Abstract
Deactivation of light-activated rhodopsin (metarhodopsin II) involves, after rhodopsin kinase and arrestin interactions, the hydrolysis of the covalent bond of all-trans-retinal to the apoprotein. Although the long-lived storage form metarhodopsin III is transiently formed, all-trans-retinal is eventually released from the active site. Here we address the question of whether the release results in a retinal that is freely diffusible in the lipid phase of the photoreceptor membrane. The release reaction is accompanied by an increase in intrinsic protein fluorescence (release signal), which arises from the relief of the fluorescence quenching imposed by the retinal in the active site. An analogous fluorescence decrease (uptake signal) was evoked by exogenous retinoids when they non-covalently bound to native opsin membranes. Uptake of 11-cis-retinal was faster than formation of the retinylidene linkage to the apoprotein. Endogenous all-trans-retinal released from the active site during metarhodopsin II decay did not generate the uptake signal. The data show that in addition to the retinylidene pocket (site I) there are two other retinoid-binding sites within opsin. Site II involved in the uptake signal is an entrance site, while the exit site (site III) is occupied when retinal remains bound after its release from site I. Support for a retinal channeling mechanism comes from the rhodopsin crystal structure, which unveiled two putative hydrophobic binding sites. This mechanism enables a unidirectional process for the release of photoisomerized chromophore and the uptake of newly synthesized 11-cis-retinal for the regeneration of rhodopsin.
During its function as a photoreceptor, the visual pigment rhodopsin undergoes changes through a cycle of uptake of 11-cis-retinal and release of photoisomerized chromophore, all-trans-retinal (reviewed in Ref. 1). In rhodopsin, 11-cis-retinal is bound to the opsin apoprotein by a Schiff base linkage to Lys296. In the dark ground state, the rhodopsin conformation with a λmax of 500 nm is stabilized by a salt bridge between the protonated Schiff base and the Glu113 counterion. The activated state metarhodopsin II (Meta II, λmax = 380 nm) arises from light-induced 11-cis/all-trans-retinal isomerization and conformational changes that break the salt bridge but retain the all-trans-retinylidene linkage in the original binding site (reviewed in Refs. 2-5). Thereby, photoisomerization is coupled to the protein conformational change that leads to a G-protein-coupled signal cascade, eventually setting off neuronal signaling (1).
Initial deactivation of Meta II begins with the interaction of active rhodopsin with its receptor kinase, phosphorylation of the receptor, and a tight binding of arrestin to the still activated phosphorylated form of the receptor (6, 7). Full deactivation occurs when rhodopsin is regenerated. This requires the hydrolysis of the all-trans-retinylidene linkage and release of all-trans-retinal from the active site (1). Critical steps include the nucleophilic attack of water on the retinylidene bond within the hydrophobic binding site of rhodopsin and the diffusion of the hydrolyzed chromophore out of the binding pocket. Formation of opsin accompanies a significant increase in intrinsic Trp fluorescence after release of all-trans-retinal from the active site (8, 9). The retinal remains associated with opsin membranes and is converted by endogenous NADPH-dependent retinol dehydrogenase (RDH,1 reviewed in Ref. 1) to all-trans-retinol without further change in the intrinsic protein fluorescence (8). However, the stability of the complex between arrestin and phosphorylated Meta II may depend on the RDH activity (10, 11). In addition, during the Meta II decay a storage form of rhodopsin, metarhodopsin III (Meta III), is generated. Meta III and opsin are formed in parallel (8).
Studies on the effect of active site methylation (12) and palmitoylation of opsin on transducin (Gt) activation, and the retinoid competition on regeneration (13, 14), led to the proposal of a secondary binding site for all-trans-retinal within the opsin molecule. In this study, we demonstrate a site-directed retinal migration from the active site to an exit site during Meta II decay. Once this process has occurred, 11-cis-retinal is taken up into an entrance site and moved to the active site for rhodopsin regeneration. This ligand channeling mechanism is based firmly on the crystal structure of rhodopsin (15, 16), which allows identification of the putative secondary binding sites of the chromophore.
EXPERIMENTAL PROCEDURES
Rod Disc Membrane Preparations—Rod outer segments (ROS) were prepared from frozen bovine retinas (17). Isolation of disc membranes from ROS was carried out under dim red light by a repetitive extraction of soluble and membrane-associated proteins with low ionic strength buffer (5 mm Pipes, pH 7.0, 1 mm EDTA) (18). Rhodopsin concentration was determined in solubilized samples (1% LDAO) from the difference in absorption at 500 nm before and after the complete bleaching using extinction coefficient ε = 40,600 m−1 cm−1. Opsin membranes were prepared from illuminated disc membranes, in the presence of hydroxylamine (10 mm), by extraction of the photoisomerized chromophore with bovine serum albumin (fatty acid-free) (13, 14). Opsin concentration was determined in solubilized samples (1% n-dodecyl-β-maltoside, DM) from its absorption at 280 nm using extinction coefficient ε = 81,200 m−1 cm−1 (19). All membrane preparations were resuspended in isotonic BTP-buffer (130 mm NaCl, 1 mm MgCl2, and 20 mm bis-Tris-propane, pH 7.5) and stored at −80 °C with 0.3 m sucrose. Membrane suspensions were sonicated briefly prior to the fluorescence measurements to reduce turbidity. Samples were solublized at low concentrations of opsin in membranes (∼1 μm) with 0.1% DM in the BTP buffer.
Illumination Protocol—Complete bleaching (15 s at 495 nm) of rhodopsin membranes under conditions favoring Meta II was carried out at pH 6.0 (20 °C), and pH was adjusted to the desired value immediately after photoactivation (8). To reach a complete decay of Meta II, the sample was incubated for 60 min.
