Abstract
Two factors are mainly responsible for the stability of the DNA double helix: base pairing between complementary strands and stacking between adjacent bases. By studying DNA molecules with solitary nicks and gaps we measure temperature and salt dependence of the stacking free energy of the DNA double helix. For the first time, DNA stacking parameters are obtained directly (without extrapolation) for temperatures from below room temperature to close to melting temperature. We also obtain DNA stacking parameters for different salt concentrations ranging from 15 to 100 mM Na+. From stacking parameters of individual contacts, we calculate base-stacking contribution to the stability of A•T- and G•C-containing DNA polymers. We find that temperature and salt dependences of the stacking term fully determine the temperature and the salt dependence of DNA stability parameters. For all temperatures and salt concentrations employed in present study, base-stacking is the main stabilizing factor in the DNA double helix. A•T pairing is always destabilizing and G•C pairing contributes almost no stabilization. Base-stacking interaction dominates not only in the duplex overall stability but also significantly contributes into the dependence of the duplex stability on its sequence.
INTRODUCTION
Stability of the DNA double helix with respect to separation of complementary strands is known to depend on the base composition of the duplex (1–4). A classical study of Marmur and Doty (1) on DNA polymer stability gives a linear relationship between the G•C content of the polymer and its melting temperature. The simplest explanation of the linear dependence is that A•T- and G•C-pairs differ in stability independently of their neighbors. Accordingly, base-stacking interactions have been thought to constitute only a small correction to the major effect of the differences in G•C and A•T pair stabilities (5). Nearest-neighbor stability parameters have been introduced to account for sequence effects in DNA stability. These parameters are obtained from the analysis of the melting data for DNA polymers (5), DNA oligomers (6–8) and DNA dumbbells (9,10), and present the cumulative (base pairing and stacking) contribution of each dinucleotide stack to the overall stability of the molecule. In fact, DNA melting experiments do not allow separation of the two contributions.
Partitioning of base pairing and stacking contributions to DNA stability not only delivers a new aspect in the fundamental understanding of DNA structure and energetics, but also it has significant implications in a number of biological processes. Fluctuations in local helical conformation of DNA, the phenomenon known as DNA breathing, lead to infrequent events of base pair opening thus making normally buried groups available for modification and interaction with proteins (11,12). Fluctuational base pair opening implies disruption of hydrogen bonds between the complementary bases and flipping the base out of the helical stack disrupting two contacts. Heterogeneous stacking at these contacts determine sequence dependence of the base pair fluctuational motility. Moreover, single-stranded break (a nick) in the DNA double helix is stabilized by stacking interactions between base pairs flanking the lesion; these interactions are sequence-dependent (13). In the cell, DNA nicks are substrates for DNA damage-detecting and DNA-repair proteins (14–17). In vitro, DNA nick-based approaches are often used in DNA detection and amplification protocols (18,19). Also, stabilization achieved through coaxial stacking interactions has been put into effect to improve the efficiency of short primer hybridization for standard sequencing protocols (20) and within the format of sequencing by hybridization approach (21). By using partially double-stranded probe with a 5 nt overhang, the latter method takes advantage of the enhancement of the stability of the short duplex formed between the probe overhang and single-stranded target by coaxial stacking with the duplex interface of the probe. Another biotechnology application involving nicked DNA is the formation of padlock probes via ligation of a nick formed by two termini of artificial oligonucleotide hybridized to single-stranded or locally opened duplex DNA (22–24).
A number of theoretical studies addressed characterization of stacking and base pairing contributions to the duplex stability (25–30). Experimentally, energetics of base-stacking interactions in nucleic acids has been evaluated by studying the effect of dangling (unpaired) terminal bases on the overall stability of duplexes (31–35) and in the coaxial stacking hybridization experiments where binding of a short oligonucleotide to the single-stranded DNA template is assisted by stacking interaction with the nearby duplex interface (36–41). Both of these approaches rely on thermal denaturation measurements of short duplex molecules.
We have recently introduced an entirely new approach for characterization of stacking interactions in the DNA double helix (13). For this purpose we study DNA molecules with solitary nicks positioned in a strictly defined sequence context. We subject these molecules to PAGE. Structurally, introduction of a break to one of the strands of DNA perturbs only slightly the double helix conformation with stacking and base pairing at the nick site remaining intact (42–49). Nicked molecules have been shown to move somewhat slower during PAGE than intact molecules of the same size; this retardation is enhanced at higher temperatures (14,50,51).
The model of the DNA nick underlying our approach involves an equilibrium between the two conformations as shown in Figure 1a (13). One conformation is very close to that of the intact double helix where stacking between the base pairs flanking the nick is conserved. We assume that no optimization of stacking interactions—like in case of duplexes with dangling nucleotides (52)—occurs at the site of the nick in DNA. The other conformation corresponds to complete loss of stacking at the nick site thus inducing a kink in DNA. The fast equilibration between stacked/straight and unstacked/bent conformations of the nick directly affects the mobility of DNA molecule during PAGE leading to a differential retardation characteristic to a particular dinucleotide carrying the nick, i.e. KL. For each contact, stacking free energy parameter, , is ascribed governing the distribution of the molecules between the two conformations. Using PAGE mobility data of DNA molecules with solitary nicks flanked by all possible combinations of the base pairs we extract stacking parameters of the DNA double helix (13). Using heterogeneous stacking parameters and a well-known Marmur–Doty (1) dependence of DNA stability on its content, we have previously arrived at a revised set of the nearest-neighbor stability parameters which is in full agreement with the DNA melting data (13).
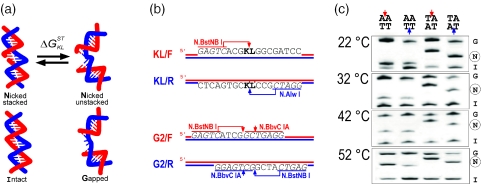
Figure 1.
Urea-enhanced gel electrophoresis of nicked and gapped DNA. (a) Nicked DNA molecule undergoes stacked-unstacked transition governed by stacking parameter . Stacked state is close to intact molecule; unstacked state is approximated by a molecule with a gap. (b) DNA molecules with solitary nicks and gaps in forward (F) or reverse (R) strand were obtained by enzymatic digestion. Nicking enzymes and their recognition sites are shown in one color; cleavage sites are indicated by arrowheads. In KL/F and KL/R fragments nick is positioned within KL/K′L′ dinucleotide stack. Fragments with 2 nt-long gaps, G2/F and G2/R, are the result of sequential digestion by two nicking enzymes as shown. (c) Effect of temperature of PAGE on the separation of intact (I), nicked (N) and gapped (G) DNA fragments. Nicked stacks are shown at the top of each lane; arrowheads point to the location of the nick. Gapped and nicked fragments resolved in one lane have lesions in the same strand. PAGE was conducted in the presence of 2.3 M urea at temperatures indicated to the left of each panel.
Here we measure, for the first time, temperature dependence and salt dependence of base-stacking contribution to the DNA duplex stability. Contributions of A•T and G•C pairing are estimated from the comparison of A•T- and G•C-containing polymer stability parameters with the stacking terms. We find that throughout the temperature range employed, base-stacking interactions stabilize DNA double helix. Temperature dependence of the base-stacking term fully determines the temperature dependence of the DNA stability parameter. Base pairing term is destabilizing in case of A•T and somewhat stabilizing in case of G•C pairs. Differential contribution of base-stacking in A•T- and G•C-containing contacts is responsible for 50% of the dependence of DNA stability on its G•C content. Salt conditions affect stacking parameters leading to stabilization upon an increase in sodium concentration. In this case, we also find that salt dependence of stacking term determines the salt dependence of the DNA stability term.
