273-279. [DOI] [PubMed] [Google Scholar]
- 243.Ticehurst, J. R., V. R. Racaniello, B. M. Baroudy, D. Baltimore, R. H. Purcell, and S. M. Feinstone. 1983. Molecular cloning and characterization of hepatitis A virus cDNA. Proc. Natl. Acad. Sci. USA 80:5885-5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Ticehurst, J. C., J. I. Cohen, and R. H. Purcell. 1988. Analysis of molecular sequences demonstrates that hepatitis A virus (HAV) is a unique picornavirus, p. 33-35. In A. J. Zuckerman (ed.), Viral hepatitis and liver disease. Alan R. Liss, New York, N.Y.
- 245.Tjon, G. M. S., C. J. Wijkmans, R. A. Coutinho, A. G. Koek, J. A. R. van den Hoeka, A. C. A. P. Leenders, P. M. Schneeberger, and S. M. Bruisten. 2005. Molecular epidemiology of hepatitis A in Noord-Brabant, The Netherlands. J. Clin. Virol. 32:128-136. [DOI] [PubMed] [Google Scholar]
- 246.Tsai, Y. L., M. D. Sobsey, L. R. Sangermano, and C. J. Palmer. 1993. Simple method of concentrating enteroviruses and hepatitis A virus from sewage and ocean water for rapid detection by reverse transcriptase-polymerase chain reaction. Appl. Environ. Microbiol. 59:3488-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Tsai, Y. L., B. Tran, L. R. Sangermano, and C. J. Palmer. 1994. Detection of poliovirus, hepatitis A virus, and rotavirus from sewage and ocean water by triplex reverse transcriptase PCR. Appl. Environ. Microbiol. 60:2400-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Tsarev, S. A., S. U. Emerson, M. S. Balayan, J. Ticehurst, and R. H. Purcell. 1991. Simian hepatitis A virus (HAV) strain AGM 27: comparison of genome structure and growth in cell culture with other HAV strains. J. Gen. Virol. 72:1677-1683. [DOI] [PubMed] [Google Scholar]
- 249.Van Steenbergen, J. E., V. G. Tjon, A. v. d. Hoek, A. Koek, R. A. Coutinho, and S. M. Bruisten. 2004. Two years' prospective collection of molecular and epidemiological data shows limited spread of hepatitis A virus outside risk groups in Amsterdam, 2000-2002. J. Infect. Dis. 189:471-482. [DOI] [PubMed] [Google Scholar]
- 250.Vento, S., T. Garofano, C. Renzini, F. Cainelli, F. Casali, G. Ghironzi, T. Ferraro, and E. Concia. 1998. Fulminant hepatitis associated with hepatitis A virus superinfection in patients with chronic hepatitis C. N. Engl. J. Med. 338:286-290. [DOI] [PubMed] [Google Scholar]
- 251.Villar, L. M., E. Lampe, A. Meyer, and A. M. Gaspar. 2004. Genetic variability of hepatitis A virus isolates in Rio de Janeiro: implications for the vaccination of school children. Braz. J. Med. Biol. Res. 37:1779-1787. [DOI] [PubMed] [Google Scholar]
- 252.Vong, S., A. E. Fiore, D. O. Haight, J. Li, N. Borgsmiller, W. Kuhnert, F. Pinero, K. Boaz, T. Badsgard, C. Mancini, O. V. Nainan, S. Wiersma, and B. P. Bell. 2005. Vaccination in the county jail as a strategy to reach high risk adults during a community-based hepatitis A outbreak among methamphetamine drug users. Vaccine 23:1021-1028. [DOI] [PubMed] [Google Scholar]
- 253.Wang, Z. Q., L. Ma, H. Gao, and X. G. Dong. 1986. Propagation of hepatitis A virus in human diploid fibroblast cells. Acta Virol. 30:463-467. [PubMed] [Google Scholar]