Fluorescence Measurements—Emission spectra of intrinsic Trp fluorescence (SPEX Fluorolog 1680 instrument) were recorded between 310 and 400 nm using excitation at 295 nm and in BTP buffer, pH 6.5, at 20 °C. Samples were bovine serum albumin-stripped opsin membranes or solubilized opsin (0.1% DM) at a final concentration of 1 μm in 750 μl. In some samples, ethanolic solutions of retinoids (5 μl) were added in a stoichiometric amount to opsin. The excitation and emission slit bandpass were 0.4 and 7 nm, respectively. Low light intensities were used to prevent bleaching reactions of retinal or rhodopsin in the samples. For time-resolved records, fluorescence emission was monitored at 330 nm under the conditions outlined above. The fluorescence intensity was measured in counts per second (cps).
Protein and Chromophore Modeling—A rhodopsin model suitable for simulations was built using the coordinates deposited in the Protein Data Bank under 1HZX identifier (15). Hydrogen atoms were added in Insight II (v.2000, Accelrys, San Diego, CA). Missing loops of rhodopsin were created using Modeler module (20) of Insight II. Verifications of created loops and the whole structure were done using Profile-3D module (21) by calculating the compatibility between the sequence and the structure.
RESULTS
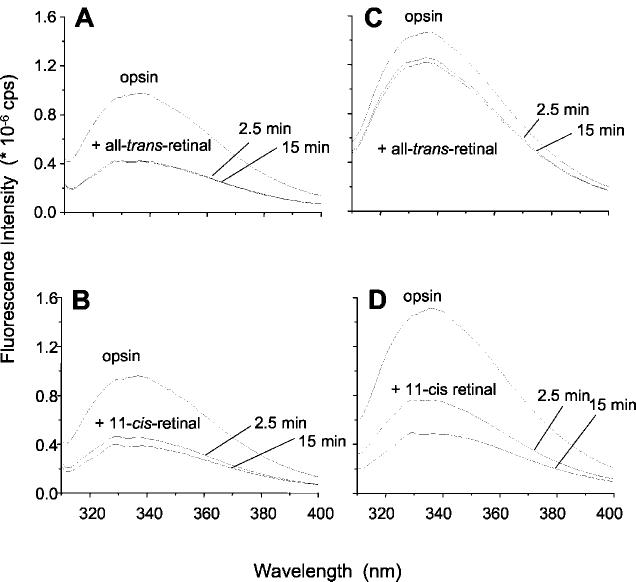
Quenching of the Intrinsic Protein Fluorescence upon Uptake of Retinal into Opsin—Changes in intrinsic Trp fluorescence are monitors of interactions between 11-cis-retinal or all-trans-retinal and opsin (8, 9, 22, 23). Addition of all-trans-retinal to membranes containing opsin significantly quenched the Trp fluorescence emission (Fig. 1A). This change coincided with the observation that all-trans-retinal formed a non-covalent complex with opsin (11-14, 24-26). When the membranes were solubilized in n-dodecyl-β-maltoside (DM), all-trans-retinal had only a minor effect on opsin fluorescence, suggesting that the binding of the retinal to opsin is detergent-sensitive (Fig. 1C). Addition of 11-cis-retinal to either membrane-bound or solubilized opsin significantly quenched Trp fluorescence (Fig. 1, B and D). 11-cis-Retinal regenerated rhodopsin under both conditions, as evidenced by an increase in absorption at 500 nm (data not shown), thereby quenching the intrinsic Trp fluorescence. These experiments unveiled that binding of all-trans-retinal or 11-cis-retinal leads to a decrease in intrinsic protein fluorescence emission. However, only 11-cis-retinal can regenerate rhodopsin.
Fig. 1.
Fluorescence emission spectra of opsin: effect of retinal isomers. All-trans-retinal (A) or 11-cis-retinal (B) was added to a final concentration of 1 μm to a preparation of photoreceptor disc membranes (1 μm opsin). The fluorescence quenching is seen with both 11-cis-retinal and all-trans-retinal, indicating the uptake of both isomers into the membrane-bound apoprotein. Spectra C and D are from solubilized membranes (0.1% n-dodecyl-β-maltoside). All spectra were recorded at λex = 295 nm, pH 6.5, 20 °C, after 2.5 and 15 min, as indicated.
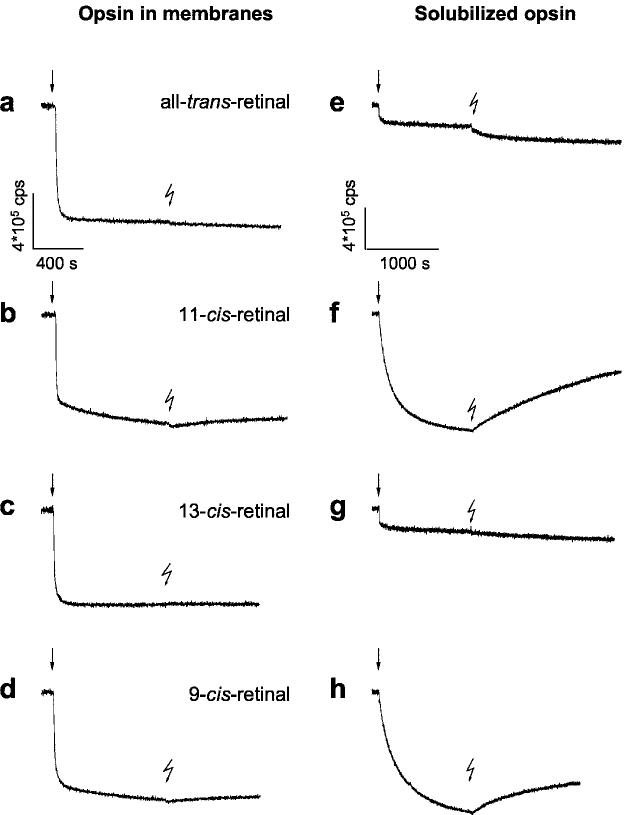
11-cis-Retinal Uptake Precedes Schiff Base Formation—The interaction of retinal isomers with opsin was further analyzed by monitoring the time-dependent changes of intrinsic Trp fluorescence (Fig. 2). The complex formation between all-trans-retinal and membrane-bound opsin proceeded within seconds (t½ ∼5 s; Fig. 2, trace a). As expected (14), subsequent illumination of the opsin·all-trans-retinal complex did not result in any significant change in fluorescence.