MATERIALS AND METHODS
DNA
The 300 bp-long DNA molecules used in this study are PvuII/PvuII (all enzymes used in this study were purchased from New England Biolabs, Ipswich MA) restriction fragments of pUC19 derivatives described before (13). Briefly, pKL (were K and L are DNA bases) carries recognition sites for nicking endonucleases each of which introduces a single nick to the forward (N.BstNB I) or reverse (N.Alw I) strand of KL/K′L′ dinucleotide stack (or simply KL; K′ and L′ are complementary to K and L) yielding KL/F and KL/R fragments, respectively (Figure 1b). The nick is located 104 bp apart from the terminus of PvuII/PvuII fragment. Molecules with a single 2 nt gap in place of a nick are obtained by digesting pG2F and pG2R by N.BbvC IA followed by N.BstNBI digestion. As shown in Figure 1b, nicking sites of these nicking endonucleases are 2 nt apart and located on the forward strand of G2/F and reverse strand of G2/R. To remove 5′ phosphoryl groups, nicked and gapped molecules were incubated with Alkaline Phosphatase (CIP). Following treatment with enzymes, all DNA samples were purified by phenol extraction, precipitated with ethanol and resuspended in TE buffer [10 mM Tris (pH 7.4) and 1 mM EDTA].
Gel electrophoresis
Nicked, gapped and intact DNA fragments 300 bp in length were concurrently subjected to electrophoresis through 6.7% (w/v) polyacrylamide gels run at 17 V/cm with 1× TBE [90 mM Tris (pH 8.0), 90 mM boric acid and 1 mM EDTA] as a running buffer. We have shown before that under this set of PAGE conditions stacked-unstacked equilibrium at the nick site of DNA is not affected by specific experimental design (13). Specifically, Figure 5 of (13) demonstrates that the presence of the gel matrix and force applied to the molecule during PAGE do not affect apparent free energy parameters of stacked-unstacked equilibrium at the DNA nick. We also conducted PAGE in the presence of increasing concentrations of sodium by adding NaCl up to 85 mM to the gel and to the running buffer; the strength of electric field for these experiments was reduced to 12 V/cm. The gel electrophoresis was performed using Protean II xi Cell (BioRad, Hercules, CA) where the gel-plate sandwich is mounted against the temperature-controlling core connected to a circulating water bath set to a desired temperature. The temperature during PAGE was measured by a thermocouple (Oakton, Vernon Hills, IL) with a type T wire probe inserted directly into the gel. DNA band patterns were visualized by ethidium bromide staining and detected by the CCD camera.
Figure 5.
Effect of phosphorylation state of 5′ nt at the nick site on stacked-unstacked equilibrium. (a) Stacking parameters measured in 1× TBE at 37°C of A•T-containing (top panel) and G•C-containing (bottom panel) nicked dinucleotide stacks before (circles) and after (triangles) dephosphorylation are compared. White and gray fills are used for fragments with the nick in the forward and reverse strand, respectively. Nicked dinucleotide stacks with a purine at a 5′-side of the nick are underscored (see text). (b) Salt dependence of ΔGST values of nicked contacts indicated to the right of each panel before (circles) and after (triangles) dephosphorylation. Total concentration of sodium assuming 1× TBE to be equivalent to 15 mM Na+ is indicated, see Figure 4.
Relative mobilities of DNA fragments with a nick, μ, were calculated as a ratio of the distance migrated by the nicked fragments to the distance migrated by the intact fragment during PAGE. All experiments were conducted at least in triplicates. To enhance the separation of the molecules with solitary nicks from the intact molecules we added various concentrations of urea (0.4 to 8 M) to the gel thus allowing subsequent extrapolation of measured parameters to zero urea (13). We also conducted PAGE in the presence of dimethylformamide (DMF) in place of urea. In this case, a smaller range of concentrations was used since the gels became fragile at concentrations of DMF above 1.5 M.
Melting experiments
Two synthetic 65 nt-long complementary DNA strands (MWG Biotech Inc., High Point, NC) were annealed in TE buffer appended with a desired concentration of NaCl (from 20 to 200 mM) at 0.5 µM of each strand. The same duplex was also prepared in the 1×TBE buffer. The purity of the duplex was checked by PAGE. Melting curves of DNA duplexes were collected using a CARY Bio100 spectrophotometer equipped with the peltier thermocontroller (Varian Instruments, Walnut Creek, CA). Both denaturation and renaturation curves were collected at slow heating/cooling rate of at 0.08 °/min; no hysteresis between heating and cooling curves has been observed. Melting temperature, TM, was estimated as a temperature at the midpoint of the duplex-to-coil transition.
In melting studies we determined stabilities of four DNA duplexes, which differed in G•C content, χ (DNA sequences are listed in Supplementary Table 1). Salt dependence of A•T-containing duplex stability (χ = 0%) was measured directly for 22 mM < [Na+] < 202 mM (i.e. 20–200 mM NaCl plus 1 mM EDTA). Salt dependence of more stable G•C-containing duplex was obtained by extrapolating TM's measured for four duplexes with χ = 0%, χ = 14%, 32% and 41.5% to χ = 100%. Corresponding plots are shown in Figure 4a.
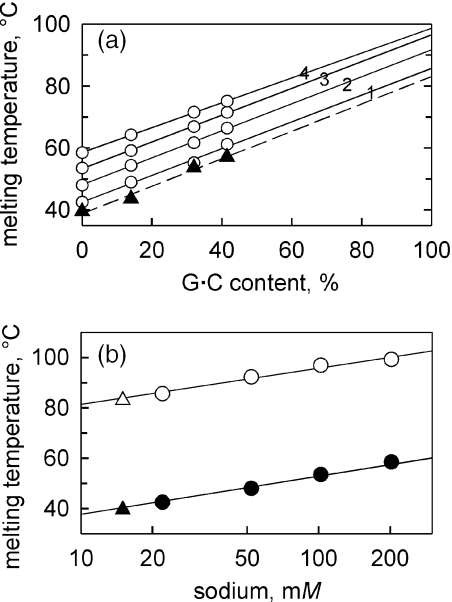
Figure 4.
Stability of DNA under different salt conditions. (a) Dependence of TM of 65 bp-long DNA duplexes on their G•C content in 1× TBE (triangles, dashed line) and in 10 mM TE (pH 7.4) (circles, solid lines) appended with NaCl at 20 mM (line 1), 50 mM (line 2), 100 mM (line 3) and 200 mM (line 4). Linear extrapolation to χ = 100% is used to estimated TM of G•C-containing DNA. (b) Stability of A•T-containing (open symbols) and G•C-containing (closed symbols) 65 bp-long duplex DNA as a function of sodium concentration (circles) and in 1×TBE (triangles). TM's of A•T-containing DNA were measured directly; TM's of G•C-containing DNA were estimated by extrapolation shown in (a). For both duplexes, stability in 1× TBE corresponds to the stability in the presence of 15 mM Na+ (as plotted).
RESULTS
Stacked-unstacked equilibrium at DNA nick in the presence of a denaturant
During gel electrophoresis, a 300 bp-long DNA molecule with a solitary nick migrates slower with respect to the DNA fragment without the lesion (Figure 1c). Retardation of nicked fragments depends on the identity of the base pairs flanking the nick like in case of the molecules with nicks flanked by As and Ts shown in Figure 1c. Differential retardation of nicked DNA is due to specific interactions characteristic to each nicked dinucleotide stack. Quantitatively, equilibrium between stacked/closed and unstacked/open conformations at the nick site is governed by stacking free energy, ΔGST, so that:
| 1 |
where Nclosed and Nopen are occupancies of stacked and unstacked conformations at the DNA nick, respectively, R is the universal gas constant and T is the absolute temperature.