- 254.Wang, C. H., S. Y. Tschen, U. Heinricy, M. Weber, and B. Flehmig. 1996. Immune response to hepatitis A virus capsid proteins after infection. J. Clin. Microbiol. 34:707-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Wasley, A., T. Samandari, T., and B. P. Bell. 2005. Incidence of hepatitis A in the United States in the era of vaccination. JAMA 294:194-201. [DOI] [PubMed] [Google Scholar]
- 256.Wattanasri, N., K. Ruchusatsawat, and S. Wattanasri. 2005. Phylogenetic analysis of hepatitis A virus in Thailand. J. Med. Virol. 75:1-7. [DOI] [PubMed] [Google Scholar]
- 257.Weber, D. J., S. L. Barbee, M. D. Sobsey, and W. A. Rutala. 1999. The effect of blood on the antiviral activity of sodium hypochlorite, a phenolic, and a quaternary ammonium compound. Infect. Control Hosp. Epidemiol. 20:821-827. [DOI] [PubMed] [Google Scholar]
- 258.Weinberg, M., J. Hopkins, L. Farrington, L. Gresham, M. Ginsberg, and B. P. Bell. 2004. Hepatitis A in Hispanic children who live along the United States-Mexico border: the role of international travel and food-borne exposures. Pediatrics 114:e68-e73. [DOI] [PubMed] [Google Scholar]
- 259.Weitz, M., B. M. Baroudy, W. L. Maloy, J. R. Ticehurst, and R. H. Purcell. 1986. Detection of a genome-linked protein (VPg) of hepatitis A virus and its comparison with other picornaviral VPgs. J. Virol. 60:124-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Wheeler, C., T. Vogt, G. L. Armstrong, G. Vaughan, A. Weltman, O. V. Nainan, V. Dato, G. Xia, K. Waller, J. Amon, T. M. Lee, A. Highbaugh-Battle, C. Hembree, S. Evenson, M. A. Ruta, I. T. Williams, A. Fiore, and B. P. Bell. 2005. An outbreak of hepatitis A associated with green onions. N. Engl. J. Med. 353:890-897. [DOI] [PubMed] [Google Scholar]
- 261.Wiedmann, M., S. Boehm, W. Schumacher, C. Swysen, and M. Zauke. 2003. Evaluation of three commercial assays for the detection of hepatitis A virus. Eur. J. Clin. Microbiol. Infect. Dis. 22:129-130. [DOI] [PubMed] [Google Scholar]
- 262.Xia, G., Y. Yi, K. Guo, and H. Tian. 1997. Comparison of the Chinese LJ strain structural gene with HM175, MBB, LA strains and the expression of hepatitis A virus antigen by LJ/HM175 recombinant vaccinia virus. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 11:208-211. [PubMed]
- 263.Yao, G. 1991. Clinical spectrum and natural history of viral hepatitis A in a 1988 Shanghai epidemic, p. 76-78. In F. B. Hollinger, S. M. Lemon, and H. S. Margolis (ed.), Viral hepatitis and liver disease. Williams & Wilkins, Baltimore, Md.
- 264.Yotsuyanagi, H., S. Iino, K. Koike, K. Yasuda, K. Hino, and K. Kurokawa. 1993. Duration of viremia in human hepatitis A viral infection as determined by polymerase chain reaction. J. Med. Virol. 40:35-38. [DOI] [PubMed] [Google Scholar]
- 265.Yotsuyanagi, H., K. Koike, K. Yasuda, K. Moriya, Y. Shintani, H. Fujie, K. Kurokawa, and S. Iino. 1996. Prolonged fecal excretion of hepatitis A virus in adult patients with hepatitis A as determined by polymerase chain reaction. Hepatology 24:10-13. [DOI] [PubMed] [Google Scholar]
- 266.Zhou, Y. J., M. K. Estes, X. Jiang, and T. G. Metcalf. 1991. Concentration and detection of hepatitis A virus and rotavirus from shellfish by hybridization tests. Appl. Environ. Microbiol. 57:2963-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]