Fig. 2.
Time-dependent fluorescence changes in opsin apoprotein evoked by retinal isomers. Changes in intrinsic protein fluorescence were recorded upon addition of retinals (1 μm) to opsin (1 μm) at 20 °C (pH 6.5). Fluorescence emission of opsin in native membrane (left, a–d) and in 0.1% n-dodecyl-β-maltoside (right, e–h) was recorded at 330 nm with λex = 295 nm. The rapid initial quenching by retinal addition (arrows) is termed the uptake signal. Each sample was illuminated for 15 s with green light (OG 495) after incubation, as indicated by the flash symbol. The fluorescence change after illumination was assigned to the release of the photoisomerized chromophore during decay of the activated metarhodopsin II (8, 9)
The uptake signal, as we term the rapid decrease in intrinsic protein fluorescence upon addition of retinoids, induced by addition of 11-cis-retinal to opsin membranes is biphasic (Fig. 2, trace b). The initial fast phase (t½ ∼6 s) is similar to the uptake signal seen with all-trans-retinal. The slower decrease in fluorescence intensity appeared to follow regeneration of rhodopsin (data not shown). This is confirmed by the fact that subsequent illumination led to an expected increase in fluorescence (Fig. 2, trace b) (8, 9), termed here the “release signal,” that is specific only to retinoids that regenerate pigments such as 9-cis-retinal (trace d) (27, 28), and not 13-cis-retinal (trace c), or all-trans-retinal (trace a). The results demonstrate that both all-trans-retinal and 11-cis-retinal associate very rapidly with opsin to form opsin·retinal complexes. In the case of 11-cis-retinal, the complex is the precursor of the regenerated pigment.
In solubilized samples, the uptake signal with 11-cis-retinal is slower (t½ ∼140 s; Fig. 2, trace f). The signal likely correspond to regeneration of rhodopsin, in which the retinal bound in the original pocket, quenches Trp fluorescence. Consistently, a substantial fluorescence change was induced by illumination of the sample after completion of the uptake signal, showing that a considerable fraction of opsin was regenerated to light-sensitive rhodopsin under these conditions. Virtually no light-induced fluorescence changes are observed after incubation of solubilized opsin with all-trans-retinal or 13-cis-retinal (Fig. 2, traces e and g), while 9-cis-retinal (Fig. 2, trace h) had similar properties to those of 11-cis-retinal.
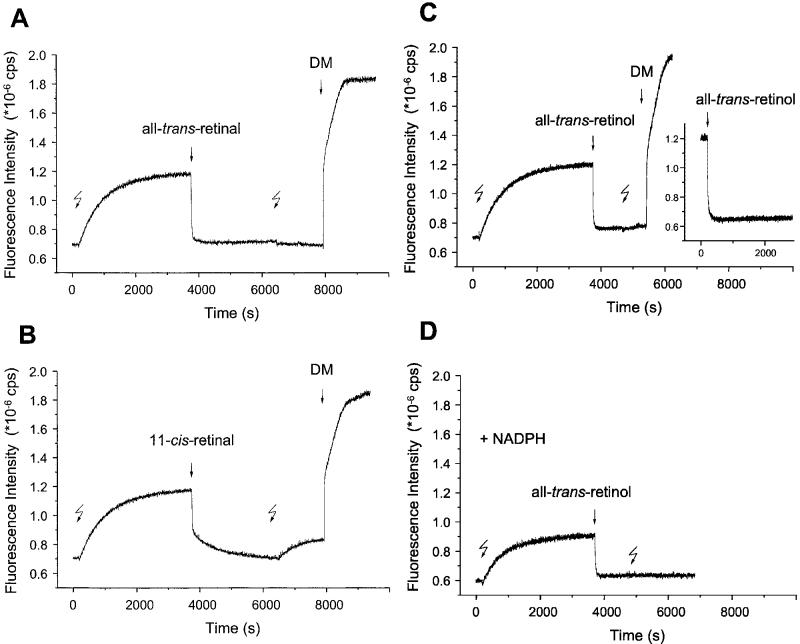
Uptake of Retinal After Decay of Meta II—The decay of Meta II is accompanied by a significant increase in fluorescence, both in detergent (8, 9) and in native membranes (8). This is the release signal, reflecting the release of all-trans-retinal from the active site. Here, Meta II formed in membranes was allowed to decay for 60 min without removal of the endogenous photoisomerized chromophore. Subsequent addition of exogenous all-trans-retinal at 1:1 ratio to opsin (Fig. 3A) produced a normal uptake signal, although an amount equimolar to opsin was already present in the membranes. The signal had the same amplitude as was measured with purified opsin without endogenous retinal (Fig. 2, trace a). A second illumination after addition of exogenous all-trans-retinal and incubation had no effect on opsin fluorescence. However, when the membranes were solublized with DM, a biphasic increase in fluorescence intensity was observed (Fig. 3A). The amplitude of the fast initial jump is comparable with the amplitude of the uptake signal, suggesting that the detergent causes dissociation of the complex between opsin and the exogenously added all-trans-retinal. Subsequently, the fluorescence increased to a high Trp emission intensity, which is typical for solubilized opsin (see Fig. 1). Consistently, the release signal from a solubilized light-activated rhodopsin sample was larger (data not shown).