At a given temperature, due to fast equilibration between the two states, mobility of nicked DNA is a weighted average of the mobility of molecules in closed state and mobility of molecules in open state (13). Since stacked conformation of the DNA nick is very close to the DNA molecule without the lesion μclosed is given by the mobility of the intact DNA fragment. The presence of a single-stranded stretch in a DNA molecule with a short gap prevents stacking interactions between the base pairs flanking the gap thus forcing the molecule into the open state. We use the mobility of molecules with a single 2 nt-long gap to estimate μopen. Thus stacking parameter ΔGST describing stacked-unstacked equilibrium at a nick site can be calculated directly from the mobility data using Equations 1 and 2 (13):
| 2 |
Since the perturbations brought about by the nick are often minor we introduce urea to the gel in order to enhance the separation of nicked molecules from intact molecules (Figure 2c) (13,51). Data obtained in urea-enhanced PAGE is then extrapolated to zero urea concentration resulting in stacking parameters for each nicked dinucleotide stack in the absence of urea, (Figure 2d). The use of denaturant during PAGE is an imperative part of our experimental approach since it allows characterization of even subtle differences in nicked molecule mobilities. To ensure that the nature of the denaturant does not affect the resulting stacking parameters we also conducted PAGE in the presence of varying concentrations of DMF (Figure 2d). Additionally, for the least stable stacks, we can measure stacking parameters directly at zero urea without extrapolating the mobility data collected in the presence of denaturant; this parameter is also plotted in Figure 2d. DMF-enhanced and urea-enhanced PAGE yield quantitatively similar dependencies of apparent stacking parameters on the concentration of the denaturant. This finding is quite unexpected and is most likely due to a narrow range of DMF concentrations used in PAGE; an increase in DMF concentration could yield more significant differences in apparent values measured in the presence of the two denaturants. We are unable to conduct these experiments since the gels containing more than 1.5 M DMF are too fragile to handle. Extrapolation of the data obtained in DMF-enhanced and urea-enhanced PAGE experiments to zero denaturant concentration is consistent with values measured directly in the absence of denaturant.
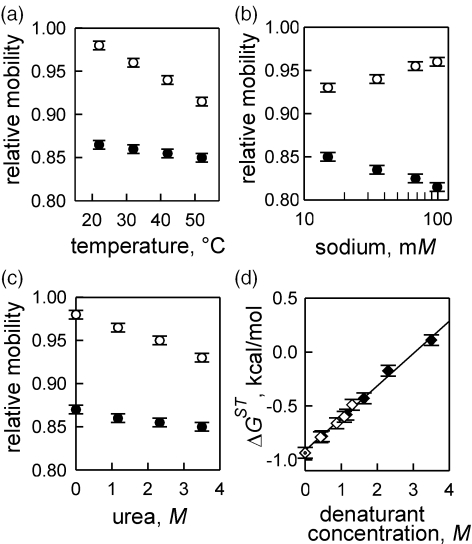
Figure 2.
Effect of ambient conditions on PAGE of nicked (open circles) and gapped (closed circles) molecules. Relative mobilities of AA/F and G2/F fragments are measured (a) at different temperatures, PAGE conditions: 1× TBE, 2.3 M urea; (b) as a function of ionic strength, PAGE conditions: 37°C, 3.5 M urea, 1× TBE appended with NaCl (total concentration of sodium assuming 1× TBE to be equivalent to 15 mM Na+ is indicated, see Figure 4); (c) in the presence of urea, PAGE conditions: 37°C, 1× TBE. (d) Effect of denaturant concentration on stacking parameters of AA/F calculated from mobility data using Equations 1 and 2. PAGE was carried out in 1× TBE at 42°C in the presence of varying concentrations of urea (closed diamonds), DMF (open diamonds) or in the absence of the denaturant (dotted diamond).
Effect of temperature on stacked-unstacked equilibrium
Our further experiments involve conducting urea-enhanced PAGE at different temperatures to obtain temperature dependence of DNA stacking parameters. Molecules with solitary nicks flanked by A•T pairs shown in Figure 1c, display a defined dependence of the electrophoretic shift towards the molecules with a 2 nt gap when temperature increases from 22 to 52°C. Relative electrophoretic mobilities of nicked AA/F fragment and gapped G2/F fragment are plotted as a function of temperature in Figure 2a. A 30 degrees increase in temperature leads to gradually decreasing PAGE mobility of the nicked fragment while mobility of the gapped fragment remains largely unaffected. In terms of occupancies of the two states of the nicked site, the shift in mobility is equivalent to the shift in the equilibrium towards the open conformation of the nicked stack. Similar results were obtained for the G•C-containing contacts. More narrow temperature range has been employed in this case since molecules with nicks located in more stable G•C-containing stacks do not resolve well enough from intact molecules at temperatures under 30°C.
Stacking free energies were calculated from the mobility data for all four A•T- and four G•C-containing nicked stacks using Equations 1 and 2. For each stack, molecules with solitary nicks located on the forward strand, KL/F and on the reverse strand, KL/R, were examined. Our further analysis is grounded on the assumption that the stacking free energies of the stacked-unstacked equilibrium at the site of the DNA nick are close to the stacking parameters of the intact DNA double helix (13). In this case we calculate the by averaging (i) the parameters obtained for KL/F and KL/R and (ii) the parameters obtained for the stacks equivalent due to dyad symmetry of the double helix (in our case there are two pairs of stacks: AA/TT and TT/AA pair and GG/CC and CC/GG pair). Individual stacking parameters are listed in Supplementary Table 2 and are plotted as a function of temperature in Figure 3a. Error bars on this plot correspond to the scatter of individual stacking parameters around the mean. As expected, throughout the temperature range employed, values for both A•T- and G•C-containing contacts increase (become less stable) gradually with increasing temperature.
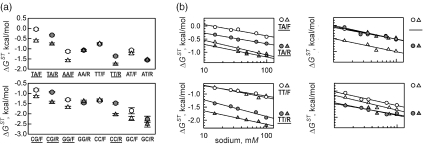
Figure 3.
Effect of ambient conditions—temperature (a) and ionic strength (b)—on DNA stacking parameters for A•T- and G•C-containing contacts. Dinucleotide stacks are shown at the top of each panel. Error bars represent the scatter range of the experimentally determined values of nicked stacks (see text). This data is tabulated in Supplementary Tables 2 and 3.
Effect of salt concentration on stacked-unstacked equilibrium
To address the effect of ionic strength on stacked-unstacked equilibrium at the site of DNA nick we conducted urea-enhanced PAGE at different salt concentrations by adding NaCl to the gel and to the running buffer. Salt dependence of relative PAGE mobilities of AA/F nicked fragment and corresponding G2/F gapped fragment is shown in Figure 2b. Unlike in case of increasing temperature (Figure 2a), opposite tendencies are displayed by the two molecules with increasing salt concentration. Mobility of the fragment with a gap decreases. The decrease is likely due to more efficient screening of phosphate charges reducing electrostatic repulsion between the two double-stranded segments on either side of the gap thus making the gap more flexible (53). Similar effect of the salt concentration is expected at the nick site in open conformation. Mobility of the nicked fragment increases corresponding to the salt-assisted shift towards stacked state of the nick.
To facilitate the comparison of DNA stacking parameters we determine experimentally, with the DNA stability parameters we have to account for the difference in salt conditions. Our PAGE-based experiments were conducted in a standard gel electrophoresis buffer 1× TBE. In order to find concentration of NaCl equivalent to PAGE conditions with respect to DNA stability, we conducted melting experiments, i.e. we determined TM of duplex DNA in 1× TBE and in the presence of various concentrations of NaCl (Figure 4). For A•T-containing DNA, melting temperatures under different salt conditions were determined directly. A DNA molecule consisting only of guanines and cytosines is much more stable and melts around or above 100°C depending on the concentration of NaCl. Melting temperatures of G•C-containing DNA were obtained by extrapolating TM's measured for four DNA duplexes with different G•C-contents (χ = 0, 14, 32 and 41.5%) to χ = 100% (Figure 4a). Thermal stabilities of A•T- and G•C-containing DNA duplexes in the presence of NaCl and in 1× TBE are compared in Figure 4b. Melting temperatures of both molecules in 1× TBE correspond to the TM of these molecules in the presence of 15 mM [Na+].
We studied eight A•T-containing and eight G•C-containing nicked dinucleotide stacks. Like in the experiments on the temperature dependence of stacking parameters described above, for each KL contact we examined KL/F and KL/R molecules and the stacking parameter was obtained as their average in case of unique contacts (AT, TA, GC and CG). In case of stacks with dyad symmetry, we also average values of equivalent stacks. Individual stacking parameters are plotted in Figure 3b and listed in Supplementary Table 3. Note that all values are plotted as a function of the ‘effective’ concentration of sodium, i.e. the fist point at [Na+] = 15 mM corresponds to the experiment conducted in 1× TBE, the last point at [Na+] = 100 mM corresponds to 1× TBE appended with 85 mM of NaCl. All values become more negative gradually as concentration of sodium increases.
Effect of phosphate at the nick site on stacked-unstacked equilibrium
Site-specific nicks in KL/F and KL/R fragments were introduced enzymatically. Nicking enzymes, like any restriction enzymes, cleave DNA strand producing a 5′ phosphate and a 3′ hydroxyl termini, which translate into two negative charges at the nick site of the double helix. To estimate the effect of the phosphate negative charges on the stacked-unstacked equilibrium at the DNA nick we considered DNA molecules where the phosphate has been removed from the nick site. Dephosphorylated nicks carry no charge.
The molecules with dephosphorylated nicks and gaps were studied by urea-enhanced PAGE in the same manner as the molecules with phosphates. As expected, treatment of DNA fragments with Alkaline Phosphatase, which results in the removal of terminal phosphates as well as dephosphorylation of the gap, had no effect on PAGE mobility of intact and gapped fragments (data not shown). ΔGST values obtained for molecules with nicked stacks before and after dephosphorylation are compared in Figure 5a. A defined trend is observed for both A•T- and G•C-containing contacts—there is a significant ∼0.4–0.6 kcal/mol shift towards more negative energies upon removal of a phosphate from the nick site only if phosphorylated 5′ nt is a purine nucleotide, like in case of AA/F and TT/R, but not in case of AA/R or TT/F. When the phosphate is located on a pyrimidine nucleotide, its removal has no effect on ΔGST values. This trend holds for 15 out of 16 nicked fragments shown in Figure 5a; the only exception is GC/F. The origin of such diverse behavior cannot be addressed by our approach and it deserves a separate detailed study. We can only suppose that removal of the phosphate from purine nucleotides allows rearrangement—in a sequence-independent manner—of the local structure benefiting stacked conformation. Alternatively, differential effect of phosphate positioning on purines or pyrimidines might be entropic in origin due to larger conformational flexibility of purine nucleotides without 5′ phosphates (54).
Note that ΔGST values of nicked stacks with and without phosphate display similar dependences on salt concentration (Figure 5b). For nicks flanked by A•T pairs, this is the case for AA/R, TT/F, AT/F and AT/R fragments which display no change in ΔGST values upon removal of the phosphate from 5′-T nt and for AA/F, TT/R, TA/F and TA/R fragments which shift towards more negative ΔGST values upon dephosphorylation of 5′-A nt. This result indicates that the phosphate charge at the nick does not contribute to the salt dependence of the measured stacking component; differences seen in Figure 5a between the nicked stacks with or without phosphate on 5′-purine nucleotides are not due to electrostatic interactions. In our further analysis we consider only molecules carrying phosphates at the nicks.
DISCUSSION
The nature of the free energy parameters governing stacked-unstacked equilibrium at DNA nick
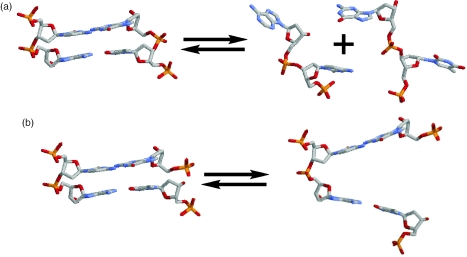
We obtain stacking free energy parameters by studying the equilibrium between stacked and unstacked form of DNA nick schematically presented in Figure 1a. The DNA molecule in the stacked state is very close the molecule without the lesion (42–49). Strictly speaking, the conformation of the unstacked state is unknown and our analysis is based on the assumption that unstacked state is close to the molecule with a short gap in place of a nick. Indeed, presence of a single-stranded gap precludes stacking interactions between two helical interfaces. Note, that this is not the case for 1 nt gaps—it appears that stacking between the base pairs flanking the gap is restored to some degree leading to anisotropic, directional bending of the molecule reducing the size of the gapped cavity (48,55). Molecules with longer gaps however, have been shown to possess isotropic bending flexibility which manifests itself in the absence of helical periodicity in electrophoretic mobility and cyclization kinetics measurements (50,53). Molecules with gaps 2, 3 and 4 nt in length migrate very closely during PAGE revealing similarity of their effective conformations (13,50). Additional factors [i.e. sequence of single-stranded linker (56)] come into effect once the gap size is longer than persistent length of single chains—in this case, the gap is likely to act as a hinge.
Thus, we use molecules with 2 nt-long gaps positioned in forward or reverse strand to approximate the open state of the nick; the site of the gap is thought of as a point of increased isotropic bending flexibility, which leads to retardation of the molecule during PAGE (57). Perturbations of DNA fragment at the gap sites are largely unaffected by ambient conditions—temperature, salt and denaturant concentration (Figure 2a–c).
Nicked DNA undergoes transition between the stacked (closed or intact-like) and unstacked (open or gapped-like) states (13). Note, that like in case of short gaps, perturbation at the nick site has been shown to be isotropic with no preferential directionality (50,58). The partitioning of molecules between these conformations determines the electrophoretic mobility of the nicked fragment and is the basis for measuring stacking parameters using Equations 1 and 2.
For further analysis we assume that the parameters describing the equilibrium at the nick site of DNA that we measure are in fact the same stacking parameters that stabilize intact double helix and determine the first term on the right-hand side of Equation 3:
| 3 |
where ΔGKL is the total melting free energy parameter (per 1 base pair) of a long DNA molecule (neglecting the end effects) and ΔGBP terms are the free energy contributions due to pairing between complementary bases (5). There are two ΔGBP parameters and for A•T and G•C pairs, respectively.
How accurate is the above assumption? Experimentally, the assumption proves to stand a critical test when we compare the values obtained for KL/F and KL/R series: these values indeed are very close [see Figure 4b in ref. (13)]. The values for dinucleotide stacks equivalent due to dyad axis of symmetry of the double helix (e.g. AA/TT and TT/AA) are close as well.
Let us examine the stacked-unstacked equilibrium at the nick of the dinucleotide stack and compare it to the helix-to-coil transition of a DNA doublet. The schematic in Figure 6 depicts these two transitions. Hydrogen bonding between complementary bases and stacking of pairs along the helical axis are disrupted upon melting; duplex melting to single strands is accompanied by the gain in conformational entropy, release of counterions and change in interactions with the solvent. Strictly speaking, partitioning of these effects between ΔGST and ΔGBP terms is not known. For example, base pairing terms in Equation 3 include breakage of hydrogen bonds between bases and formation of hydrogen bonds with water molecules rearranging the water structure around DNA chains. Likewise, base-stacking interactions have electrostatic and hydrophobic components.
Figure 6.