Fig. 3.
Time-dependent fluorescence changes evoked by retinoids after decay of active metarhodopsin II. Decay of light-activated metarhodopsin II (Meta II, 1st flash) and release of all-trans-retinal from the active site caused an increase in the intrinsic protein fluorescence emission (initial slow release signal). Subsequent addition of all-trans-retinal (1 μm)(A), 11-cis-retinal (1 μm) (B), or all-trans-retinol (1 μm) (C and D) to membranes containing the Meta II decay product results in the full reversal of opsin fluorescence to its initial level. The addition of all-trans-retinol to opsin apoprotein evokes a uptake signal (C, inset), which is very similar to that obtained after complete Meta II decay (C). Due to rhodopsin regeneration, a second increase in fluorescence is seen for the 11-cis-retinal-treated sample (B) upon a second illumination (2nd flash), but not for all-trans-retinal- or all-trans-retinol-treated samples. The maximal emission intensity was observed upon membrane solubilization with 0.1% dodecyl-β-maltoside (DM) as shown for samples A and B. All-trans-retinal release signal is also seen in the presence of NADPH, and subsequent addition of all-trans-retinol again leads to a normal uptake signal (D).
A large, biphasic uptake signal was observed when 11-cis-retinal was used instead of all-trans-retinal (Fig. 3B). These results are similar to the observation made for opsin depleted of endogenous chromophore (see Fig. 2, trace b). A second illumination evoked a small release signal. The addition of detergent increased the fluorescence to the same maximum level as was seen with the all-trans isomer. No significant uptake signals were observed upon addition of retinoids to dark-adapted rhodopsin membranes (data not shown).
The observation of a normal uptake signal after Meta II decay demonstrates that there is a rapid complex formation between the exogenously added all-trans-retinal/11-cis-retinal and the decay product of Meta II. The retinal released in the course of Meta II decay does not diffuse freely. If this were the case, it would necessarily equilibrate, thus reducing or even canceling the uptake signal. Even if we assume that part of the released chromophore bound to binding sites other than on opsin, the same would apply to the exogenously provided chromophore. Consistently, all measured uptake signals (including that with empty apoprotein and with the Meta II decay product) had the same amplitude. Moreover, the absolute level of fluorescence intensity of purified empty opsin was similar to the Meta II decay product (i.e. opsin containing endogenous all-trans-retinal). These data argue for two independent binding sites for retinals on the opsin molecule (entrance and exit sites, see “Discussion”), in addition to the original binding pocket bearing the 11-cis-retinylidene/all-trans-retinylidene linkage.
A similar protocol was employed with all-trans-retinol. The uptake signal was again observed both with decayed Meta II and isolated opsin apoprotein (Fig. 3C), demonstrating that all-trans-retinol quenches opsin fluorescence like the retinal isomers. The release signal of all-trans-retinal is also seen in the presence of NADPH (Fig. 3D). The amplitude of the signal is reduced due to the quenching effect of NADPH but is similar when normalized to initial fluorescence intensity (see Ref. 8). This result suggests that all-trans-retinol enzymatically formed during Meta II decay, like endogenous all-trans-retinal, may not be entirely free to diffuse.
Crystal Structure of Rhodopsin Offers Potential Binding Sites for Retinoids—In the crystal structure of rhodopsin, two heptane-1,2,3-triol (HTO) molecules were identified that bind to two hydrophobic cavities (Fig. 4a). One of them is located in a large cavity, which also easily accommodates 11-cis-retinal (311 Å3) and all-trans-retinal (309 Å3), close to the palmitoylation site within the transmembrane segment (termed site II). For the all-trans-retinal, the contact residues include Met317, Val318, and Leu321 of helix 8 around the polyene chain and Leu50, Ile54 (helix 1), Val304, and Met308 (helix 7) around the β-ionone ring (Fig. 4b). Cys322 and Leu328 of the C-terminal loop are close to the oxygen atom of retinal. Site II for the cis-retinoid consists of Phe52, Pro53, Phe56, Leu57, Gln64, and Tyr60 of helix 1. Thr 320 and Lys339 of the C-terminal loop are close to the oxygen atom of retinal. Leu321 divides the cis- from the trans-site and is covered by palmitoyl chains. It is plausible that these sites are occupied by detergent when membranes are solubilized, competing out the retinoid effects as demonstrated in the study. The second site is located on the cytoplasmic surface of rhodopsin (site III) (Fig. 4c). It encompasses Asn315, Cys316, Thr319 (helix 8), Lys325, Asn326, Leu328, and Gly329 (C-terminal loop) and residues around the β-ionone include Ala333, Thr336, Val337, Ser338, Glu341, and Thr342. The size of site II is large enough to accommodate all-trans-retinal and 11-cis-retinal individually or together (Fig. 4b), while the smaller site III can only bind one all-trans-retinal molecule (Fig. 4c). Site III is different from the HTO-binding site located in the main cavity on the rhodopsin cytoplasmic surface, which is more hydrophilic than site III. In agreement with the modeling, all-trans-retinal is likely to bind to the more hydrophobic cavity termed site III.
Fig. 4.