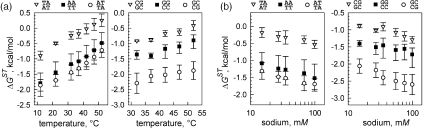
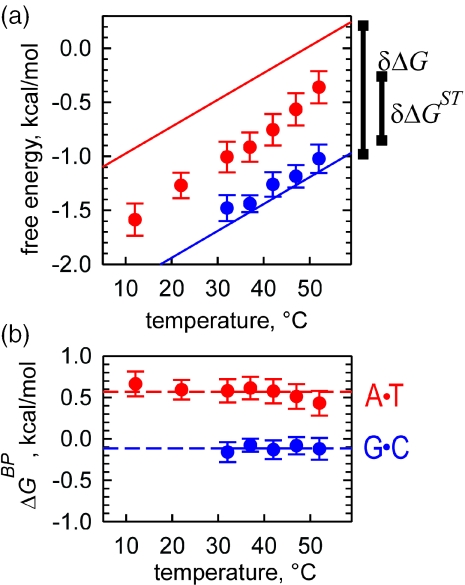
Overall stability, stacking and base pairing contributions for DNA polymers at different temperatures (a) Temperature dependence of the stacking contributions to the stability of A•T- (red circles) and G•C- (blue circles) containing polymers. Straight solid line of the same color gives the temperature dependence of the stability parameter of corresponding polymer calculated using Equations 5 and 6 at [Na+] = 15 mM. (b) Temperature dependence of A•T (red) and G•C (blue) base pairing parameters calculated as a difference between stability and stacking terms using data in (a). Horizontal broken lines correspond to mean values of kcal/mol and kcal/mol.
The stacked-unstacked transition of a nicked DNA doublet involves loss of stacking interactions while hydrogen bonding between the complementary bases is preserved (Figure 6). It also involves removal of structural constraints imposed in the double helix making most of the conformational space of the intact sugar-phosphate backbone accessible. This part of backbone conformational entropy gain is close to the gain per one strand upon duplex melting. The conformation about the glycosidic bond in an open stack, however, is preserved as in duplex form. Nicked strand is lacking O3′-P linkage; the absence of this constraint allows for additional conformations when second-order interactions (interactions depending on two torsional angles) are repealed (59). Note that the backbone conformational entropy change is independent of the particular sequence and does not affect heterogeneity of values. We assume that conformation of base pairs flanking the nick is preserved as in stacked duplex. Like in case of helix-to-coil transition, other factors contributing to transition parameters come from interactions with solvent and counterions.
We assign free energy difference to stacked-unstacked equilibrium at the nick site of DNA—this parameter describes interaction of two neighboring base pairs and is sequence-dependent. As noted above, it also includes a part of conformational entropy change and other relevant interactions. These factors are universal for all neighboring pairs and do not contribute to heterogeneity of measured . We assume further that this parameter represents the stacking term of intact DNA duplex in Equation 3. Note that this assumption explicitly implies that stacked/closed state of the nicked stack is equivalent to the stack in duplex without the lesion and no optimization of the base pair stacking occurs upon introduction of the nick like in case of dangling unpaired nucleotides (52). If this assumption is valid, then parameters of stacked-unstacked equilibrium at the nicks positioned on one or the other strand of the each dinucleotide stack will be close. Indeed, we have shown previously that stacking parameters for KL/F and KL/R molecules are similar (13). Parameters obtained for stacks equivalent due to dyad axis of symmetry, i.e. AA and TT, GG and CC, are also similar. Note that marked difference is observed in case of coaxial stacking parameters determined in melting experiments (39).
Stability of DNA polymer under different ambient conditions
Let us consider a DNA polymer comprised of adenines and thymines. The set of nearest-neighbor stability parameters of all A•T-containing dinucleotides, i.e. ΔGAT, ΔGAA = ΔGTT, ΔGTA, fully describes the contacts found in this polymer. According to Equation 3, stability parameter per 1 bp, ΔGA•T, of the A•T-polymer with a random sequence is given by [see ref. (13)]:
| 4 |
Stacking contribution to the polymer stability is the first term of the right-hand side of Equation 4; the second term presents the base pairing contribution. Experimentally determined stacking term is the mean of stacking parameters we obtained for all A•T-containing contacts. Stability parameter for G•C-containing polymer, ΔGG•C, is calculated analogously from stacking, and base pairing contributions:
| 5 |
DNA stability parameters in terms of melting temperatures, and , are salt-dependent and are given by the following empirical equations for A•T- and G•C-containing polymers, respectively (60):
| 6 |
where and are in degrees Kelvin. Equation 6 translates into free energy difference stability parameters using ΔS° = −24.85 cal/mol K (61) and Equation 7.
| 7 |
Stability parameters of A•T-containing and G•C-containing polymers calculated for [Na+] = 15 mM using Equations 6 and 7 are plotted in Figure 7a. Salt dependences of these parameters calculated at 37°C are presented in Figure 8a.
Figure 7.
Overall stability, stacking and base pairing contributions for DNA polymers at different salt conditions. (a) Salt dependence of the stacking term in the stability of A•T- (red circles) and G•C- (blue circles) containing polymers. Straight solid lines of the same color correspond to the salt dependence of the DNA polymer stability calculated using Equations 5 and 6 at 37°C. (b) Salt dependence of the base pairing parameters for A•T (red) and G•C (blue) pairs calculated at each concentration as a difference between stability term and stacking term using data from (a). Horizontal broken lines correspond to mean values of and .
Figure 8.
Schematic representation of (a) helix-to-coil transition of a DNA dinucleotide stack and (b) stacked-unstacked equilibrium at a nick site.
Temperature dependence of stacking and base pairing contributions to DNA stability
Stability of a DNA polymer comprised only of A's and T's arranged in a random order is given by two terms—the stacking term and the base pairing term—that appear on the right-hand side of Equation 4. Stacking term is calculated as an average of stacking parameters of A•T-containing contacts, which we determine in PAGE experiments conducted under certain set of ambient conditions (temperature and salt concentration). Stacking term of the G•C-containing polymer is obtained analogously.
Stacking contributions to the stability of A•T- and G•C-containing polymers for different temperatures are plotted in Figure 7a. Comparison of the temperature dependence of stacking terms with the temperature dependence of stability terms for both polymers reveals remarkable similarity between the two. Linear regression analysis of the temperature dependences of ΔGST values gives mean slope of kcal/molK, which is very close to the slope of temperature dependence of stability parameters described by Equations 6 and 7. It is evident that temperature dependence of stacking component fully determines the temperature dependence of DNA stability parameters throughout the temperature range employed in our experiments.
According to Equations 4 and 5, the difference between the melting stability of the polymer and the stacking term gives the contribution of A•T or G•C base pairing. The plots shown in Figure 7b present the base pairing term calculated for different temperatures by subtracting the experimentally determined stacking term from the polymer stability parameter. With good accuracy, the base pairing term is independent of temperature within the temperature range used in our experiments. The horizontal lines give the mean values of base pairing parameters at 15 mM Na+ of and . We conclude that stability of the DNA polymer with respect to strand separation is mainly determined by stacking interactions with base pairing being destabilizing (A•T pairs) or contributing almost nothing (G•C-pairs) (Table 1).
Table 1.
Stacking and base pairing contributions to DNA polymer stability under different ambient conditionsa
| [Na+], mM | Temperature, °C | A•T-containing polymer | G•C-containing polymer | ||
|---|---|---|---|---|---|
| 15 | 32/52 | −1.01/−0.36b | 0.57 | −1.48/−1.02b | −0.11 |
| 15/100 | 37 | −0.92/−1.32b | 0.61 | −1.44/−1.83b | −0.01 |
aAll free energy parameters are given in kcal/mol.
bThe two values correspond to the temperature or sodium concentration range as indicated.
Furthermore, throughout the temperature range used in our experiments, heterogeneity of base-stacking interactions of A•T- and G•C-containing contacts, , is responsible for a substantial part of the difference in the stabilities of A•T- and G•C-polymers, (Figure 7a). In the presence of 15 mM Na+, heterogeneous stacking accounts for δΔGST = 0.6 kcal/mol out of δΔG = 1.2 kcal/mol stability difference of A•T- and G•C-containing polymers. Thus, the dependence of DNA stability on the G•C content is not only due to the fact that G•C pairs are stronger than A•T pairs (Figure 7b) but also is due to the stronger stacking component of G•C-containing contacts (Figure 7a). This understanding of stacking domination constitutes a paradigm shift in the view on DNA stability.
Salt dependence of stacking and base pairing contributions to DNA polymer stability
Stacking terms of A•T- and G•C-containing polymers are calculated from stacking parameters obtained in PAGE experiments that have been carried out under different salt conditions (Figure 8a). Salt dependences of the stability parameters of both polymers at 37°C calculated using Equation 6 and 7, are also shown in Figure 8a. The two dependencies are very similar indicating that salt dependence of stacking term determines salt dependence of DNA stability parameters throughout the range of salt concentration used in our experiments. In case of A•T-containing contacts, salt dependence of stacking component reproduces salt dependence of DNA stability parameters with a great degree of accuracy. Salt dependences of stacking terms of A•T- and G•C-containing contacts are similar to . The dependence of stacking parameters on salt concentration was unexpected. In fact, in our original publication we assumed that base pairing term is fully responsible for ionic strength dependence of DNA melting stability (13). Figure 8a shows that this assumption is incorrect and base pairing term plotted in Figure 8b is virtually independent of salt concentration. Horizontal lines correspond to the values of base pairing parameters averaged over the whole range of salt concentrations: and (Table 1).
What is the origin of the ionic strength dependence of stacking parameters measured in PAGE experiments? It is clear that stacking is a net phenomenon involving hydrophobic, electrostatic and dispersion components (28,29,62–65). There is no agreement between researchers, however, what is the dominant force in this interaction. Luo et al. (29) emphasize a dominant role of nonelectrostatic interactions. Gellman and co-workers (65,66) report that neither solvophobic effects nor dispersion forces are important in aromatic stacking; stabilization is achieved through attractive interactions between positive and negative partial charges on bases. Guckian et al. found that hydrophobic effects dominate stacking but dispersion forces and electrostatic interaction between partial charges contribute substantially to the total base-stacking as well. This is particularly true for natural bases, which have significant charge localization (28,67). In either case, electrostatic component of base-stacking interaction (substantial or not) will depend on the ambient ionic strength through local screening of partial charges at the solvent accessible positions. In the context of the DNA double helix, however, this effect is likely to be overwhelmed by the salt-dependent interactions between highly negatively charged DNA backbones as described by the polyelectrolyte theory of DNA stability (68,69). Similarity between salt dependences of DNA stacking parameters measured in PAGE experiments and DNA stability parameters indicates that electrostatic component contributes exclusively to of Equation 3. Note, that charge of the phosphate at the 5′ nt of the nicked stack does not affect salt dependence of measured parameters.
CONCLUSIONS
Separation of the two contributions to thermal stability of the DNA double helix is achieved by studying PAGE of DNA molecules with solitary nicks and gaps. For the first time, the dependence of DNA stacking parameters on ambient conditions (temperature and salt concentration) is determined. Throughout the temperature and salt concentration range of our experiments, base-stacking interactions are always stabilizing for both A•T- and G•C-containing contacts in the DNA double helix. In fact, DNA stability is mainly determined by base-stacking interactions. G•C pairing does not contribute to stabilization of DNA duplex, while A•T pairing is always destabilizing. This finding presents a paradigm shift in the understanding of the interplay of the forces stabilizing DNA double helix. For all temperatures heterogeneity of stacking interactions in A•T- and G•C-containing contacts accounts for at least half of heterogeneity in the stability of A•T- and G•C-polymers; the other half is due to the difference in the energetics of A•T and G•C base pairing. The data on separation of stacking and base pairing contributions have made it possible to describe sequence-dependent fluctuational opening of the DNA double helix (70).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The authors thank Dr R.M. Georgiadis for providing access to spectrophotometer and Ms J. Ruemmele for technical assistance with melting experiments. The authors thank Dr Y. Mamasakhlisov and Dr P. E. Nielsen for fruitful discussions. Funding to pay the Open Access publication charges for this article was provided by NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Marmur J., Doty P. Determination of the base composition of dioxyribonucleic acid from its thermal melting temperature. J. Mol. Biol. 1962;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- 2.Wartell R.M., Benight A.S. Thermal denaturation of DNA molecules. Phys. Rep. 1985;126:67–107. [Google Scholar]
- 3.SantaLucia J., Jr, Hicks D. The thermodynamics of DNA structural motifs. Annu. Rev. Biophys. Biomol. Struct. 2004;33:415–440. doi: 10.1146/annurev.biophys.32.110601.141800. [DOI] [PubMed] [Google Scholar]
- 4.Williams M.C., Rouzina I. Force spectroscopy of single DNA and RNA molecules. Curr. Opin. Struct. Biol. 2002;12:330–336. doi: 10.1016/s0959-440x(02)00340-8. [DOI] [PubMed] [Google Scholar]
- 5.Vologodskii A.V., Amirikyan B.R., Lyubchenko Y.L., Frank-Kamenetskii M.D. Allowance for heterogeneous stacking in the DNA helix-coil transition theory. J. Biomol. Struct. Dyn. 1984;2:131–148. doi: 10.1080/07391102.1984.10507552. [DOI] [PubMed] [Google Scholar]
- 6.SantaLucia J., Jr, Allawi H.T., Seneviratne P.A. Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry. 1996;35:3555–3562. doi: 10.1021/bi951907q. [DOI] [PubMed] [Google Scholar]
- 7.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugimoto N., Nakano S., Yoneyama M., Honda K. Improved thermodynamic parameters and helix initiation factor to predict stability of DNA duplexes. Nucleic Acids Res. 1996;24:4501–4505. doi: 10.1093/nar/24.22.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doktycz M.J., Goldstein R.F., Paner T.M., Gallo F.J., Benight A.S. Studies of DNA dumbbells. I. Melting curves of 17 DNA dumbbells with different duplex stem sequences linked by T4 endloops: evaluation of the nearest-neighbor stacking interactions in DNA. Biopolymers. 1992;32:849–864. doi: 10.1002/bip.360320712. [DOI] [PubMed] [Google Scholar]