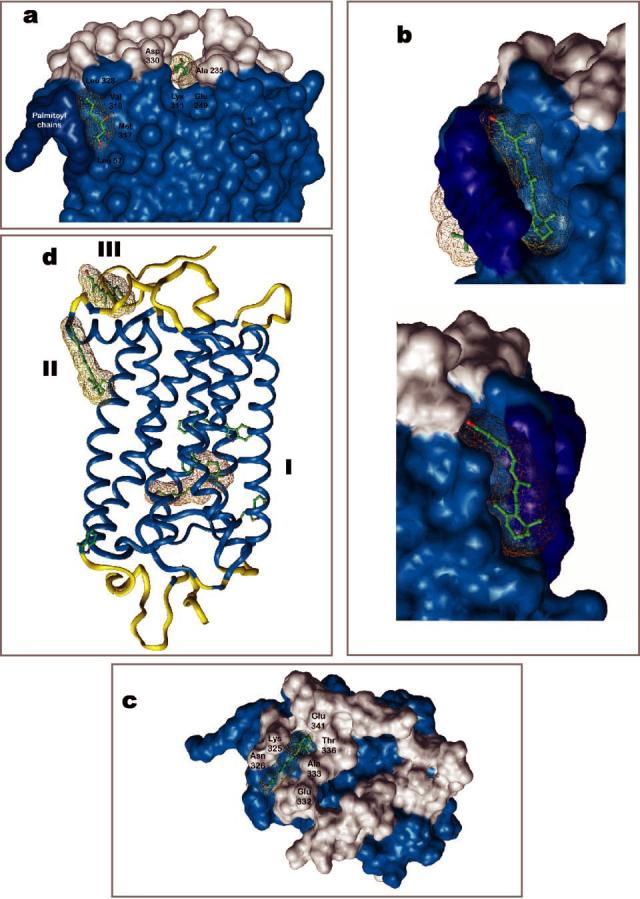
Putative binding sites of retinals on rhodopsin. Two heptane-1,2,3-triol (HTPO)-binding sites were identified in the crystal structure of rhodopsin (15, 16), one site is located close to the palmitoylation site (site II) and the second in the vicinity of C terminus and the 3rd cytoplasmic loop. Additional four HTO molecules are bound to extracellular half of rhodopsin (not shown) (a). The large site II can accommodate all-trans-retinal, 11-cis-retinal, or both together (b). Site III is located closer to the surface and could be the exit site for all-trans-retinal (c). In all figures, the transmembrane segments are colored in light blue and palmitoyl chains in dark blue. Site I is the original binding site for the chromophore, 11-cis-retinylidene, which undergoes photoisomerization. All three sites and the side chains of Trp residues are depicted in panel d.
DISCUSSION
Based on intrinsic opsin fluorescence data, we have analyzed the conversions linked to the uptake and release of retinoids by rhodopsin. We used preparations of isolated disc membranes, a simplified system devoid of the complex metabolic machinery of regeneration but providing the correct lipid host and thus a valid model to study retinoid release (8) and uptake. The evidence presented in our studies shows that rhodopsin contains putative peripheral binding sites for retinoids in addition to the well known retinylidene pocket (site I in Fig. 4d). The data suggest a channeling of the retinal ligand through an entrance site (site II), the active site I, and an exit site (site III). Sites II and III are only functional in an undisturbed lipid environment in the absence of detergent, because binding of retinoids is based on hydrophobic interactions.
During Decay of Meta II, all-trans-Retinal Is Released from the Active Site to an Exit Site—A release of the chromophore from the original binding site (site I) can be observed by an accompanying change in the intrinsic Trp fluorescence of the apoprotein (this study and Refs. 8 and 9). According to current understanding, all-trans-retinal is released to the cytoplasm and then reduced to all-trans-retinol by a membrane-bound RDH (1). A fraction of all-trans-retinal may escape into the intradiscal space, where it is removed to the cytoplasm by an ABCR transporter protein (reviewed in Refs. 1 and 29).
A first surprising result of the current analysis is that opsin, embedded in the lipid host of the membrane, does not release its photoisomerized chromophore into the bulk lipid phase. Instead, opsin keeps the chromophore bound in a site termed the “exit” site, preventing all-trans-retinal to diffuse freely. Endogenous all-trans-retinal does not migrate to the entrance site and does not cancel the release signal, while exogenously added retinoids bind to the entrance site, suppressing Trp fluorescence. Moreover, the persistent occupation of the exit site by endogenous all-trans-retinal is not merely due to a higher affinity of the exit site compared with the entrance site. If this were the case, exogenously added retinoid (1:1 ratio of retinoid to opsin) would preferentially bind to the exit site, thus not causing a quenching of fluorescence. The data in Fig. 3 show that after completion of the release signal, the binding signal is the same as with opsin, i.e. with empty active and release sites (Fig. 2). We conclude that the exit site remains closed for added retinoids, and then filling of the entrance site does not require emptying of the exit site.
Reduction of all-trans-Retinal Can Occur in the Exit Site—Although all-trans-retinol evokes an uptake signal when exogenously added, release signal of all-trans-retinal in the presence of NADPH and subsequent uptake signal of exogenous retinol were again of the same size (Fig. 3, C and D). This indicates that during Meta II decay and subsequent enzymatic reduction, neither all-trans-retinal nor -retinol became free (see above). We may infer that, at least under the experimental conditions, even all-trans-retinol remained bound to the exit site. This would in turn allow the conclusion that reduction involves opsin·RDH complex formation. Although we have not yet attempted to study the complex with arrestin, reduction of the opsin-bound photoisomerized chromophore would open the possibility that both arrestin and RDH bind. This observation would explain why arrestin does not inhibit the visual cycle in vivo (30).