- 10.Owczarzy R., Vallone P.M., Goldstein R.F., Benight A.S. Studies of DNA dumbbells VII: evaluation of the next-nearest-neighbor sequence-dependent interactions in duplex DNA. Biopolymers. 1999;52:29–56. doi: 10.1002/(SICI)1097-0282(1999)52:1<29::AID-BIP2>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 11.Frank-Kamenetskii M.D. How the double helix breathes. Nature. 1987;328:17–18. doi: 10.1038/328017a0. [DOI] [PubMed] [Google Scholar]
- 12.Gueron M., Kochoyan M., Leroy J.L. A single mode of DNA base-pair opening drives imino proton exchange. Nature. 1987;328:89–92. doi: 10.1038/328089a0. [DOI] [PubMed] [Google Scholar]
- 13.Protozanova E., Yakovchuk P., Frank-Kamenetskii M.D. Stacked-unstacked equilibrium at the nick site of DNA. J. Mol. Biol. 2004;342:775–785. doi: 10.1016/j.jmb.2004.07.075. [DOI] [PubMed] [Google Scholar]
- 14.Le Cam E., Fack F., Menissier-de Murcia J., Cognet J.A., Barbin A., Sarantoglou V., Revet B., Delain E., de Murcia G. Conformational analysis of a 139 base-pair DNA fragment containing a single-stranded break and its interaction with human poly(ADP-ribose) polymerase. J. Mol. Biol. 1994;235:1062–1071. doi: 10.1006/jmbi.1994.1057. [DOI] [PubMed] [Google Scholar]
- 15.de Murcia G., Menissier de Murcia J. Poly(ADP-ribose) polymerase: a molecular nick-sensor. Trends Biochem. Sci. 1994;19:172–176. doi: 10.1016/0968-0004(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 16.Petrucco S., Volpi G., Bolchi A., Rivetti C., Ottonello S. A nick-sensing DNA 3′-repair enzyme from Arabidopsis. J. Biol. Chem. 2002;277:23675–23683. doi: 10.1074/jbc.M201411200. [DOI] [PubMed] [Google Scholar]
- 17.Timson D.J., Singleton M.R., Wigley D.B. DNA ligases in the repair and replication of DNA. Mutat. Res. 2000;460:301–318. doi: 10.1016/s0921-8777(00)00033-1. [DOI] [PubMed] [Google Scholar]
- 18.Nuovo G.J. In situ strand displacement amplification: an improved technique for detection of low copy nucleic acids. Diagn. Mol. Pathol. 2000;9:195–202. doi: 10.1097/00019606-200012000-00004. [DOI] [PubMed] [Google Scholar]
- 19.Van Ness J., Van Ness L.K., Galas D.J. Isothermal reactions for the amplification of oligonucleotides. Proc. Natl Acad. Sci. USA. 2003;100:4504–4509. doi: 10.1073/pnas.0730811100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotler L.E., Zevin-Sonkin D., Sobolev I.A., Beskin A.D., Ulanovsky L.E. DNA sequencing: modular primers assembled from a library of hexamers or pentamers. Proc. Natl Acad. Sci. USA. 1993;90:4241–4245. doi: 10.1073/pnas.90.9.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broude N.E., Sano T., Smith C.L., Cantor C.R. Enhanced DNA sequencing by hybridization. Proc. Natl Acad. Sci. USA. 1994;91:3072–3076. doi: 10.1073/pnas.91.8.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nilsson M., Malmgren H., Samiotaki M., Kwiatkowski M., Chowdhary B.P., Landegren U. Padlock probes: circularizing oligonucleotides for localized DNA detection. Science. 1994;265:2085–2088. doi: 10.1126/science.7522346. [DOI] [PubMed] [Google Scholar]
- 23.Kuhn H., Demidov V.V., Frank-Kamenetskii M.D. Rolling-circle amplification under topological constraints. Nucleic Acids Res. 2002;30:574–580. doi: 10.1093/nar/30.2.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hardenbol P., Baner J., Jain M., Nilsson M., Namsaraev E.A., Karlin-Neumann G.A., Fakhrai-Rad H., Ronaghi M., Willis T.D., Landegren U., et al. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat. Biotechnol. 2003;21:673–678. doi: 10.1038/nbt821. [DOI] [PubMed] [Google Scholar]
- 25.Danilov V.I., Tolokh I.S. Nature of the stacking of nucleic acid bases in water: a Monte Carlo simulation. J. Biomol. Struct. Dyn. 1984;2:119–130. doi: 10.1080/07391102.1984.10507551. [DOI] [PubMed] [Google Scholar]
- 26.Cieplak P., Kollman P.A. Calculation of the free energy of association of nucleic acid bases in vacuo and water solutions. J. Am. Chem. Soc. 1988;110:3734–3739. [Google Scholar]
- 27.Dang L.X., Kollman P.A. Molecular dynamics simulations study of the free energy of association of 9-methyladenine and 1-methylthymine bases in water. J. Am. Chem. Soc. 1990;112:503–507. [Google Scholar]
- 28.Hunter C.A. Sequence-dependent DNA structure. The role of base stacking interactions. J. Mol. Biol. 1993;230:1025–1054. doi: 10.1006/jmbi.1993.1217. [DOI] [PubMed] [Google Scholar]
- 29.Luo R., Gilson H.S., Potter M.J., Gilson M.K. The physical basis of nucleic acid base stacking in water. Biophys. J. 2001;80:140–148. doi: 10.1016/S0006-3495(01)76001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sponer J., Jurecka P., Hobza P. Accurate interaction energies of hydrogen-bonded nucleic acid base pairs. J. Am. Chem. Soc. 2004;126:10142–10151. doi: 10.1021/ja048436s. [DOI] [PubMed] [Google Scholar]
- 31.Burkard M.E., Kierzek R., Turner D.H. Thermodynamics of unpaired terminal nucleotides on short RNA helixes correlates with stacking at helix termini in larger RNAs. J. Mol. Biol. 1999;290:967–982. doi: 10.1006/jmbi.1999.2906. [DOI] [PubMed] [Google Scholar]
- 32.Bommarito S., Peyret N., SantaLucia J., Jr Thermodynamic parameters for DNA sequences with dangling ends. Nucleic Acids Res. 2000;28:1929–1934. doi: 10.1093/nar/28.9.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petersheim M., Turner D.H. Base-stacking and base-pairing contributions to helix stability: thermodynamics of double-helix formation with CCGG, CCGGp, CCGGAp, ACCGGp, CCGGUp and ACCGGUp. Biochemistry. 1983;22:256–263. doi: 10.1021/bi00271a004. [DOI] [PubMed] [Google Scholar]
- 34.Freier S.M., Alkema D., Sinclair A., Neilson T., Turner D.H. Contributions of dangling end stacking and terminal base-pair formation to the stabilities of XGGCCp, XCCGGp, XGGCCYp, and XCCGGYp helixes. Biochemistry. 1985;24:4533–4539. doi: 10.1021/bi00338a008. [DOI] [PubMed] [Google Scholar]
- 35.Freier S.M., Kierzek R., Jaeger J.A., Sugimoto N., Caruthers M.H., Neilson T., Turner D.H. Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl Acad. Sci. USA. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lane M.J., Paner T., Kashin I., Faldasz B.D., Li B., Gallo F.J., Benight A.S. The thermodynamic advantage of DNA oligonucleotide ‘stacking hybridization’ reactions: energetics of a DNA nick. Nucleic Acids Res. 1997;25:611–617. doi: 10.1093/nar/25.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vasiliskov V.A., Prokopenko D.V., Mirzabekov A.D. Parallel multiplex thermodynamic analysis of coaxial base stacking in DNA duplexes by oligodeoxyribonucleotide microchips. Nucleic Acids Res. 2001;29:2303–2313. doi: 10.1093/nar/29.11.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pyshnyi D.V., Pyshnaya I., Levina A., Goldberg E., Zarytova V., Knorre D., Ivanova E. Thermodynamic analysis of stacking hybridization of oligonucleotides with DNA template. J. Biomol. Struct. Dyn. 2001;19:555–570. doi: 10.1080/07391102.2001.10506763. [DOI] [PubMed] [Google Scholar]
- 39.Pyshnyi D.V., Goldberg E.L., Ivanova E.M. Efficiency of coaxial stacking depends on the duplex structure. J. Biomol. Struct. Dyn. 2003;21:459–468. doi: 10.1080/07391102.2003.10506940. [DOI] [PubMed] [Google Scholar]
- 40.Walter A.E., Turner D.H., Kim J., Lyttle M.H., Muller P., Mathews D.H., Zuker M. Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc. Natl Acad. Sci. USA. 1994;91:9218–9222. doi: 10.1073/pnas.91.20.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walter A.E., Turner D.H. Sequence dependence of stability for coaxial stacking of RNA helixes with Watson–Crick base paired interfaces. Biochemistry. 1994;33:12715–12719. doi: 10.1021/bi00208a024. [DOI] [PubMed] [Google Scholar]
- 42.Pieters J.M., Mans R.M., van den Elst H., van der Marel G.A., van Boom J.H., Altona C. Conformational and thermodynamic consequences of the introduction of a nick in duplexed DNA fragments: an NMR study augmented by biochemical experiments. Nucleic Acids Res. 1989;17:4551–4565. doi: 10.1093/nar/17.12.4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shore D., Baldwin R.L. Energetics of DNA twisting. I. Relation between twist and cyclization probability. J. Mol. Biol. 1983;170:957–981. doi: 10.1016/s0022-2836(83)80198-3. [DOI] [PubMed] [Google Scholar]
- 44.Erie D.A., Jones R.A., Olson W.K., Sinha N.K., Breslauer K.J. Melting behavior of a covalently closed, single-stranded, circular DNA. Biochemistry. 1989;28:268–273. doi: 10.1021/bi00427a037. [DOI] [PubMed] [Google Scholar]
- 45.Snowden-Ifft E.A., Wemmer D.E. Characterization of the structure and melting of DNAs containing backbone nicks and gaps. Biochemistry. 1990;29:6017–6025. doi: 10.1021/bi00477a020. [DOI] [PubMed] [Google Scholar]
- 46.Aymami J., Coll M., van der Marel G.A., van Boom J.H., Wang A.H., Rich A. Molecular structure of nicked DNA: a substrate for DNA repair enzymes. Proc. Natl Acad. Sci. USA. 1990;87:2526–2530. doi: 10.1073/pnas.87.7.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh S., Patel P.K., Hosur R.V. Structural polymorphism and dynamism in the DNA segment GATCTTCCCCCCGGAA: NMR investigations of hairpin, dumbbell, nicked duplex, parallel strands, and i-motif. Biochemistry. 1997;36:13214–13222. doi: 10.1021/bi970819y. [DOI] [PubMed] [Google Scholar]
- 48.Roll C., Ketterle C., Faibis V., Fazakerley G.V., Boulard Y. Conformations of nicked and gapped DNA structures by NMR and molecular dynamic simulations in water. Biochemistry. 1998;37:4059–4070. doi: 10.1021/bi972377w. [DOI] [PubMed] [Google Scholar]
- 49.Kozerski L., Mazurek A.P., Kawecki R., Bocian W., Krajewski P., Bednarek E., Sitkowski J., Williamson M.P., Moir A.J., Hansen P.E. A nicked duplex decamer DNA with a PEG(6) tether. Nucleic Acids Res. 2001;29:1132–1143. doi: 10.1093/nar/29.5.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mills J.B., Cooper J.P., Hagerman P.J. Electrophoretic evidence that single-stranded regions of one or more nucleotides dramatically increase the flexibility of DNA. Biochemistry. 1994;33:1797–1803. doi: 10.1021/bi00173a024. [DOI] [PubMed] [Google Scholar]
- 51.Kuhn H., Protozanova E., Demidov V.V. Monitoring of single nicks in duplex DNA by gel electrophoretic mobility-shift assay. Electrophoresis. 2002;23:2384–2387. doi: 10.1002/1522-2683(200208)23:15<2384::AID-ELPS2384>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 52.Turner D.H., Sugimoto N., Kierzek R., Dreiker S.D. Free energy increments for hydrogen bonds in nucleic acid base pairs. J. Am. Chem. Soc. 1987;109:3783–3785. [Google Scholar]
- 53.Du Q., Vologodskaia M., Kuhn H., Frank-Kamenetskii M., Vologodskii A. Gapped DNA and cyclization of short DNA fragments. Biophys. J. 2005;88:4137–4145. doi: 10.1529/biophysj.104.055657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petersheim M., Turner D.H. Proton magnetic resonance melting studies of CCGGp, CCGGAp, ACCGGp, CCGGUp and ACCGGUp. Biochemistry. 1983;22:269–277. doi: 10.1021/bi00271a006. [DOI] [PubMed] [Google Scholar]
- 55.Guo H., Tullius T.D. Gapped DNA is anisotropically bent. Proc. Natl Acad. Sci. USA. 2003;100:3743–3747. doi: 10.1073/pnas.0737062100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mills J.B., Vacano E., Hagerman P.J. Flexibility of single-stranded DNA: use of gapped duplex helices to determine the persistence lengths of poly(dT) and poly(dA) J. Mol. Biol. 1999;285:245–257. doi: 10.1006/jmbi.1998.2287. [DOI] [PubMed] [Google Scholar]
- 57.Kahn J.D., Yun E., Crothers D.M. Detection of localized DNA flexibility. Nature. 1994;368:163–166. doi: 10.1038/368163a0. [DOI] [PubMed] [Google Scholar]
- 58.Furrer P., Bednar J., Stasiak A.Z., Katritch V., Michoud D., Stasiak A., Dubochet J. Opposite effect of counterions on the persistence length of nicked and non-nicked DNA. J. Mol. Biol. 1997;266:711–721. doi: 10.1006/jmbi.1996.0825. [DOI] [PubMed] [Google Scholar]
- 59.Cantor C.R., Schimmel P.R. Biophysical Chemistry Part I: The Conformation of Biological Macromolecules. San Francisco: W. H. Freeman; 1980. [Google Scholar]
- 60.Frank-Kamenetskii M.D. Simplification of the empirical relationship between melting temperature of DNA, its GC content and concentration of sodium ions in solution. Biopolymers. 1971;10:2623–2624. doi: 10.1002/bip.360101223. [DOI] [PubMed] [Google Scholar]
- 61.Delcourt S.G., Blake R.D. Stacking energies in DNA. J. Biol. Chem. 1991;266:15160–15169. [PubMed] [Google Scholar]
- 62.Guckian K.M., Schweitzer B.A., Ren R.X.-F., Sheils C.J., Tahmassebi D.C., Kool E.T. Factors contributing to aromatic stacking in water: evaluation in the context of DNA. J. Am. Chem. Soc. 2000;122:2213–2222. doi: 10.1021/ja9934854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Friedman R.A., Honig B. A free energy analysis of nucleic acid base stacking in aqueous solution. Biophys. J. 1995;69:1528–1535. doi: 10.1016/S0006-3495(95)80023-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sponer J., Leszczynski J., Hobza P. Electronic properties, hydrogen bonding, stacking, and cation binding of DNA and RNA bases. Biopolymers. 2001;61:3–31. doi: 10.1002/1097-0282(2001)61:1<3::AID-BIP10048>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 65.Newcomb L.F., Gellman S.H. Aromatic stacking interactions in aqueous-solution: evidence that neither classical hydrophobic effects nor dispersion forces are important. J. Am. Chem. Soc. 1994;116:4993–4994. [Google Scholar]
- 66.Mckay S.L., Haptonstall B., Gellman S.H. Beyond the hydrophobic effect: attractions involving heteroaromatic rings in aqueous solution. J. Am. Chem. Soc. 2001;123:1244–1245. doi: 10.1021/ja003256d. [DOI] [PubMed] [Google Scholar]
- 67.Packer M.J., Dauncey M.P., Hunter C.A. Sequence-dependent DNA structure: dinucleotide conformational maps. J. Mol. Biol. 2000;295:71–83. doi: 10.1006/jmbi.1999.3236. [DOI] [PubMed] [Google Scholar]
- 68.Frank-Kamenetskii M.D., Anshelevich V.V., Lukashin A.V. Polyelectrolyte model of DNA. Sov. Phys. Uspekhi. 1987;30:317–330. [Google Scholar]
- 69.Bond J.P., Anderson C.F., Record M.T., Jr Conformational transitions of duplex and triplex nucleic acid helices: thermodynamic analysis of effects of salt concentration on stability using preferential interaction coefficients. Biophys. J. 1994;67:825–836. doi: 10.1016/S0006-3495(94)80542-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krueger A., Protozanova E., Frank-Kamenetskii M.D. Sequence-dependent base pair opening in DNA double helix. Biophys. J. 2006 doi: 10.1529/biophysj.105.078774. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.