Retinoids Are Taken Up into an Entrance Site—The release of all-trans-retinal and also the uptake of retinoids accompany a change in intrinsic protein fluorescence. The quench effect upon uptake is observed in native disc membranes, but not in the presence of detergent (Fig. 1). The uptake signal is observed with both all-trans-retinal and 11-cis-retinal when they are added to opsin devoid of any endogenous retinoid (Figs. 1 and 2) or after the decay of Meta II (Fig. 3). For a 1:1 ratio of retinal to opsin, the signal is of approximately the same size as the release signal. An obvious explanation would be that the added retinal re-fills the active site during Schiff base formation. However, this cannot be the case because the fluorescence change is 20 times faster than rhodopsin regeneration. The kinetics indicate a separate complex formation step, different from the formation of ground state rhodopsin. This would still leave open the possibility that the retinoids enter the active site and form a non-covalent association inside the pocket (site I). In such a model, an assumption would be that in a subsequent slower reaction, 11- or 9-cis-retinal would form the Schiff base from this pre-bound state. However, excess of all-trans-retinal or all-trans-retinol do not inhibit formation of rhodopsin (14, 31, 32). This observation excludes any significant occupation of the active site by all-trans-retinal during uptake into opsin and proves that, for all-trans-retinal, the uptake signal cannot be interpreted as an uptake into the active site. Consistent with these observations, all-trans-retinal and 13-cis-retinal, which do not regenerate rhodopsin because they do not fit the active site of opsin (33), also evoke a detergent-sensitive uptake signal (Fig. 2). Because the uptake yielded a signal with the same amplitude and similarly fast kinetics, not only with all-trans-but also with 11-cis- and 9-cis-retinal, we infer that 11-cis-retinal also passes through an “entrance” site(s) before it migrates to the active site and forms the Schiff base. In the case of 11-cis-retinal, it is channeled to the active site, causing a transient activation of opsin (34) before the 11-cis-reinylidene linkage is formed. This reaction could be aided by two residues, Glu181 and Glu113, which may act as general bases.
Ligand Channeling in Opsin—The scheme in Fig. 5A interprets the findings based on the notion that retinals are unidirectionally passed through the three sites identified above (ligand-channeling hypothesis). The reaction pathway starts with rhodopsin (activation state), in which 11-cis-retinal is covalently bound via a protonated Schiff base to the active site and the intrinsic Trp fluorescence is fully quenched. Light absorption leads through retinal isomerization to Meta II (active state). Despite the activating conversion, the net change in fluorescence between rhodopsin and Meta II is very small (8). A large change in Trp fluorescence (relief from fluorescence quenching) occurs in the subsequent Meta II decay. In parallel to Meta III (storage state) formation, exit and entrance sites are now set to function. Channeling all-trans-retinal into the exit site and opening the entrance for various retinoids generates a native form of opsin, the opsin (exit) state. In this state, the RDH enzyme has access to its all-trans-retinal substrate and can form all-trans-retinol with NADPH as a cofactor. The cycle is closed when fresh 11-cis-retinal binds to the entrance site, to form the opsin·11-cis-retinal complex (entrance state), which enables regeneration of rhodopsin. The driving force for retinal channeling is provided by the light reaction, which is also the link between the two ducts for cis- and trans-retinal, respectively (Fig. 5A), and secures the unidirectionality of the overall process.
Fig. 5.
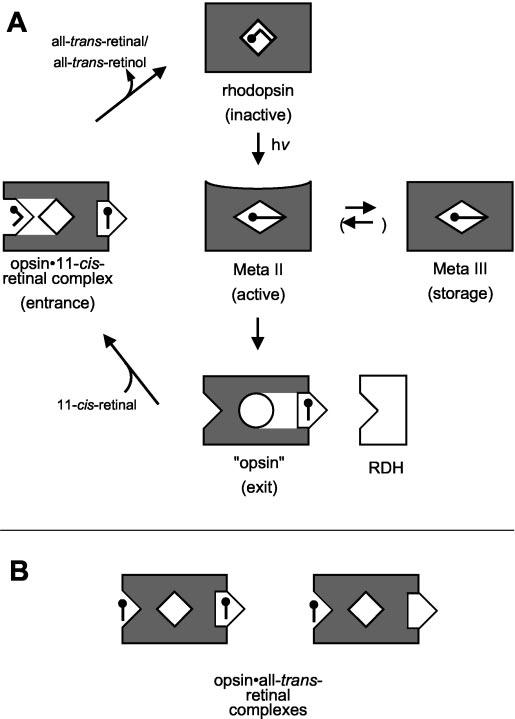
Intramolecular ligand channeling in rhodopsin. A, in the light-sensitive ground state (rhodopsin, top) the chromophore 11-cis-retinal is covalently attached to Lys296 within the binding pocket (site I) of the apoprotein opsin. Photoisomerization to all-trans-retinal results in formation of the active intermediate metarhodopsin II (Meta II, center). Meta II decays to a storage form (Meta III, right) or into the apoprotein opsin with the photolyzed all-trans-retinal bound in the exit site (site III; opsin, bottom). In this state, the retinal does not quench intrinsic Trp fluorescence. Opsin eventually returns to the ground state via a transiently formed opsin·11-cis-retinal·complex, which contains both retinal isomers bound to the entrance (site II) and exit (site III). RDH has access to its retinal substrate while bound to the exit site of opsin. Opsin·all-trans-retinal complexes (B) are formed with the stripped apoprotein (right) or the Meta II decay product (left) in the presence of free diffusible all-trans-retinal.
When fresh 11-cis-retinal binds to the entrance, the exit site can release all-trans-retinal or all-trans-retinol. In this condition, the all-trans species may also bind to the entrance site (of another opsin molecule) (14). The complex formed in vitro (Fig. 5B), for which an activity toward the G-protein (12, 14, 25) and rhodopsin kinase (35) has been demonstrated, may be identified with the state with all-trans-retinal in the entrance site. It is important to note that all-trans-retinal bound to the entrance site does not inhibit the regeneration of rhodopsin (14). In agreement with the modeling (see below), this implies a more complex structure of the entrance domain, with a site to which all-trans-retinal can bind without inhibition of the duct for 11-cis-retinal permeation. A consequence of the above reaction scheme is that empty opsin is unlikely to exist in situ. Only harsh treatment in vitro, e.g. by washing with bovine serum albumin or treatment with hydroxylamine, can fully strip the exit site. Finally, it must be recognized that all-trans-retinal added in excess forms both covalent and non-covalent complexes with opsin as documented in previous studies (11, 12).
Assignment of Ligand Channeling to Rhodopsin Structure—The ligand channeling found in this study is reminiscent of the tunneling of substrate found in some hydrophobic ligand-binding proteins, such as phosphatidyl-transfer protein (36). Most likely, the selectivity comes about through a specific opsin conformation that arises when all-trans-retinal is released from the active site. This conformational change is a late consequence of photon absorption and cannot be induced once opsin is stripped of endogenous all-trans-retinal and collapsed to a new conformation. Such a transformation would allow unidirectional uptake and release of the chromophore. In a previous investigation of the opsin·all-trans-retinal complex (14), we had found that all-trans-retinal-induced opsin activity toward the G-protein depended on palmitoylation of Cys322 and Cys323. When combining this with the present results, one would identify site II as the entrance, and would suggest site III as the exit. In native opsin, only 11-cis-retinal can pass the tunnel to the active site I. Only for certain mutants, including E113Q (37), the all-trans isomer has access to site I.
According to modeling, either all-trans- or 11-cis-retinal can bind to site II in rhodopsin, but it would take opsin to allow all-trans- or 11-cis-retinal to induce the conformational change seen in the fluorescence uptake signal. The detailed mechanism by which the conformational change translates into a fluorescence change remains to be elucidated. There is a large distance between the entrance site and Trp265, which resides in site I and is the interaction partner of the β-ionone in the dark state. Therefore, we must envisage an indirect mechanism for the quenching related to retinal uptake. There is an additional change in fluorescence when 11-cis-retinal passes from the entrance to the active site. 11-cis-retinal may reorient itself during Schiff base formation, thus tightly immobilizing Trp265 (38).
CONCLUSIONS
Using intrinsic Trp fluorescence, we analyzed how a ligand finds its way through a native receptor protein. The ligand (11-cis-/all-trans-retinal) is passed over through rhodopsin from entrance to active to exit sites. Although these sites can already be assigned to the structure of the inactive ground state, the structure of the tunnel, which allows the ligand to pass, is not known. The data indicate that after the passage, the ligand remains bound to the putative exit site (site III). In this way, reduction to retinol can occur through the action of a retinol-binding protein, RDH. Although these results could be specific to the visual system, the mechanism may be more universal for the large family of G-protein-coupled receptors.
Acknowledgments
We thank Jana Engelmann for excellent technical assistance. Computational tasks were performed in part at the ICM Computer Center in Warsaw, Poland.
Footnotes
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (SFB 366), the Fonds der Chemischen Industrie (to K. P. H.), National Institutes of Health Grant EY09339, a grant from Research to Prevent Blindness, Inc. (RPB) to the Dept. of Ophthalmology at the University of Washington, and grants from the Foundation Fighting Blindness, Inc., the E. K. Bishop Foundation (to K. P.), and National Institutes of Health Grant GM-63020 (to D. C. T.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The abbreviations used are: RDH, retinol dehydrogenase; BTP, bis-Tris-propane; Gt, photoreceptor G-protein, transducin; HTO, heptane-1,2,3-triol; Meta II, metarhodopsin II (photoactivated rhodopsin); Meta III, metarhodopsin III; DM, n-dodecyl-β-maltoside; Pipes, 1,4-piperazinediethanesulfonic acid; ROS, rod outer segment.
REFERENCES
- 1.McBee JK, Palczewski K, Baehr W, Pepperberg DR. Prog. Retin. Eye Res. 2001;20:469–529. doi: 10.1016/s1350-9462(01)00002-7. [DOI] [PubMed] [Google Scholar]
- 2.Okada T, Ernst OP, Palczewski K, Hofmann KP. Trends Biochem. Sci. 2001;26:318–324. doi: 10.1016/s0968-0004(01)01799-6. [DOI] [PubMed] [Google Scholar]
- 3.Hofmann KP. Novartis Found Symp. 1999;224:158–175. doi: 10.1002/9780470515693.ch10. discussion 175–180. [DOI] [PubMed] [Google Scholar]
- 4.Filipek S, Stenkamp RE, Teller DC, Palczewski K. Annu. Rev. Physiol. 2003;65:851–879. doi: 10.1146/annurev.physiol.65.092101.142611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeGrip WJ, DeLange F, Klaassen CH, Verdegem PJ, Wallace-Williams S, Creemers AF, Bergo V, Bovee PH, Raap J, Rothschild KJ, DeGroot HJ, Lugtenburg J. Novartis Found Symp. 1999;224:102–118. doi: 10.1002/9780470515693.ch7. discussion 118–123. [DOI] [PubMed] [Google Scholar]
- 6.Palczewski K. Eur. J. Biochem. 1997;248:261–269. doi: 10.1111/j.1432-1033.1997.00261.x. [DOI] [PubMed] [Google Scholar]
- 7.Pulvermüller A, Palczewski K, Hofmann KP. Biochemistry. 1993;32:14082–14088. doi: 10.1021/bi00214a002. [DOI] [PubMed] [Google Scholar]
- 8.Heck M, Schädel SA, Maretzki D, Bartl FJ, Ritter E, Palczewski K, Hofmann KP. J. Biol. Chem. 2003;278:3162–3169. doi: 10.1074/jbc.M209675200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farrens DL, Khorana HG. J. Biol. Chem. 1995;270:5073–5076. doi: 10.1074/jbc.270.10.5073. [DOI] [PubMed] [Google Scholar]
- 10.Palczewski K, Jäger S, Buczylko J, Crouch RK, Bredberg DL, Hof-mann KP, Asson-Batres MA, Saari JC. Biochemistry. 1994;33:13741–13750. doi: 10.1021/bi00250a027. [DOI] [PubMed] [Google Scholar]
- 11.Hofmann KP, Pulvermüller A, Buczylko J, Van Hooser P, Palczewski K. J. Biol. Chem. 1992;267:15701–15706. [PubMed] [Google Scholar]
- 12.Jäger S, Palczewski K, Hofmann KP. Biochemistry. 1996;35:2901–2908. doi: 10.1021/bi9524068. [DOI] [PubMed] [Google Scholar]
- 13.Sachs K, Maretzki D, Hofmann KP. Methods Enzymol. 2000;315:238–251. doi: 10.1016/s0076-6879(00)15847-1. [DOI] [PubMed] [Google Scholar]
- 14.Sachs K, Maretzki D, Meyer CK, Hofmann KP. J. Biol. Chem. 2000;275:6189–6194. doi: 10.1074/jbc.275.9.6189. [DOI] [PubMed] [Google Scholar]
- 15.Teller DC, Okada T, Behnke CA, Palczewski K, Stenkamp RE. Biochemistry. 2001;40:7761–7772. doi: 10.1021/bi0155091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 17.Papermaster DS. Methods Enzymol. 1982;81:48–52. doi: 10.1016/s0076-6879(82)81010-0. [DOI] [PubMed] [Google Scholar]
- 18.Kühn H, Wilden U. Methods Enzymol. 1982;81:489–496. doi: 10.1016/s0076-6879(82)81066-5. [DOI] [PubMed] [Google Scholar]
- 19.Surya A, Foster KW, Knox BE. J. Biol. Chem. 1995;270:5024–5031. doi: 10.1074/jbc.270.10.5024. [DOI] [PubMed] [Google Scholar]
- 20.Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- 21.Bowie JU, Luthy R, Eisenberg D. Science. 1991;253:164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- 22.Ebrey TG. Photochem. Photobiol. 1972;15:585–588. doi: 10.1111/j.1751-1097.1972.tb06269.x. [DOI] [PubMed] [Google Scholar]
- 23.Ebrey TG. Proc. Natl. Acad. Sci. U. S. A. 1971;68:713–716. doi: 10.1073/pnas.68.4.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fukada Y, Yoshizawa T. Biochim. Biophys. Acta. 1981;675:195–200. doi: 10.1016/0304-4165(81)90226-9. [DOI] [PubMed] [Google Scholar]
- 25.Cohen GB, Oprian DD, Robinson PR. Biochemistry. 1992;31:12592–12601. doi: 10.1021/bi00165a008. [DOI] [PubMed] [Google Scholar]
- 26.Cohen GB, Yang T, Robinson PR, Oprian DD. Biochemistry. 1993;32:6111–6115. doi: 10.1021/bi00074a024. [DOI] [PubMed] [Google Scholar]
- 27.Kandori H, Shichida Y, Yoshizawa T. Biochemistry (Mosc.) 2001;66:1197–1209. doi: 10.1023/a:1013123016803. [DOI] [PubMed] [Google Scholar]
- 28.Kawamura S, Wakabayashi S, Maeda A, Yoshizawa T. Vision Res. 1978;18:457–462. doi: 10.1016/0042-6989(78)90057-3. [DOI] [PubMed] [Google Scholar]
- 29.Sun H, Nathans J. J. Bioenerg. Biomembr. 2001;33:523–530. doi: 10.1023/a:1012883306823. [DOI] [PubMed] [Google Scholar]
- 30.Palczewski K, Van Hooser JP, Garwin GG, Chen J, Liou GI, Saari JC. Biochemistry. 1999;38:12012–12019. doi: 10.1021/bi990504d. [DOI] [PubMed] [Google Scholar]
- 31.Matsumoto H, Yoshizawa T. Vision Res. 1978;18:607–609. doi: 10.1016/0042-6989(78)90212-2. [DOI] [PubMed] [Google Scholar]
- 32.Daemen FJ. Nature. 1978;276:847–848. doi: 10.1038/276847a0. [DOI] [PubMed] [Google Scholar]
- 33.Jang GF, Kuksa V, Filipek S, Bartl F, Ritter E, Gelb MH, Hofmann KP, Palczewski K. J. Biol. Chem. 2001;276:26148–26153. doi: 10.1074/jbc.M102212200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kefalov VJ, Crouch RK, Cornwall MC. Neuron. 2001;29:749–755. doi: 10.1016/s0896-6273(01)00249-5. [DOI] [PubMed] [Google Scholar]
- 35.Buczylko J, Saari JC, Crouch RK, Palczewski K. J. Biol. Chem. 1996;271:20621–20630. doi: 10.1074/jbc.271.34.20621. [DOI] [PubMed] [Google Scholar]
- 36.Roderick SL, Chan WW, Agate DS, Olsen LR, Vetting MW, Rajashankar KR, Cohen DE. Nat. Struct. Biol. 2002;9:507–511. doi: 10.1038/nsb812. [DOI] [PubMed] [Google Scholar]
- 37.Sakmar TP, Franke RR, Khorana HG. Proc. Natl. Acad. Sci. U. S. A. 1989;86:8309–8313. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saam J, Tajkhorshid E, Hayashi S, Schulten K. Biophys. J. 2002;83:3097–3112. doi: 10.1016/S0006-3495(02)75314-9. [DOI] [PMC free article] [PubMed] [Google Scholar]